Abstract
Activation of circulating platelets by exposed vessel wall collagen is a primary step in the pathogenesis of heart attack and stroke, and drugs to block platelet activation have successfully reduced cardiovascular morbidity and mortality. In humans and mice, collagen activation of platelets is mediated by glycoprotein VI (GPVI), a receptor that is homologous to immune receptors but bears little sequence similarity to known matrix protein adhesion receptors. Here we present the crystal structure of the collagen-binding domain of human GPVI and characterize its interaction with a collagen-related peptide. Like related immune receptors, GPVI contains 2 immunoglobulin-like domains arranged in a perpendicular orientation. Significantly, GPVI forms a back-to-back dimer in the crystal, an arrangement that could explain data previously obtained from cell-surface GPVI inhibition studies. Docking algorithms identify 2 parallel grooves on the GPVI dimer surface as collagen-binding sites, and the orientation and spacing of these grooves precisely match the dimensions of an intact collagen fiber. These findings provide a structural basis for the ability of an immunetype receptor to generate signaling responses to collagen and for the development of GPVI inhibitors as new therapies for human cardiovascular disease.
Introduction
Thrombus formation in the arterial vasculature is a process initiated by the interaction of several platelet receptors with collagen and collagen-associated proteins at the site of vascular injury. Initially, platelets are tethered transiently to exposed collagen when the receptor GPIbα interacts with collagen-bound von Willebrand factor (VWF).1 For stable platelet adhesion to occur, the immunoglobulin (Ig)–like receptor GPVI must bind to collagen, triggering the activation of a signaling cascade.2,3 GPVI signaling leads to inside-out activation of the platelet integrins α2β1 and αIIbβ3.4,5 Activated α2β1 binds tightly to a specific sequence in collagen to allow firm adhesion of the platelets to the site of injury,5,6 and activated αIIbβ3 mediates platelet aggregation.7 In addition, GPVI signaling stimulates secretion of platelet granule contents to activate nearby circulating platelets and propagate thrombus formation. In humans, GPVI deficiency causes a loss of platelet activation in response to collagen,2,3 and GPVI polymorphisms have been linked to increased risk of myocardial infarction.8 Remarkably, loss or inhibition of GPVI prevents arterial thrombus formation in animal models but causes only mildly prolonged bleeding times in mice and humans, suggesting that GPVI could be a prime therapeutic target for prevention of arterial thrombotic diseases such as heart attack and stroke.3
The gene for GPVI is found in the leukocyte receptor cluster (LRC) on human chromosome 19.9 The sequence of the GPVI ectodomain was predicted to form 2 Ig-like domains comprising the collagen-binding domain followed by a heavily O-glycosylated stalk.2 Like other LRC receptors, GPVI associates with the FcR γ-chain coreceptor, and signaling is mediated both indirectly through the γ-chain and directly through the GPVI cytoplasmic domain.2,3 The quaternary structure of fibrous collagen is required for GPVI activation, although GPVI can also be activated by a synthetic collagen-related peptide (CRP) containing cross-linked strands of the repeating tripeptide (POG)n, where P is proline, O is hydroxyproline, and G is glycine.10 Recently, GPVI has been shown to interact with the ectodomain of GPIbα on the surface of platelets11 and to bind to laminin, a matrix protein exposed at sites of vascular injury.12 Multimeric snake venom proteins such as convulxin can also strongly activate GPVI,13 suggesting that clustering of GPVI receptors through multiple binding events leads to activation. To better understand the molecular basis for collagen activation of platelets by GPVI, we have determined the crystal structure of the collagen-binding domain (CBD) of human GPVI and have characterized its interaction with CRP by experimental and computational methods.
