It is hard today, in 2008, when genetic changes in cancer and specifically hematologic malignancies are accepted as central players, to look back 50 years and realize how different the landscape was then. In 1957, knowledge of the correct chromosome number of man was 1 year old,1 and that DNA was a double-helix was only 4 years old.2 Chromosomal studies in cancer, started only a few years before, mainly used ascitic fluid from experimental rodent tumors.3 These studies revealed a bizarre array of chromosomal changes, including ring chromosomes, with no discernible pattern of change. The general notion was that these wild chromosomal abnormalities were a consequence of multipolar spindles and other aberrations noted by Boveri4 and were not the cause of cancer. This had practical consequences. Many scientists were studying abnormal proteins and external causes of cancer, such as chemicals including benzene, hydrocarbons, or X-rays. These were known to cause chromosomal breaks and rearrangements, but these appeared, at the time, to be random changes and further supported the notion that chromosomal changes were mere consequences of other, more direct carcinogenic events.
So what happened to change this view? One view would be that it was all technologic, and certainly improvements in procedures and staining methods have had a major impact on cytogenetics, as on other scientific endeavors (think polymerase chain reaction, for example). But the change also involved a major change in perspective and the critical question is how did that happen? In 1956, Tjio and Levon published that man had 46 chromosomes, not 48 as had been thought previously.1 They were successful because cell culture had improved and later the use of hypotonic solution, acetic acid-alcohol fixative, etc, improved the separation of chromosomes in cells so that counts were more reliable. Although Peter Nowell's5 discovery of phytohemagglutinin (PHA), which stimulated T lymphocytes to grow and divide, was a boon to clinical cytogenetics by eliminating the need for skin biopsy to obtain fibroblasts for culture, this had little or only a negative effect on leukemia cytogenetics. A normal karyotype in a 72-hour PHA-stimulated blood culture from a leukemic patient could reflect dividing normal cells, not malignant cells. David Hungerford, a cytogeneticist at Fox Chase Cancer Center, collaborated with Nowell to examine the chromosomes from bone marrows of patients with various leukemias including chronic myelogenous leukemia (CML). They found that one of the small chromosomes (either 21 or 22) was too small.6 It was not the Y chromosome because it was present in women with CML. It was not present in normal PHA-stimulated cells from individuals with CML, so it was leukemia-restricted. This was a very important but perplexing observation. Because it was unique, it immediately raised the question, was it a sign that chromosomal changes were important in leukemia or was it just an aberration? The consistency of the Philadelphia chromosome was confirmed worldwide, so there was no question of its validity. Studies of cells from patients with acute leukemia still showed an apparently random pattern of gains and losses, so the controversy about the role of chromosomal changes in cancer remained unresolved. When CML evolved to blast crisis in approximately 3 to 5 years, the chromosomal changes, mainly additions, appeared to be somewhat random, although some groups of chromosomes (6-12-X and 21-22) were more likely to be involved.7 In fact, the study of CML in blast crisis was one experimental system that provided evidence for chromosomal clonal evolution even in the 1960s and 1970s.7,8 It was noted by Whang-Peng et al9 that there appeared to be a group of “CML-like” patients who lacked the Philadelphia (Ph) chromosome (they usually had a normal karyotype) and they paradoxically had a shorter survival than Ph+ CML patients. We now know that these patients do not have CML, but rather a myeloproliferative disorder.10
