Abstract
The biological and clinical implications of p16 gene methylation in multiple myeloma (MM) are still unclear despite previous studies. In this comprehensive study, using methylation-specific PCR (MS-PCR), we show that p16 methylation is relatively common and occurs in monoclonal gammopathy of undetermined significance (MGUS; n = 17), smoldering multiple myeloma (SMM; n = 40), and MM (n = 522) at a prevalence of 24%, 28%, and 34%, respectively. However, p16 methylation does not appear to affect gene expression level. In a large cohort of patients with long-term follow-up information (n = 439), there was no difference in overall survival between patients with or without p16 methylation. We also found no association between p16 methylation and the main cytogenetic categories, although it was more common among patients with 17p13.1 deletions (p53 locus), a genetic progression event in MM. In addition, p16 methylation has no apparent effect on the cycle because there was also no difference in the plasma cell labeling index (a direct measurement of proliferation) between patients with and without p16 methylation. Our results question a major role for p16 methylation in the oncogenesis of the PC neoplasm, and we now believe p16 methylation may be a marker for overall epigenetic changes associated with disease progression, with no obvious direct biological or clinical consequences.
Introduction
The retinoblastoma (Rb) checkpoint controls G1/S transition.1 Abnormalities in this pathway caused either by loss of Rb or p16 function, or increased activity of cyclin D1 or CDK4/6 in the cell-cycle machinery, results in uncontrolled cell proliferation, invasion, and metastasis.2-4 The p16 gene is one of the most commonly affected members of this pathway. The cyclin-dependent kinase 4 inhibitor p16 gene, located at 9p21, has been shown to be inactivated in a variety of tumors by deletions, point mutations, or hypermethylation of its promoter.5-7 Furthermore, inactivation of this tumor suppressor gene can be important for initiation and maintenance of the transformed phenotype.7 Reports also suggest that loss of p16 gene function is increasingly common with advancing stages of various neoplasms, suggesting that p16 inactivation may contribute to disease progression.8 This body of evidence suggests a key role of this gene in initiation and progression during carcinogenesis.
The plasma cell (PC) neoplasms range from indolent disorders (ie, monoclonal gammopathy of undetermined significance [MGUS] and smoldering multiple myeloma [SMM]) to aggressive variants (eg, active multiple myeloma [MM] and PC leukemia); indolent forms often progress to the more advanced stages. Although the genetic/cytogenetic abnormalities responsible for the PC neoplasm are becoming better understood, the role of p16 methylation, if any, is largely unknown. Studies in MM have found no mutations or deletions, but promoter methylation has been reported with an incidence of up to 58%.9 Fewer data are available regarding inactivating abnormalities of the p16 gene in normal plasma cells, MGUS, and SMM. In this study we analyzed bone marrow samples from healthy persons and persons with MGUS, SMM, and MM for p16 promoter methylation by methylation-specific polymerase chain reaction (MS-PCR) and interstitial deletions by interphase fluorescence in situ hybridization (FISH). In addition, we sought to clarify the effect of p16 methylation on its gene expression. We also studied a large cohort of patients to determine in a definitive fashion what, if any, prognostic and clinical significance p16 methylation has in plasma cell neoplasm. We confirm that p16 hypermethylation is slightly more common with advancing stages of the disease. However, p16 methylation is not associated with reduced gene expression, increased proliferation, cytogenetic categories, or survival.
Materials and methods
Patients
Mayo patients.
We first studied 2 series of Mayo Clinic patients who had consented for their samples to be used for research purposes. These patients had a bone marrow (BM) research sample collected at the time of routine clinical procurement. In the first series, promoter methylation of p16 was studied using whole BM DNA (in 37 patients with MM, 30 with SMM, and 5 with non-IgM MGUS). In the second series, we used DNA from CD138+-purified PCs (in 46 patients with MM, 10 with SMM, and 12 with non-IgM MGUS). CD138 selection was performed using immunomagnetic bead separation (AutoMACS; Miltenyi-Biotec, Auburn, CA). In addition, cytospin slides from 37 patients with MM, 30 patients with SMM, and 20 patients with MGUS were used for the study of p16 deletion. Diagnosis and stage determination was based on standard criteria.10 For samples in which whole BM DNA was used, each BM sample contained a minimum of 1 × 106 cells, with the percentage of PCs ranging from 1% to 75%. To study normal PCs, we collected surgical waste samples from persons undergoing arthroplasty surgery at the Mayo Clinic. These plasma cells were enriched by using the CD138+ immunomagnetic bead sorting. This study and the collection of tissues were approved by the Mayo Clinic Institutional Review Board. Informed consent was provided according to the Declaration of Helsinki.