Materials and methods
Cloning, expression, refolding, and purification of GPVI
Human GPVI cDNA was prepared as described.14 The DNA sequence encoding the CBD (residues Q1-T183) was amplified by polymerase chain reaction (PCR) using CACCGAAAACCTGTATTTTCAGGGCCAGAGTGGACCGCTCCCC (the tobacco etch virus protease [TEVp] cleavage site is underlined) and CTATGTGACCACAAGCTCCAGCGGGTCGCTGGG as the sense and antisense primers, respectively. The PCR product was inserted into the pDEST17 vector encoding an N-terminal (His)6–tag using the GATEWAY system (Invitrogen, Carlsbad, CA), and transformed into Escherichia coli strain Tuner(DE3) (Novagen, Madison, WI). Cultures were grown to an A600 of 0.8 and induced for 4 hours at 37°C with 0.1 mM isopropyl β-d-thiogalactopyranoside. The recombinant GPVI CBD was solubilized from inclusion bodies using denaturant buffer (6 M guanidine hydrochloride, 20 mM NaH2PO4, 10 mM imidazole, 1 mM dithiothreitol, pH 7.3) and purified under denaturing conditions by immobilized-metal affinity chromatography (IMAC) using Ni-NTA agarose (QIAGEN, Valencia, CA).
The denatured and reduced protein was refolded by rapid dilution with vigorous stirring in refolding buffer (1 M l-arginine, 2 mM EDTA, 5 mM reduced l-glutathione, 0.5 mM oxidized l-glutathione, and 100 mM Tris-HCl, pH 8.8) at 4°C for 16 hours. The refolded protein was dialyzed against TEVp cleavage buffer (100 mM NaCl, 2 mM CaCl2, and 20 mM Tris-HCl, pH 8.0), cleaved overnight at room temperature, and further purified by IMAC and size exclusion chromatography using a HiLoad 26/60 Superdex 75 column (Amersham Biosciences, Piscataway, NJ) equilibrated with TBS buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.4). The recombinant GPVI was judged to be more than 90% pure by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with a yield of 0.5 to 1.0 mg per 1 L LB. CRP used for binding experiments was purchased from Peptide International (Louisville, Kentucky) as a non–cross-linked (POG)10 polypeptide.
Crystallization and structure determination
Purified GPVI was crystallized by mixing 0.4 μL GPVI (5 mg/mL in TBS) with 0.4 μL crystallization buffer (1 M ammonium sulfate and 5% MPD) in sitting drop crystallization plates. Small needle-shaped crystals appeared in 3 days and were improved by microseeding and macroseeding techniques. For seeding, 2 μL protein solution (10 mg/mL in TBS) was mixed with 2 μL crystallization buffer (0.9 M ammonium sulfate, 8% MPD, and 20% glycerol). After seeding, diamond-shaped platelike crystals grew to a maximum size of approximately 150 × 150 × 20 μm in one month. The crystals belong to the space group P21212 with 2 GPVI molecules in the asymmetric unit. Data were collected at 100 K with an R-AXIS IV++ image-plate detector using CuKα radiation generated by a Micromax-007 rotating anode generator (Rigaku MSC, The Woodlands, TX). A complete data set was collected to 0.24 nm (2.4 Å) resolution from 176 images (0.5° oscillation with 5-minute exposure time), processed by Mosflm, and scaled by SCALA from the CCP4 suite.15
The structure was solved by molecular replacement using Phaser1.3.1.16 The search model used was LIR-1 (pdb 1G0X17 ) after truncating loops, which yielded 2 clear solutions. All crystallographic refinements were performed with CNS18 using the maximum-likelihood target function. Rigid body refinement was followed by several cycles of torsion-angle simulated annealing, positional refinement, individual B-factor refinement, and manual model rebuilding. Both 2Fo – Fc and Fo – Fc electron density maps were used to manually rebuild the model with XtalView.19 When the value of the R-factor dropped to 24%, solvent molecules and ions were gradually included. The electron density was well defined for the overall structure except for 2 residues of the N-terminus for molecule A and residues 99 to 107 and 130 to 137 for molecule B. The final model contained 182 and 167 residues for molecules A and B, respectively. Criteria for inclusion of solvent and ion molecules included height and shape of the electron density peaks and appropriate coordination by GPVI residues. Data collection and refinement statistics are reported in Table 1.