Chromosomal reports related to leukemia and various myeloproliferative diseases continued to be published in the 1960s,11 but all studies were hampered by the technical limitations of the time. Chromosomes were generally stained with Giemsa and they had a relatively uniform staining pattern. In a normal cell, the 46 chromosomes were grouped together with those of similar size and shape and each group was identified by capital letters (A through G). A few chromosomes had distinctive shapes (1, 2, 3, 16, and Y), but most did not. Thus, when cells had an abnormal chromosomal number, one could often determine the group to which it belonged, such as Group C (6-12-X), but not which specific chromosome was abnormal. Because Group C chromosomes were often present in excess or were missing, it was frustrating not to know whether the same chromosome was involved in different patients. Banding of human chromosomes was introduced in 1969 and 1970. With banding, we know now that a gain of chromosome 8 or, less often, a loss of chromosome 7 are the most frequent changes.12-14 Two banding methods were used, one involving quinacrine mustard (Q-banding) using fluorescence microscopy applied to human chromosomes by Lore Zech in Caspersson's laboratory15 and the other involving Giemsa (G-banding).16,17 At a meeting in Paris in 1971,18 cytogeneticists agreed on a uniform system of nomenclature for banded chromosomes. They continued the convention of labeling the short arm of chromosomes as “p” and the long arm as “q,” established in Chicago in 1966.19 The bands were numbered from the centromere of each chromosome; chromosomes were numbered in order of decreasing size, except that the chromosome present in trisomy in Down syndrome had been called “21,” even though it turned out to be smaller than no. 22. Rather than cause confusion by correcting it, it was agreed that it would remain no. 21. It is not hyperbole to say that these techniques revolutionized cytogenetics, especially cancer cytogenetics. For various technical reasons, G-banding is now the most widely used. A series of committees has ensured a uniform system of chromosomal nomenclature.20
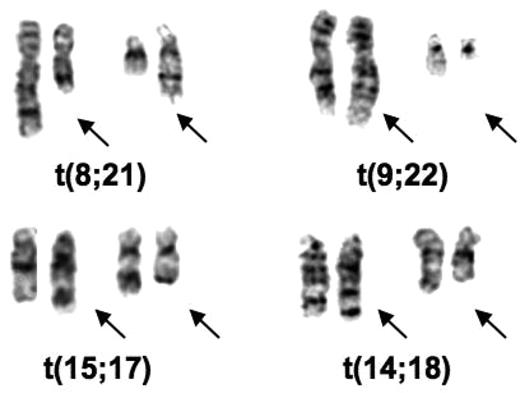
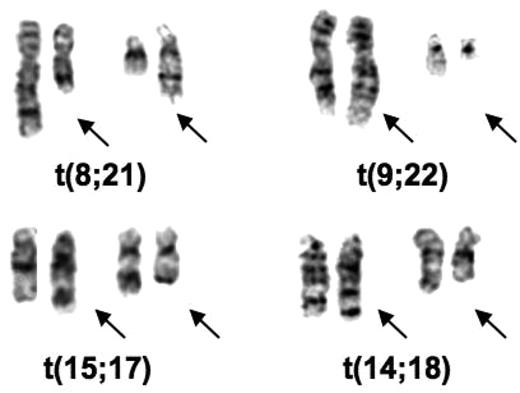
I was in England in 1970 and 1971 and learned the Q-banding method. I immediately applied it to cells from some elderly male patients with 45 chromosomes to prove that the chromosome lost from their bone marrow cells was a Y chromosome.21 On returning to the University of Chicago, I had to find someone to buy a fluorescence microscope (cost approximately $3000) for this procedure and then I began a study of our leukemic samples. We had recently received samples from 2 patients with an unusual chromosomal abnormality: 46 chromosomes, but missing chromosomes in 2 groups that were replaced by chromosomes in 2 other groups. When I used banding to analyze the chromosomes in these patients they both had the 8;21 translocation [t(8;21)(q22;q22)] (Figure 1). It was the first recurrent translocation in leukemia described. I wrote a brief letter and sent it to the New England Journal of Medicine in the summer of 1972, which rejected it. Being impatient, I then sent the paper to a journal (Annales de Genetique) whose editor (Jean de Grouchy) I knew.22 It was accepted and I have learned from publishers of Citation Index that it was one of the most cited papers published by that journal.
Partial karyotypes of common translocations discovered by Rowley. The translocations appear in the order in which they were discovered.
Partial karyotypes of common translocations discovered by Rowley. The translocations appear in the order in which they were discovered.