Easter Cooperative Oncology Group (ECOG) patients.
We also studied patients enrolled in the ECOG clinical trial E9486/E9487 (n = 561) that has been described in detail elsewhere.11 Briefly, patients with newly diagnosed MM were randomly assigned to receive treatment with conventional chemotherapy variations (vincristine, bischloroethylnitrosourea, melphalan, cyclophosphamide, prednisone [VBMCP] versus VBMCP plus high-dose cyclophosphamide, versus VBMCP plus interferon). A total of 439, based on sample availability, were included in our current study and appeared to be no different from the larger cohort of patients when all relevant biological and prognostic factors are considered (data not shown). Pertinent clinical and prognostic features are available for the majority of the patients. For this cohort, again, DNA from unsorted whole bone marrow was used.
Methylation-specific PCR
Extracted DNA was subjected to sodium bisulphite modification using a CpGenome DNA Modification Kit (Intergen, New York, NY). Universal methylated human male genomic DNA (Intergen) was used as the positive control. Genomic DNA purified from peripheral blood of healthy voluntary donors was used as a negative control. Primer sequences and PCR conditions were previously described.12
Interphase FISH
Deletion of the p16 locus on chromosome 9p21 was studied by using interphase fluorescent in situ hybridization with immunofluorescent detection of the clonotypic cytoplasmic-immunoglobulin light-chain (cIg-FISH) as previously reported by us.13 We used a chromosome- and locus-specific (CEP9/p16) dual-color probe set (Vysis, Applied Biosystems, Downers Grove IL). We scored the samples as follows: greater than 25% of neoplastic cells with only one signal were considered positive for deletion; those with 10% to 25% abnormal cells were equivocal; and those with less than 10% abnormal cells were considered normal.14 Two observers scored each slide.
Gene expression profiling (GEP)
To study the effect of promoter methylation of p16 on its mRNA expression, we used expression data obtained from samples that were also studied by GEP. GEP was performed (46 MM, 10 SMM, and 12 MGUS) using the Affymetrix U133A chip (Affymetrix, Santa Clara, CA) on those samples with purified CD138+ PCs.
RNA isolation, purification, and microarray hybridization have been previously reported.15 Gene expression intensity values were generated using the Affymetrix MAS 5.0 software; log transformed, normalized to the median, and analyzed using GeneSpring 7 (Agilent Technologies, Palo Alto, CA). As the same gene, CDNK2A, gives rise to p16 and p14, involved in the Rb and p53 pathways, respectively, through alternate splicing, we matched the target sequence of the probes for CDNK2A available on the U133A chip with the sequence of p16 and p14 using ClustalW, a web-based bioinformatics tool from the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/). Specifically, we wanted to identify the probes with target sequence specific for p16. Probe 209644_x_at has sequence specific for p14, whereas 207039_at targets the 3′ UTR that is common to both p14 and p16. We then extracted the expression values for these probes, and the difference in the expression value between these 2 probes is used as an approximation for p16 expression.