Analytical ultracentrifugation
Sedimentation velocity and equilibrium experiments were carried out at 20°C in a Beckman XL-I ProteomeLab analytical ultracentrifuge (Beckman Coulter, Palo Alto, CA) using absorbance optics, as described.20 For sedimentation velocity experiments, samples of GPVI or mixtures with CRP were spun at 48 000 rpm. Sedimentation coefficients were determined using the program SEDFIT.21 For sedimentation equilibrium experiments, mixtures of 10 μM CRP and 10, 40, and 80 μM GPVI were spun at speeds of 16 000, 19 000, 29 000, 35 000, and 48 000 rpm. Data files were trimmed and analyzed by global fitting using the programs WinREEDIT and WinNONLIN (Jeff Lary, University of Connecticut, Storrs, CT). Values of sw, the weight-average sedimentation coefficient determined by SEDFIT, were fitted to a single-site binding isotherm using SEDPHAT.22
Docking of CRP to GPVI
We used the PatchDock server23 (http://bioinfo3d.cs.tau.ac.il/PatchDock) to predict the binding orientation of CRP on GPVI. A truncated CRP with the sequence (POG)5 was created from the crystal structure of (POG)4POA(POG)5 (pdb 1CAG24 ) and used as the ligand, with GPVI D1 as the receptor. Six of the top 10 solutions showed CRP bound in a putative binding groove adjacent to the C′E loop and neighboring several residues implicated in collagen and CRP binding (K41, K59, R60, and R16625,26 ). Three of these solutions bound in one orientation (ie, with the N-termini of CRP closer to GPVI residue F54), and the other 3 solutions bound in the opposite orientation (ie, with the C-termini closer to F54), consistent with the pseudo-2-fold symmetry within CRP.
We also used the FTDock program from the 3D-Dock software package to dock CRP to GPVI.27 Because FTDock does not recognize hydroxyproline, a modified CRP with the sequence (PPG)5 was created from the crystal structure of (PPG)10, (pdb 1K6F28 ). The grid-based shape complementarity search was performed using 148 grid units in each dimension (grid point spacing: 0.07 nm [0.7 Å]). The docking solutions were sorted by surface complementarity and the top solution was also found in the same surface groove in D1 identified by PatchDock. Furthermore, filtering the solutions by proximity to K59 reveals a second solution within the binding groove, but in the opposite orientation.
Calculation of estimated 2D Kd at platelet surface
Correlating the reported GPVI receptor density on the platelet surface29 with an estimated Kd for dimerization of soluble GPVI CBD was carried out according the approach of Dustin et al.30,31 The interaction of 2 membrane-embedded receptors is estimated to occur with a loss of 3 degrees of freedom (compared with their interaction as soluble receptors), due to their restriction to 2-dimensional diffusion within the plane of a lipid bilayer.32 The 3D Kd was converted into ΔG and corrected by a factor of –1.5 RT to account for the 2D restriction of the receptors. The corrected 3D Kd* was then converted from molar units to receptors per cubic micrometer. Finally, the 2D Kd was estimated based on the equation 2D Kd =σ (3D Kd*), where σ is the confinement region corresponding to the distance the receptor extends outward from the membrane. For GPVI, the confinement region was calculated to be 22 nm by adding the height of the CBD (5.2 nm) and the estimated height of the 65-residue O-glycosylated stalk (16.9 nm, based on the reported dimensions of 0.26 nm per residue for the O-glycosylated stalk of CD833 ).
Other computational methods
Generation of collagen fiber models was carried out by applying crystallographic symmetry to the CRP structures (pdb 1CAG, 1CGD, and 1BKV) using the program O.34 Analysis of buried surface areas and interdomain angles were calculated using Areaimol from the CCP4 suite15 and Dom_Angle, respectively.35 Domain boundaries were defined as residues 0 to 89 for D1 and residues 90 to 183 for D2. Figures were generated with PyMOL (DeLano Scientific, San Francisco, CA),36 Molscript,37 and Raster3D.38
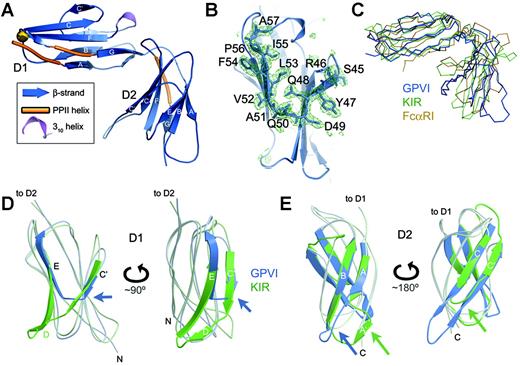
Structure of the GPVI CBD. (A) Ribbon diagram of GPVI. The N-terminal domain is labeled D1, and the C-terminal domain is D2. The predicted N-glycosylation site at N72 is shown by a gold ball. (B) Simulated annealing electron density omit map showing the C′E region of the CBD that differs from related immune receptors, with a ball-and-stick model of the deleted region superimposed. (C) Superposition of GPVI (blue), p58 KIR40 (green), and FcαRI41 (yellow). The orientation is similar to that shown in panel A. (D) Superposition of D1 of GPVI (blue) and p58 KIR40 (green) shown in 2 orientations, highlighting the unusual C′E region in GPVI (blue arrows). (E) Superposition of D2 of GPVI (blue) and p58 KIR40 (green) in 2 orientations, highlighting the lack of an A′ strand and the extended CC′ hairpin in GPVI (note blue/green arrows).