I was most intrigued by the identity of the extra chromosomes in CML in blast crisis and the question whether the extra C group chromosomes (6-12-X) were the same or different in different patients. I worked only part-time, so I took the photographs of the Q-banded and standard Giemsa-stained chromosomes from 2 patients home to analyze; each had 48 chromosomes with 2 extra C group chromosomes. With banding it became apparent that both patients had an extra chromosome 8. In the course of the analysis I identified the Ph chromosome, a deletion of chromosome 22, but I also noted some extra pale material at the end of chromosome 9 (Figure 1). I immediately analyzed the chronic phase sample from both patients by Q-banding, I could see the added material on the end of chromosome 9 (9q+) in these samples. I confirmed the translocation [t(9;22)(q34;q11)] in 2 more patients, both in blast crisis, and sent the paper to Nature. It was rejected with some reasonable comments and some truly wrong (for example, suggesting that this translocation could be due to an unusual chromosomal variant that was normal). By this time, I had studied 7 patients, and had analyzed PHA-stimulated peripheral blood T lymphocytes that had a normal karyotype. I revised the paper, including the additional data; it was accepted and finally was published in 1973.23 At the time, I was trying to find out what biologic process could account for recurring translocations in 2 different leukemias. No one had any precedent. Besides, not knowing what could cause translocations, it was not clear why these translocations would cause leukemia.
Since that time, hundreds of translocations have been identified not only in leukemia and lymphoma, but in sarcomas,24 some carcinomas, especially now in prostate cancer,25 and small cell lung cancer.26 In the middle 1970s and early 1980s, I was convinced that recurring chromosomal abnormalities, especially translocations, were important. As a consequence, when I was invited to speak at the Neoplastic Diseases portion of the Education Session at the American Society of Hematology (ASH), I treated the audience as if I were a missionary. These sessions were held Sunday morning and I took advantage of the timing to emphasize that although they did not believe it, they should do a karyotype analysis on all their patients with acute leukemia (first session), and later I sent the same message regarding lymphoma (second session). By 1979 we had described the 14;18 translocation [t(14;18)(q32;q21)] in small noncleaved cell lymphoma,27 providing evidence of chromosomal differences in specific types of lymphoma. Lore Zech had previously identified the 8;14 translocation in Burkitt lymphoma.28
The next important development related to translocations was cloning the 8;14 translocation in Burkitt lymphoma in 1982. This was done independently by Carlo Croce's group29 and Phil Leder's group30 using gene probes for the immunoglobulin heavy chain (IGH) gene at 14q32 and MYC at 8q24. The fact that a translocation involved a known oncogene convinced many scientists that translocations were important whereas other scientists became convinced that oncogenes really mattered because they were involved in translocations. Because oncogenes were hot in the 1980s and subsequently, the fact that a number of translocations involved oncogenes encouraged molecular biologists to become more interested in cytogenetics, or at least the results of cytogenetic analysis based on banding. Uncloned translocations have been a critical source of finding new and important genes. For example, relatively recently, cloning the genes in the rare 5;12 translocation in a myeloproliferative disease identified the PDGFR-TEL (ETV6) translocation.31 The identification of TEL led to the identification of 12;21 (p13;q22) translocation involving TEL and AML1, which turns out to be the most common translocation in childhood acute lymphoblastic leukemia (ALL).32 It had been overlooked because the ends of 12p and 21q have a similar pale appearance on banding and they could not be distinguished in banded chromosomes from most patients.
Translocations have been important from the biologic as well as the therapeutic point of view. Depending on one's perspective, the biologic discoveries could be as important as the therapeutic importance of translocations. Based on their functional categories, it was clear that genes involved in translocations affect many different pathways within the cell. A good example is cloning the 14;18 translocation,33 which revealed the involvement of IGH on chromosome 14 (band q32) and a new gene, BCL2, on chromosome 18 (band q21; Figure 1). The discovery of BCL2 has opened up a whole new field related to apoptosis.34,35 It was critical because the importance of programmed cell death in the biology of cancer cells was not appreciated before the translocation breakpoint was cloned and the function of BCL2 was clarified.