Quantitative real-time RT-PCR
To confirm the gene expression findings, we also analyzed 23 of the Mayo patient samples (7 methylated, 11 unmethylated, and 5 unmethylated normal marrows) by quantitative real-time reverse transcriptase (RT-PCR). For cDNA synthesis of sorted CD138+ cells, 1 μg total RNA was reverse transcribed in a 20-μL volume reaction mixture, using a First Strand cDNA Kit from Amersham Bioscience (HP 7 9NA; Chalfont St Giles, United Kingdom). Primers used to amplify only p16's exon 1 (not amplifying p14) were as follows: 5′-gttacggtcggaggccgatcc-3′ forward and 5′-gagcagcagcagctccgc-3′ for the reverse, and were obtained from IDT (Integrated DNA Technologies, Coralville, IA). These primers cover nucleotides 163 through 183 in exon 1 and nucleotides 218 through 205 in exon 2 (positions from L27211 cDNA sequence for p16INK4A). To perform the detection, we used the following probe: 5′ VIC-catgacctggatcggcctcgga-TAMRA-3′, from Applied Biosystems (Foster City, CA). PCR reactions were performed with an automated fluorometer ABI PRISM7700 Sequence Detection System (Applied Biosystems) using 96-well optical plate. Each sample was analyzed in triplicates. Fifty nanograms of total RNA was used in 20 μL reaction mixture containing 12.5 μL TaqMan qPCR MasterMix (Branchburg, NJ), 300 nM of each primer, and 200 nM TaqMan probe. Amplification was carried out as follows: denaturation for 10 minutes at 95°C followed by 45 cycles at 95°C for 15 seconds, and 60°C for 60 seconds.
Complimentary DNA from the HeLa cell line was used as positive control to verify the amplification efficiency, and cDNA from PANC-1 cell lines served as negative control and did not result in product amplification, confirming the specificity of these reactions for p16 detection. Relative quantification of the p16 was done by normalizing the amplification signals with the GAPDH signal (as “housekeeping gene”) using the comparative ΔΔCT methods describe previously by Livak and Schmittgen.16 Fold change in transcript levels between unmethylated MM, methylated MM, and unmethylated normal control were compared.
Statistical analysis
The Fisher exact test or chi-square test was used to examine the relation between categorical parameters. Student t test was used for comparing continuous variables. Overall survival was calculated from the date of diagnosis until the patient's death or last visit. Survival probability curves were analyzed and plotted according to the method of Kaplan and Meier and compared using the log-rank test.
Results
Methylation is more common than deletion of p16 in MM
Among the Mayo samples, aberrant methylation was found in 20% (1 of 5) of MGUS, 30% (9 of 30) of SMM, and 41% (15 of 37) of MM samples studied using whole bone marrow DNA. The prevalence of p16 methylation appeared to be no different when the source material was CD138+ cells, 25% (3 of 12) of MGUS, 20% (2 of 10) of SMM, and 30% (14 of 46) of MM samples. Of the 439 ECOG patients studied, 149 MM samples (36%) were positive for p16 methylation.
When the analysis of Mayo and ECOG samples were considered together, MS-PCR showed that p16 gene hypermethylation was present in 1 (12.5%) of 8 normal bone marrow samples, 4 (24%) of 17 MGUS samples, 11 (28%) of 40 SMM samples, and 178 (34%) of 522 MM samples. The difference between asymptomatic (MGUS/SMM) and symptomatic stage (MM) of PC neoplasm is modest, although it achieves significance (chi-square = 5.077, P = .02).
FISH demonstrated the presence of a hemizygous deletion in only 10% (4 of 37) of MM patients. In addition, 10 cases had equivocal results for deletion with the percentage of abnormal cells ranging from 20% to 30% (4 MGUS, 2 SMM, 4 MM). In 2 patients with MM, there was coexistence of p16 methylation and deletion.
p16 methylation is associated with deletion of the p53 locus
No significant associations between p16 methylation and the major cytogenetic categories of MM were found. In particular, no association was found between p16 methylation and t(11;14)(q13;q32), t(4;14)(p16.3;q32), t(14;16)(q32;q23), or deletion/monosomy of chromosome 13 (Δ13). The only positive association was of p16 methylation with 17p13.1 deletions (p53 locus) (Fisher exact test P = .002). Of 31 patients with 17p13.1 deletions, 20 (65%) had p16 methylation (n = 267) (Table 1).