Structure of the GPVI CBD. (A) Ribbon diagram of GPVI. The N-terminal domain is labeled D1, and the C-terminal domain is D2. The predicted N-glycosylation site at N72 is shown by a gold ball. (B) Simulated annealing electron density omit map showing the C′E region of the CBD that differs from related immune receptors, with a ball-and-stick model of the deleted region superimposed. (C) Superposition of GPVI (blue), p58 KIR40 (green), and FcαRI41 (yellow). The orientation is similar to that shown in panel A. (D) Superposition of D1 of GPVI (blue) and p58 KIR40 (green) shown in 2 orientations, highlighting the unusual C′E region in GPVI (blue arrows). (E) Superposition of D2 of GPVI (blue) and p58 KIR40 (green) in 2 orientations, highlighting the lack of an A′ strand and the extended CC′ hairpin in GPVI (note blue/green arrows).
Results
Crystal structure of the collagen-binding domain of GPVI
The crystal structure of the GPVI CBD was solved by molecular replacement using data to a resolution of 0.24 nm (2.4 Å). The CBD is composed of 2 Ig-like domains oriented 90° apart, similar to other LRC receptors such as FcαRI and the leukocyte Ig-like (LILR or LIR) and killer-cell Ig-like (KIR) receptor families17,39-41 (Figure 1A,C; for a sequence alignment see Figure S1, available on the Blood website by clicking on the Supplemental Figure link at the top of the online article). Both the N-terminal and C-terminal domains (D1 and D2, respectively) of GPVI most closely resemble the domains of p58 KIR,40 with rms deviations of 0.17 and 0.16 nm (1.7 and 1.6 Å) over 83 and 82 residues, respectively. There were 2 GPVI molecules per asymmetric unit, with D1-D2 interdomain angles ranging from 90° to 92° and with D1-D2 interfaces that buried 8.55 to 8.77 nm2 (855-877 Å2).2 These interdomain angles and interface areas are comparable with those observed for FcαRI, LIR-1, and p58 KIR, although the interface area for GPVI is somewhat less extensive. As in many LRC receptors, a conserved cis proline in D1 (P14) introduces a bend after the first β strand, creating a distinct A′ strand that forms hydrogen bonds with the G strand. As a result, the Ig fold of D1 is I-type, formed by 2 β sheets composed of the ABE and A′GFCC′ strands. D1 also contains a short 310 helix and 2 stretches of polyproline type II helix. Within D1, the most significant differences between GPVI and other LRC receptors occur in the C′ and E strands and intervening C′E loop, which adopt unusual conformations due to an 11-residue deletion in GPVI (Figure 1B,D). This deletion creates a shallow, primarily hydrophobic groove on the surface of D1 bordered by charged and polar residues including K41, K59, R60, and R166, which have been implicated in collagen or CRP binding.25,26 There is a single predicted N-glycosylation site at residue N72 in the FG loop of D1; the N-glycan would be expected to extend outward from the end of D1, as seen for FcαRI.41
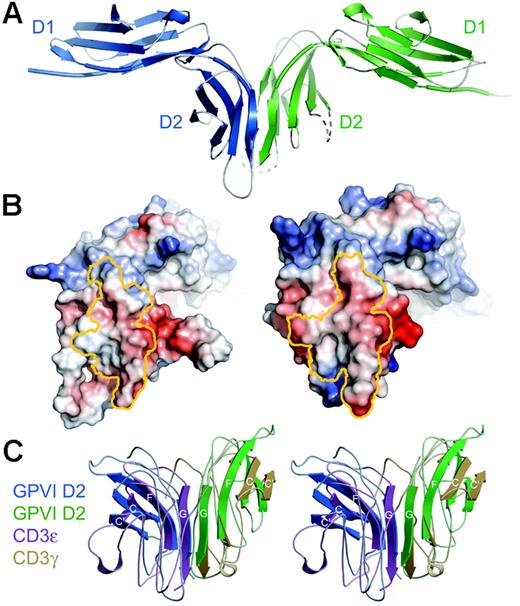
Dimeric conformation of GPVI in the crystal. (A) Structure of the GPVI dimer in the asymmetric unit. (B) Electrostatic potential of GPVI mapped onto the surface of each GPVI protomer, shown in an “open-book” view. Residues contributing to the dimer interface on each protomer have been outlined in yellow. The potential map has been contoured from –100 to +100 kT. (C) Stereoview of a superposition of the dimers formed by GPVI D2 (blue/green) and the CD3ϵγ heterodimer42 (purple/yellow). For clarity, only those β-strands in the continuous β-sheet are shown as ribbons; all others are shown as coils.