Studying translocations has resulted in a number of important discoveries. First, they are of diagnostic importance. In fact, the current World Health Organization (WHO) classification system of AML36 includes specific karyotypes. For example, using t(8;21) provides a more precise classification than the French-American-British (FAB) because, although the vast majority are M2, a few could fall in other subgroups. Similarly, inv(16)(p13q22) is primarily M4E0 but a few samples could fall in other categories and acute promyelocytic leukemia (APL) is restricted to translocations involving the RARA and PML genes in the t(15;17)(q22;q12). Although there are rare translocations that involve RARA with other partner genes and although the leukemic cells have features of APL, some of the leukemias may not respond to APL therapy. Early and correct diagnosis of APL with a t(15;17) is essential because all-trans retinoic acid (ATRA) with or without arsenic trioxide and chemotherapy is a potentially life-saving treatment.37 The WHO classification has also stated that the BCR-ABL fusion, which occurs as a result of the t(9;22), is essential for the diagnosis of chronic myeloid leukemia (CML).
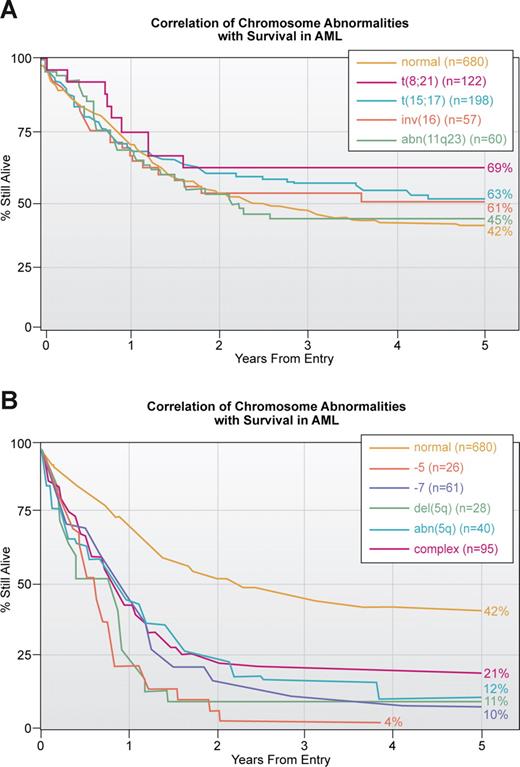
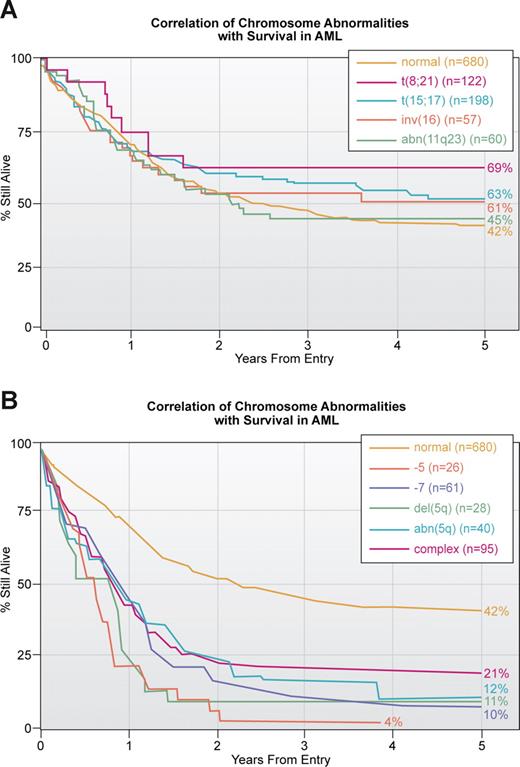
A second important application of our knowledge of translocations and other chromosomal abnormalities in acute leukemia, lymphoma and other hematologic malignancies is prognosis. Ideally, every patient who is suspected of having a malignant or premalignant hematologic disorder should have a cytogenetic analysis of bone marrow cells. For various reasons, including financial, this is more easily accomplished in Europe than in the United States. The presence of some translocations, for example t(15;17) or inv(16), is associated with a high response to therapy and a long survival, whereas other translocations such as a break at 11q23 involving the MLL gene is generally associated with a poorer outcome38,39 (Figure 2A). Similarly complex karyotypes with 3 or more abnormalities or loss of parts of chromosome 5 and/or loss of 7 is associated with a dismal outcome (Figure 2B). Similar associations of karyotype with response to treatment are seen in other hematologic malignancies. In fact, early on the cytogenetic pattern was a major determinant of responses in ALL; with improved, more targeted therapy, this association has weakened.40
Correlation of chromosomal abnormalities with survival in AML based on an MRC study.38 (A) Correlation with translocations which is the subset with the best response to treatment. Note that of all translocations, those involving 11q23 have the poorest survival. This study, published in 1998, does not reflect the enormous improvement in survival in APL associated with treatment with ATRA and arsenic trioxide. The yellow line for those thought to have a normal karyotype is the same in this and panel B. (B) Correlation with abnormalities that have the poorest outcome. As noted in Figure 3, these abnormalities tend to be more common in older patients.