Gene expression of p16 in PC neoplasm and its relation to p16 methylation status
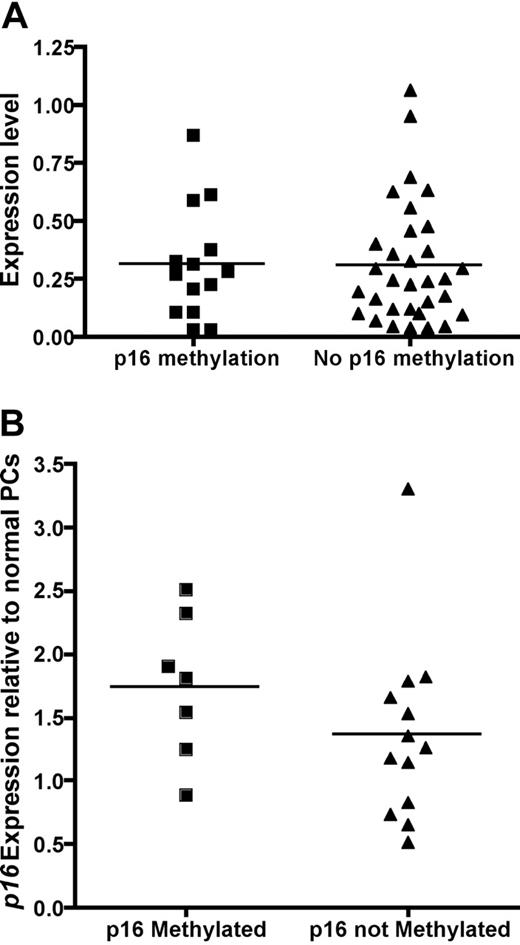
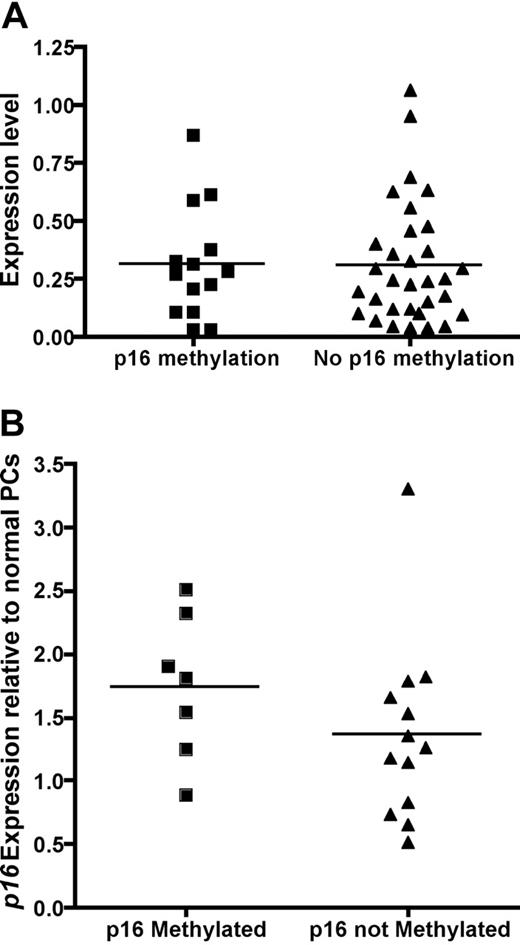
Next, we analyzed the effect of p16 methylation on the gene expression of p16 in patients with MM with both GEP and p16 methylation results. The gene expression of p16 was not different in the 14 patients with methylated p16 compared to the 32 patients with unmethylated p16 (Figure 1A). The lack of effect of p16 methylation on gene expression was confirmed by quantitative RT-PCR (Figure 1B).
Expression level of p16 is not affected by p16 methylation. (A) Gene expression profile between patients with methylated p16 (n = 14, mean normalized expression 0.31 ± 0.24) and unmethylated p16 (n = 32, mean normalized expression 0.31 ± 0.26). The normalized expression level p16 is similar when compared by Student t test (P = .49). (B) Quantitative RT-PCR was performed on 7 patients with p16 methylation, 11 without p16 methylation, and 5 normal unmethylated plasma cells samples. Fold change analysis relative to normal unmethylated plasma cells using the ΔΔCT method showed once again there is no difference in p16 expression between patients with and without p16 methylation as detected by MS-PCR (1.87 ± 0.43 versus 1.56 ± 0.59, P = .70, t test).
Expression level of p16 is not affected by p16 methylation. (A) Gene expression profile between patients with methylated p16 (n = 14, mean normalized expression 0.31 ± 0.24) and unmethylated p16 (n = 32, mean normalized expression 0.31 ± 0.26). The normalized expression level p16 is similar when compared by Student t test (P = .49). (B) Quantitative RT-PCR was performed on 7 patients with p16 methylation, 11 without p16 methylation, and 5 normal unmethylated plasma cells samples. Fold change analysis relative to normal unmethylated plasma cells using the ΔΔCT method showed once again there is no difference in p16 expression between patients with and without p16 methylation as detected by MS-PCR (1.87 ± 0.43 versus 1.56 ± 0.59, P = .70, t test).