Dimeric conformation of GPVI in the crystal. (A) Structure of the GPVI dimer in the asymmetric unit. (B) Electrostatic potential of GPVI mapped onto the surface of each GPVI protomer, shown in an “open-book” view. Residues contributing to the dimer interface on each protomer have been outlined in yellow. The potential map has been contoured from –100 to +100 kT. (C) Stereoview of a superposition of the dimers formed by GPVI D2 (blue/green) and the CD3ϵγ heterodimer42 (purple/yellow). For clarity, only those β-strands in the continuous β-sheet are shown as ribbons; all others are shown as coils.
GPVI D2, like D1, contains the conserved proline (P100) at the end of the A strand, but in D2 it adopts the trans rather than cis conformation. As a result, there is no sharp bend in the protein backbone, the A′ strand does not form, and the architecture of the subsequent AB loop is significantly altered (Figure 1E). The topology of the D2 domain is therefore a C2-type Ig fold with ABE and GFCC′ β sheets, rather than the typical I-type fold. GPVI D2 diverges somewhat from the canonical C2 fold, which features a very short C′ strand of 3 residues. Instead, GPVI D2 has significantly elongated C and C′ strands, each containing 10 residues. Both the unusual AB loop and CC′ β hairpin extend outward from the main body of D2 and appear to be quite flexible, given the lack of clear electron density for these regions in a second independent molecule within the asymmetric unit.
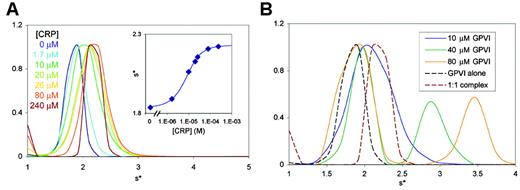
GPVI-CRP binding affinity and stoichiometry. (A) Normalized sedimentation velocity size distribution from a titration of 6.7 μM GPVI with CRP (1.7 to 240 μM). (Inset) Fitting the sw values as a function of CRP concentration to a binding isotherm yields a Kd of 5 μM. (B) Normalized sedimentation velocity size distributions for 1:1, 4:1, and 8:1 GPVI/CRP mixtures (solid lines), with the free GPVI and final 1:1 complex from panel A included for comparison (dashed lines). Note that the mixtures with 4- and 8-fold molar excess of GPVI (green and yellow lines) form complexes larger than a 1:1 complex (red dashed line).
GPVI-CRP binding affinity and stoichiometry. (A) Normalized sedimentation velocity size distribution from a titration of 6.7 μM GPVI with CRP (1.7 to 240 μM). (Inset) Fitting the sw values as a function of CRP concentration to a binding isotherm yields a Kd of 5 μM. (B) Normalized sedimentation velocity size distributions for 1:1, 4:1, and 8:1 GPVI/CRP mixtures (solid lines), with the free GPVI and final 1:1 complex from panel A included for comparison (dashed lines). Note that the mixtures with 4- and 8-fold molar excess of GPVI (green and yellow lines) form complexes larger than a 1:1 complex (red dashed line).