Correlation of chromosomal abnormalities with survival in AML based on an MRC study.38 (A) Correlation with translocations which is the subset with the best response to treatment. Note that of all translocations, those involving 11q23 have the poorest survival. This study, published in 1998, does not reflect the enormous improvement in survival in APL associated with treatment with ATRA and arsenic trioxide. The yellow line for those thought to have a normal karyotype is the same in this and panel B. (B) Correlation with abnormalities that have the poorest outcome. As noted in Figure 3, these abnormalities tend to be more common in older patients.
Another important result is the discovery of critical genes; cloning translocation breakpoints has revealed a host of previously unknown genes, among them BCR, BCL2, AML1 (RUNX1), ETO, PML, CBFB, TEL, NOTCH and MLL. Some of these were shown to have homologs in other organisms, such as BCL2 which is homologous to ced-9 in Caenorhabditis elegans or AML1 and MLL which are homologous to Drosophila orthologs runt and trithorax, respectively. Nonetheless, these genes were first identified in humans by cloning translocation breakpoints. These genes have had a major impact on cancer biology because they provided new tools with which to study mechanisms of carcinogenesis in tumors, especially the hematologic malignancies. These studies have been conducted on patient cells and cell lines; in mice, the fusion construct has been introduced into bone marrow cells and the abnormal cells injected into lethally irradiated mice, or cells with a fusion has been mixed with normal embryonic cells, forming a chimeric embryo and then examined for chimeric offspring.41 As emphasized by Mitelman and his colleagues42 although translocations in hematologic malignancies get virtually all the press, there are proportionately just as many translocations in solid tumors, just as many of them have been cloned, and they involve the same functional gene families. This is a very important misconception that should be corrected.
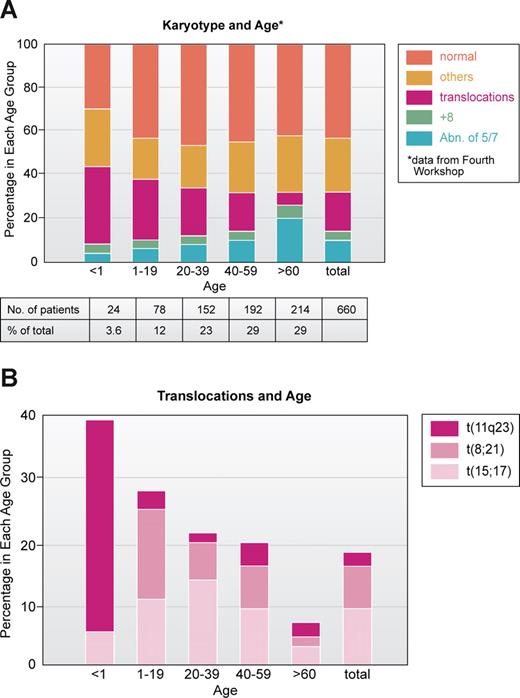
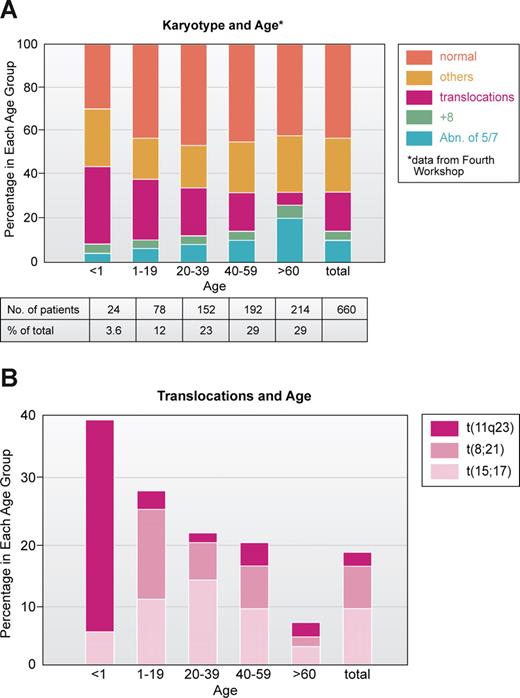
To me, one of the most interesting and unresolved questions is the age distribution of chromosomal abnormalities, especially translocations. In an analysis of 660 AML patients studied at the Fourth Workshop on Chromosomes in Leukemia,43 most patients with 11q23 translocations were infants and young children; the t(8;21) was primarily seen in children younger than 20 years old, the t(15;17) in patients ranged from 10 to 30 years, whereas patients with −5 and/or −7 were older (Figure 3A,B). Thus, it is clear from the cytogenetic point of view that patients who are young have a genetically different disease than those who are older. Except for infant MLL at 11q23, the young patients have a much higher proportion of good outcome