No significant correlation between p16 methylation and clinical features of MM
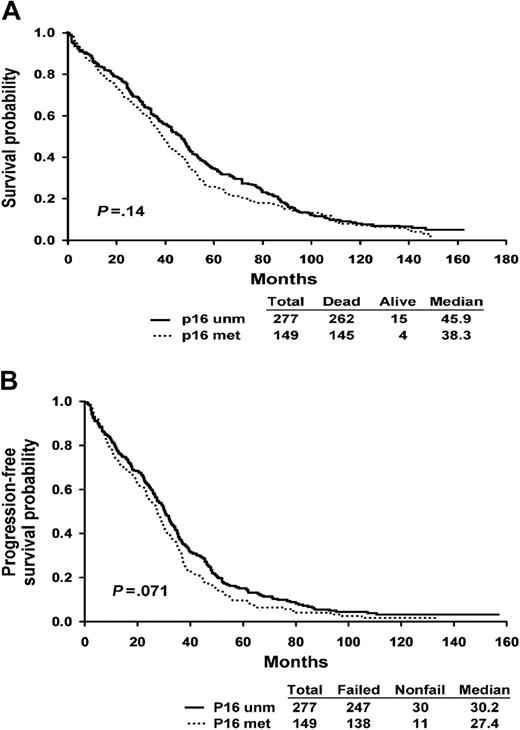
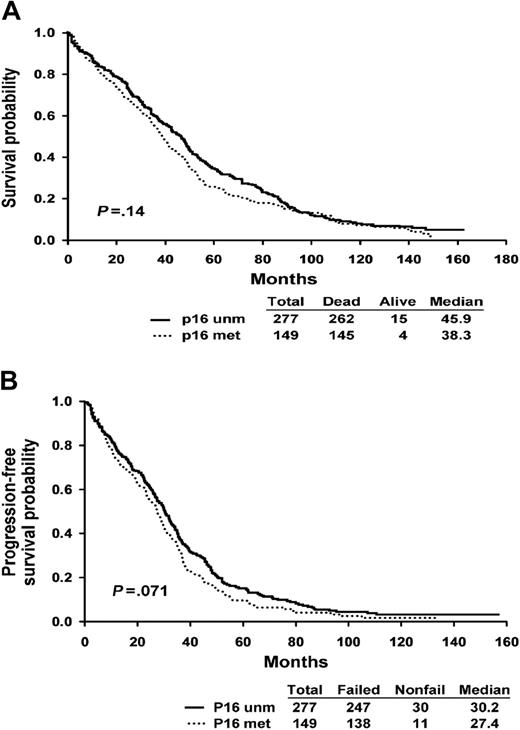
The baseline clinical characteristics of the ECOG patients are summarized in Table 2. The patients are typical of those entered into clinical trials, and there is no difference between patient with and without p16 methylation. Of note, the plasma cell labeling index is similar between patients with and without p16 methylation. Patients with p16 methylation did not have a significantly worse survival as compared with patients without the abnormality. The median survival of patients with the p16 methylation was 38.8 months as compared with 45.9 months for patients without the abnormality (P = .14, log-rank) (Figure 2A). Progression-free survival was also not significantly different among patients with and without the abnormality (27.4 versus 30.2 months, P = .071) (Figure 2B).
Survival curves according to p16 methylation status. (A) Kaplan-Meier overall survival curve shows the survival of patients stratified by the presence of p16 methylation (P = .071, log rank). (B) Progression-free survival of patients stratified by the presence of p16 methylation (P = .14, log rank).
Survival curves according to p16 methylation status. (A) Kaplan-Meier overall survival curve shows the survival of patients stratified by the presence of p16 methylation (P = .071, log rank). (B) Progression-free survival of patients stratified by the presence of p16 methylation (P = .14, log rank).