The GPVI CBD forms a dimer in the crystal
The asymmetric unit of the crystal contains a parallel, back-to-back dimer formed by the D2 domains of the 2 GPVI molecules (Figure 2A). The G strands of the D2 domains interact to create a continuous β sheet across the dimer interface. The unusual D2 architecture lacking an A′ strand is necessary for dimer formation, since the presence of A′ strands would sterically block the G strands from forming a continuous β sheet. The dimer interface buries 10.46 nm2 (1046 Å2) of accessible surface area, which is within the range expected for dimeric proteins, although lower than average for a protein of this size.43 The dimer interface shows excellent surface complementarity, with a shape complementarity index of 0.73, comparable with oligomeric proteins (Sc = 0.70-0.74) and protease-inhibitor complexes (Sc = 0.71-0.76).44 The interface is dominated by hydrophobic interactions along with hydrogen bonds contributed by the peptide backbone in the parallel G strands of both D2 domains (Figure 2B). Of interest, the GPVI dimerization mode is essentially identical to that observed for CD3ϵγ and CD3ϵδ receptor heterodimers in the T-cell receptor (TCR) complex.42,45,46 Like GPVI, CD3ϵγ and CD3ϵδ form back-to-back dimers with the G strands forming a continuous β sheet (Figure 2C). Indeed, the CD3ϵγ dimer42 superimposes on the GPVI D2 dimer with an rms deviation of only 0.2 nm (2.0 Å) over 55 residues, which is remarkable given the low sequence identity between GPVI and the CD3 receptors (20% or 14% identity between GPVI and CD3ϵ or γ, respectively).
Interaction of GPVI CBD and collagen-related peptide in solution
To understand how GPVI associates with the macromolecule collagen, we next studied the interaction between the GPVI CBD and CRP, which functionally mimics collagen in biologic assays. To analyze the affinity of the interaction under conditions favoring a 1:1 complex, sedimentation velocity analytical ultracentrifugation experiments were carried out by titrating GPVI with CRP (a non–cross-linked (POG)10 triple helix) at up to 35-fold molar excess (Figure 3A). The weight-averaged sedimentation coefficient (sw) of each dataset was plotted as a function of CRP concentration and fitted to a single-site binding isotherm, yielding a Kd of 5 μM (Figure 3A inset). This affinity is significantly tighter than that determined by surface plasmon resonance (SPR)47 ; however, the SPR experiment measured binding of GPVI to immobilized, cross-linked CRP with a different sequence than the non–cross-linked CRP described here.
Additional sedimentation velocity and sedimentation equilibrium experiments conducted using 1:1, 4:1, or 8:1 molar ratio mixtures of GPVI/CRP indicated that multiple GPVI molecules can bind to a single CRP triple helix, consistent with the presence of multiple overlapping sites for GPVI within the repeating tripeptide sequence of the CRP triple helix. The sw values indicated formation of a 1:1 complex in the first sample and formation of higher-order complexes in the 4:1 and 8:1 mixtures (Figure 3B). Sedimentation equilibrium experiments conducted in parallel on similar mixtures indicated that 2 or 3 molecules of GPVI could bind simultaneously to a CRP triple helix (data not shown).