leukemias, based on karyotype, compared with older individuals who mainly have leukemias with bad karyotypes.38,39 Why? Some might say, at least for ALL, that many of the translocations have been shown to originate in utero, so naturally the leukemia will occur early.44,45 Work of Mitelman46 and others47 also suggested that older individuals with leukemia, especially men, were exposed to mutagenic agents and that abnormalities of chromosomes 5 and 7 reflected damage from environmental factors. This certainly appears to be the case with treatment of primary malignancies. Treatment with cytotoxic drugs and/or radiation greatly increases the frequency of chromosome 5 and 7 abnormalities in AML cells, first reported in a series in 1977.48 On the other hand, the increasing use of the epipodophyllotoxins such as etoposide has been associated with balanced translocations most often involving t(11q23) or t(8;21).49
Correlation of major subsets of karyotype in AML with age based on patient data analyzed at Fourth International Workshop on Chromosomes and Leukemia.43 (A) All 660 patients. (B) Patients with specific translocations. Note that those involving 11q23, especially t(4;11) are very prominent in infants (younger than 1 year of age). Illustration by Kenneth X. Probst.
Correlation of major subsets of karyotype in AML with age based on patient data analyzed at Fourth International Workshop on Chromosomes and Leukemia.43 (A) All 660 patients. (B) Patients with specific translocations. Note that those involving 11q23, especially t(4;11) are very prominent in infants (younger than 1 year of age). Illustration by Kenneth X. Probst.
Another area to benefit from the study of translocations is basic science. Once the first translocation was cloned in 1982, then all translocations became the subjects of intense interest. Thus t(8;21) was cloned in Japan by Miyoshi et al.50 and in the United States by Drabkin's group51 in 1991; t(15;17) was cloned virtually simultaneously by groups in England,52 France,53 Italy54 and the United States55 in 1990. t(9;22) was cloned in 1984 by Heisterkamp and Groffen working with Stephenson and Grosfeld.56,57 The observation that some known oncogenes were at translocation breakpoints piqued the curiosity of virologists and molecular biologists. Knowing that an ETS family gene, AML1 (also called CBFA or RUNX1), was involved in the 8;21 translocation and that RARA was involved in the 15;17 translocations encouraged cell biologists to study them. Suddenly many people who were not cytogeneticists were paying attention to translocations. The translocations have proven much more tractable to analysis and cloning of the relevant genes than partial or whole gains or losses of chromosomes. In translocations, the breakpoint can be cloned. They are defined so they provide a reasonably precise chromosomal location for the involved gene.