Discussion
In this comprehensive study of a large number of patients with PC neoplasms, we observed that the prevalence of p16 methylation increased as the disease progressed (24% MGUS, 28% with SMM, and 34% in MM), but only in a modest fashion. Our reported prevalence of p16 methylation falls within the range of 10% to 51% reported in previous literature.9,17-26 The heterogeneity in methodology and sample sizes may explain such variation among studies. Although p16 methylation is slightly (but significantly) more prevalent in MM than SMM/MGUS and may be a marker of advance disease, our data did not confirm previous studies that suggested p16 methylation may contribute in the transformation from MGUS to MM as a substantial proportion of MGUS already harbors p16 methylation.
Interestingly, we also showed for the first time that gene expression of p16 was not affected by p16 methylation status. It has been previously described that transcriptional silencing by promoter methylation only occurs with complete methylation27 or when crucial regions of the promoter are methylated. Some studies have shown that for some genes it is not a specific pattern of methylation that is associated with gene silencing, but rather a regional increase of methylation that changes chromatin conformations that can lead to gene inactivation.28,29 The result of our study suggests that the p16 promoter methylation detected in PC neoplasm is often partial or involves noncrucial regions of the promoter and thus not associated with transcriptional silencing. This is consistent with previous studies in MM cell lines that showed p16 methylation is not always associated with transcriptional silencing.30 Furthermore, we showed that proliferation as measured by the PCLI was not different between tumors with or without p16 methylation (Table 2), suggesting a lack of functional consequence to p16 methylation in MM. Therefore, p16 methylation in association with transcriptional silencing is an infrequent finding and probably unimportant in the pathogenesis of multiple myeloma.
Methylation of p16 has been linked with poor clinical outcome in bladder tumors, colorectal cancer, and lung cancer.31,32 In our study, and unlike previous reports,33 we were unable to observe any major effect of p16 methylation on overall survival and progression-free survival. Compared with previous studies, our survival analysis was performed on a large cohort of patients uniformly treated in a clinical trial setting with long-term follow-up such that there is minimal censoring. This lack of effect on survival is again consistent with the lack of functional impact of p16 methylation. We also found no association between p16 methylation and the main cytogenetic categories of MM, except p53 deletions (likely explaining a trend toward shorter survival among patients with p16 methylation). This association is likely due to p53 being a marker of advanced disease34 and the greater prevalence of p16 methylation in more advanced disease. Recent information suggests loss of epigenetic silencing regulation is associated with derangements of the p53 pathway.35
In summary, although we showed that p16 methylation is relatively common in PC neoplasm, it neither affects gene expression nor proliferation of malignant PCs. Consistent with this, p16 methylation has no impact on clinical outcome. The increasing prevalence of p16 methylation from MGUS to MM is consistent with other studies showing increased (cancer testis antigens through hypomethylation)36,37 or decreased (E-cadherin through hypermethylation)22 expression of epigenetically regulated genes in more advanced PC neoplasm. Therefore, p16 methylation may simply be a surrogate for overall epigenetic deregulation of the myeloma genome that is associated with disease progression.
Authorship
Contribution: N.G.-P. performed the research, analyzed the results, and wrote the paper; W.J.C. analyzed the results and wrote the paper; E.B. analyzed the data; R.F.M., B.V.N., C.D.J., and P.J.K. designed the study; K.H. and G.J.A. performed the research; M.M.O., M.G., M.L., A.D., and P.R.G. designed the study and contributed patients; R.F. designed the study, analyzed the results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rafael Fonseca, Mayo Clinic Scottsdale, 13208 E Shea Blvd, Scottsdale, AZ 85259; e-mail: fonseca.rafael@mayo.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Supported in part by the Donaldson Charitable Trust, Public Health Service (grant R01 CA83724-01) (R.F.) and (grant P01 CA62242) (P.R.G.); the National Cancer Institute (SPORE P50 CA100707-01 and P01 CA62242), (ECOG grant CA21115-25C) (P.R.G. and R.F.); the Mayo Foundation (R.F. and P.R.G.), the CI-5 Cancer Research Fund-Lilly Clinical Investigator Award of the Damon Runyon–Walter Winchell Foundation (R.F. and P.R.G.); and by an International Fellowship from the Agency of Science, Technology and Research, Singapore (A*STAR) (W.J.C.).
We are grateful to Dr W. Michael Kuehl for his useful comments, revision, and suggestions.