Computational docking of CRP onto GPVI. (A) Side view of the surface of D1, illustrating the predicted binding of CRP within the binding groove. Two distinct docking algorithms placed CRP within the groove in D1 adjacent to the C′E region (shown in yellow). (B) Model of the GPVI-CRP complex predicted by the PatchDock and FTDock algorithms. The docking prediction for CRP determined by each program is shown in red with the C′E loop labeled for reference. Three crystal structures of CRP variants with their PDB IDs are illustrated above the predicted GPVI-CRP complex to show that the spacing and orientation of the docked CRP triple helices (red) match the conserved approximately 1.4 nm (14 Å) spacing observed in the CRP crystal structures, which is itself similar to that observed in intact collagen fibers. Of note, although collagen fibers have circular cross-sections, within a blood vessel the average fiber diameter is very large (∼ 30-40 nm [∼300-400 Å]) relative to the spacing between individual collagen triple helices (∼ 1.4 nm [14 Å]), such that the surface recognized by GPVI would be approximately planar. (C) Electrostatic potential of GPVI D1 mapped onto its surface, with the PatchDock CRP model superimposed. (D) Stereoview of the residues in D1 that contact CRP in one or both of the docked models. Side chains are colored according to chemical properties: yellow are hydrophobic, green are polar, and blue are positively charged. (E) Residues implicated in collagen or CRP binding are shown in color on the GPVI surface after adding modeled N-glycans. Purple residues (V34, K41, K59, and N-glycan) have been implicated in both collagen and CRP binding,25,26,48,49 green residues (L36, R60, R166) have been implicated in collagen binding only,25,48 and light-blue residues (F91, R117, Y118, F120, R139, S164) were mutated with no effect on either collagen or CRP binding.25
Computational docking of CRP onto GPVI. (A) Side view of the surface of D1, illustrating the predicted binding of CRP within the binding groove. Two distinct docking algorithms placed CRP within the groove in D1 adjacent to the C′E region (shown in yellow). (B) Model of the GPVI-CRP complex predicted by the PatchDock and FTDock algorithms. The docking prediction for CRP determined by each program is shown in red with the C′E loop labeled for reference. Three crystal structures of CRP variants with their PDB IDs are illustrated above the predicted GPVI-CRP complex to show that the spacing and orientation of the docked CRP triple helices (red) match the conserved approximately 1.4 nm (14 Å) spacing observed in the CRP crystal structures, which is itself similar to that observed in intact collagen fibers. Of note, although collagen fibers have circular cross-sections, within a blood vessel the average fiber diameter is very large (∼ 30-40 nm [∼300-400 Å]) relative to the spacing between individual collagen triple helices (∼ 1.4 nm [14 Å]), such that the surface recognized by GPVI would be approximately planar. (C) Electrostatic potential of GPVI D1 mapped onto its surface, with the PatchDock CRP model superimposed. (D) Stereoview of the residues in D1 that contact CRP in one or both of the docked models. Side chains are colored according to chemical properties: yellow are hydrophobic, green are polar, and blue are positively charged. (E) Residues implicated in collagen or CRP binding are shown in color on the GPVI surface after adding modeled N-glycans. Purple residues (V34, K41, K59, and N-glycan) have been implicated in both collagen and CRP binding,25,26,48,49 green residues (L36, R60, R166) have been implicated in collagen binding only,25,48 and light-blue residues (F91, R117, Y118, F120, R139, S164) were mutated with no effect on either collagen or CRP binding.25
Computational determination of CRP binding sites on the GPVI dimer
How does the GPVI dimer recognize CRP and fibrous collagen? Unfortunately, complexes of GPVI with CRP were found to be resistant to crystallization, most likely due to excessive heterogeneity of the complexes, which results from the ability of GPVI to bind at multiple overlapping sites along the triple helix. In order to identify collagen-binding sites on GPVI, we therefore used 2 different computational algorithms, PatchDock23 and FTDock,27 to dock CRP onto GPVI. Both docking programs positioned CRP within the shallow groove on D1 adjacent to the C′E loop (Figure 4A-B). The floor of the putative binding groove is formed by several hydrophobic residues (L53, F54, P56, L62, and Y66, and the aliphatic portion of K41), with several polar (S43, S44, Q48, Q50, S61) and basic (K41, R46, K59, R166) residues around the periphery (Figure 4B-D). This groove is unique to GPVI among LRC receptors, as it results from the 11-residue deletion in GPVI. The binding of CRP to this groove provides a structural explanation for how an immune-type receptor has evolved to bind vessel wall collagen. In these models, CRP is immediately adjacent to GPVI residues K41, K59, R60, and R166, which have been implicated as collagen and CRP-binding residues by mutational analysis, as described in “Discussion”25,26 (Figure 4E). Furthermore, previously described GPVI mutations having no effect on CRP affinity are not located within the putative binding groove25,26 (Figure 4E, light-blue surfaces). The docked CRP solutions from each program are evenly distributed between 2 nearly opposite orientations within the groove. This is consistent with the pseudo-2-fold symmetry present in CRP, caused by its imino groups (prolines and hydroxyprolines) occupying similar positions in both orientations.