A gene that has been of interest to me for more than 25 years resides at 11q23. In the 1980s, a number of translocations involving 11q23 were identified, such as t(4;11)(q21;q23) in ALL, especially in infants and young children, and t(9;11)(p221;q23) and t(6;11) (q27;q23) in AML, as well as 2 different recurring breakpoints in t(11;19) in AML and ALL (q23;p13.3 and q23;p13.1). Because of the variable phenotype, it was reasonable to assume that at least 2 different genes were involved in these 11q23 translocations. We undertook to map and then to clone the breaks in these leukemias. We used probes mapped to chromosome 11 with fluorescence in situ hybridization (FISH) by Peter Lichter in David Ward's laboratory.58 We found that the breaks in all the translocations mentioned above fell between 2 probes on 11q.59 We then obtained the yeast clones that contained yeast artificial chromosomes (YACs) for that region of chromosome 11 from Maynard Olson (Washington University, St Louis, MO) and discovered a yeast clone yB22B2 that contained a YAC with 320 Kb of human DNA that was split by all our translocations. This result surprised me but made it likely that there was just one gene involved in the 4 translocations. We called the gene MLL for mixed lineage leukemia or myeloid-lymphoid leukemia.60 Others cloned the gene first and gave it various names, but the name MLL stuck. MLL is important for a number of reasons. It is involved in 80% or more of infant leukemias39 ; Greaves and others44,45 have shown that translocations involving MLL as well as other translocations occur in utero. It has at least 60 known partner genes of which approximately 40 have been cloned.61 It is involved in one-half to two-thirds of treatment-related leukemias in cancer patients who have been previously been treated with toposisomerase II inhibitors. Why?
MLL codes for a very large protein with many domains. Like the Drosophila ortholog, trithorax, it regulates the expression of some of the Homeobox genes, notably HOX A9 and A10, and MEIS1.62 What has become apparent very recently is that several of the MLL fusion partner proteins are associated with the RNA polymerase II complex and that some of the most frequent MLL partner genes/proteins are also involved in this complex, eg AF4 (and LAF and AF5q) and AF9, or ENL and AF10.63 Although MLL itself has not been shown to be part of an RNA Pol II complex, it is recruited to the HOX genes usually with Pol II.64 What better way to make sure that a critical component is present in a large protein complex than to have that partner involved in a translocation?
Many laboratories including my own have mapped motifs in MLL65,66 and other genes (AML167 ) that might be associated with breaks. Although we have mapped the DNA breakpoints on both translocations in some detail, as well as topo II cleavage sites and DNase 1 hypersensitive sites, the conclusions have been that although there are occasional suggestive associations, such as microhomologies and alu sequences at translocation breakpoints, there is no motif that is consistent. Although various laboratories have focused on the chromatin structure at or near the breakpoints, this may not be the relevant factor. Current research in several laboratories, notably that of Peter Fraser, have focused on the structural changes that affect genes that are being actively transcribed. RNA Pol II, topoisomerase II, and other factors involved in transcription are localized in “transcription factories.” Active genes loop from chromosomal domains over long distance, often many microns, and the DNA strands are transcribed in these “factories,” roughly 200 per cell. Of relevance to the present discussion is the fact that if one considers “factories” with which the immunoglobulin heavy chain genes associate, then 25% of the time MYC is colocalized in the same factory. By comparison, random associations involving any one gene appear to occur approximately 1% of the time.68 Given the recurrence of t(8;14)(q24;q32) or MYC: IGH translocations in B cell tumors, this apparent preferential association of these 2 genes is especially relevant. If further research on other translationally associated genes shows that they, too, associate preferentially in the same “factories,” this will be a major advance in our understanding of possible mechanisms related to translocations. It might also explain the observations that genes frequently involved in chromosomal translocations are seen, albeit rarely, in apparently nonmalignant cells of normal individuals. This latter observation also reinforces earlier observations that, in many circumstances, translocations alone are not sufficient to form a fully malignant cell, but that other changes are required.69 Thus, the study of translocations continues to yield surprises.
The issue of what causes translocations has been studied extensively but the fundamental question still remains. Similarly, the questions about the functions of the different translocations are largely unresolved. For each translocation we know a fair amount—often a great deal—about the function of each partner gene and how that function is altered, but we still remain ignorant of the critical steps by which a translocation changes a normal cell into a malignant cell. Moreover, is it the first change or have other changes occurred before in the cell? What are the secondary changes that make the cell fully malignant? Work by Philip Fialkow70 many years ago suggested that the Ph chromosome arose in a clonal population, indicating that it was a secondary event. However, experiments indicate that transgenic mice with cells containing a translocation often develop leukemia or lymphoma, suggesting that the translocation may be an initial event. However, some translocations in transgenic mice can lead to a myeloproliferative state but not leukemia unless a mutogeneic agent such as N-ethyl-N-nitrosourea (ENU) is also used, suggesting that second (or more) hits are required.71 What are they?
Many of us are trying to answer these questions using different strategies. Some are using the new cytogenetic and genetic tools looking for small deletions, duplications, or mutations that had escaped notice. A recent paper from Jim Downing's laboratory reported that there are many small aberrations not detected cytogenetically in a large series of childhood ALL.72 These new leads merit vigorous investigation. In fact, this work confirms what cytogeneticists have known for some time, that banded chromosomes are fairly gross representations of the genes contained in the underlying chromosomal DNA. That fact is reflected in the use of the designation “Marker” and “?” in the nomenclature used to describe complex karyotypes. The use of spectral karyotyping73 has improved our precision dramatically as have the techniques used by Mullighan et al.72
Others have used various tools to study gene expression in different translocations to identify consistent changes that could help to identify the other genes whose altered function is critical in changing a cell with a translocation into a fully malignant cell. The most popular tool has been microarrays, which are chips containing probes from many genes to which RNA from leukemic cells has been hybridized to provide information about the expression pattern of various genes. Many different microarray platforms have been used and the results from different laboratories have often differed with regard to significant changes in expression levels of genes in the same translocation.74 Reliable detection of aberrant gene expression that focuses on one or a few genes has yet to be achieved.
Very recently a new molecule has captured the attention of many scientists, namely microRNAs (miRs). They are small RNAs (approximately 22 nucleotides long) derived from a larger RNA that is transcribed from either introns or exons of primary protein-coding genes, or intergenic regions. These small RNAs, like shRNAs (RNAi), generally degrade messenger RNAs after binding to complementary “seed” segments in the 3′ untranslated region (UTR) or interfere with translation of the mRNA into protein. Their regulation of gene expression is complex; of the 500 or so known miRs, some bind to many genes and similarly the 3′ UTR of a single mRNA can bind multiple miRs.75 Many aspects of miR biology, function, and regulation are presently unknown. In any event, miRs have been reported to have major effects in several cancers and lymphomas.76,77 We have reported that only 4 miRs are sufficient to distinguish ALL from AML78 ; in addition, we have found that fewer than 10 miRs can distinguish the common translocations in AML. Interestingly, core binding factor leukemia, t(8;21) and inv(16), form a hierarchial cluster that they do not do in microarrays.79 Lu et al80 have reported that miRs can distinguish various tumor types more accurately than can microarrays.
So what does the future hold?
In this reminiscence, I have summarized the role of cancer cytogenetics focusing on translocations, especially those in AML. I have raised questions in various parts of this paper. What are the really big issues? For me, the biggest is what do translocations do in a cell? How do they change it? If we really understood the answer to this question we could answer the next big question: what can we do about it therapeutically? Given the wide variety of functions of genes involved in translocations, these questions do not have a single answer, or imatinib would treat every tumor, which is not the case. Given the disparate functions of ABL (a tyrosine kinase) and MLL (involved in transcriptional regulation), the “one size fits all mentality” that has afflicted chemotherapy development clearly must be abandoned, as has been done by the molecularly biologically educated clinical oncology community.
As we develop more targeted therapy, we must enhance our capability precisely to diagnose not only the type of tumor but the genetic, functional and immunologic changes that are present in that particular tumor; in other words we will truly need personalized medicine! To reach these goals we will need the collaboration of clinicians with biologic and physical scientists. Translational medicine has become the new catch phrase, with good reason, because to be successful it must benefit from the skills of multiple, differently trained scientists. But, at least in the United States, we must also strive to have these new strategies available to all our citizens, especially the poorly served who are least likely to benefit from our remarkable scientific advances. This is a goal that should appeal to every compassionate physician.
Authorship
Contribution: J.D.R. wrote the paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Janet D. Rowley, University of Chicago Medical Center, 5841 S Maryland Avenue, MC 2115, Chicago, IL 60637-1470; e-mail: jrowley@medicine.bsd.uchicago.edu.