Native collagen fibers are composed of a pseudo-hexagonal array of parallel CRP-like triple helices separated by 1.3 to 1.4 nm (13 to 14 Å),50 an arrangement that is also conserved in crystal structures of soluble CRP-like peptides24 (Figure 4B top). Of interest, the 2 putative CRP-binding grooves within a GPVI dimer are essentially parallel and are separated by approximately 5.5 nm (55 Å), which is equivalent to the distance between the n and n + 4 helices in a collagen fiber. The geometric compatibility of the binding grooves with collagen helices would allow the GPVI dimer to bind simultaneously to 2 helices within a collagen fiber.
Discussion
The receptor GPVI is central to the process of collagen-mediated platelet activation and subsequent thrombus formation. The atomic structure of GPVI is therefore of interest in terms of understanding how an immune-type receptor can recognize fibrous collagen. Furthermore, the structure allows the identification of potential regions responsible for interacting with collagen, which may serve as desirable targets for inhibitory drugs. The crystallographic data presented here reveal that the GPVI CBD adopts a fold previously seen in related immune receptors of the leukocyte receptor cluster, but an 11-residue deletion in the sequence of GPVI relative to other LRC receptors creates a shallow groove on the surface of D1 that forms a putative collagen-binding site, based on docking algorithms and mutagenesis data. The CBD forms a back-to-back dimer in the crystal in which the 2 putative collagen-binding grooves are nearly parallel and separated by 5.5 nm (55 Å), a configuration that matches the orientation and dimensions of triple helices within fibrous collagen.
The dimeric GPVI conformation observed in the crystal is intriguing and may well represent the physiologically relevant form of GPVI on the platelet surface. Previous studies have shown that soluble GPVI-Fc fusions, but not monomeric soluble GPVI, inhibited platelet activation,2,47,51,52 suggesting that either a dimeric conformation or the higher avidity conferred by the Fc fusion was required to effectively compete with cell-surface GPVI for binding to collagen. This was further supported by surface plasmon resonance assays showing that the GPVI-Fc fusion bound collagen nearly 200-fold more tightly than monomeric GPVI did.47
The data presented here suggest that GPVI dimerization is a rather weak interaction that nonetheless could occur on the platelet surface. Analytical ultracentrifugation experiments indicated that the soluble GPVI CBD construct used for crystallization remained monomeric in solution at up to 100 μM (Figure 3A and data not shown). However, the construct we crystallized lacks the stalk region, which could help stabilize a dimeric conformation, as seen for CD94.53 Furthermore, the high density of GPVI at the platelet surface29 would favor dimer formation. It is well established that weak protein-protein interactions in solution occur to a significant extent when the components are restricted to 2-dimensional diffusion in a lipid bilayer.31 For example, if the Kd for GPVI dimerization in solution were 420 μM (∼ 8.5 mg/mL), this would correspond to a 2D Kd of 1260 receptors per square micrometer, which is equivalent to the GPVI density on platelets.29 Thus even very weak dimerization of the CBD in solution would be sufficient to allow significant dimerization of GPVI at the platelet surface. Of interest, activation of GPVI signaling by collagen is critically dependent on GPVI surface density; RBL cells transfected with GPVI were unresponsive to collagen unless expressed at a surface density approximating that observed on platelets.14,29 These studies, like the inhibition studies using GPVI-Fc fusion proteins, suggest that increased avidity of GPVI—or potentially the dimeric GPVI conformation described here—are required for effective interaction with collagen.
The GPVI structural data provide a framework for understanding the interaction between GPVI and collagen or CRP by allowing accurate mapping of mutagenesis results onto the surface of the GPVI dimer. The residues implicated in collagen or CRP binding fall into 2 clusters: the primary region includes basic residues on the surface of D1 including K41, K59, R60, and R16625,26 (Figure 4E). Mutation of K41 or K59 affects binding to both collagen and CRP; a K41A mutation increases the affinity of GPVI for both ligands, whereas mutation of residue K59 to the mouse equivalent (K59E) decreases affinity for both collagen and CRP.25,26 The R60A and R166A mutations reduce collagen affinity but have no effect on CRP binding.25 A second cluster of residues implicated in collagen or CRP binding is found at the distal end of D1; these residues include L36, implicated in collagen (but not CRP) binding, and both V34 and the N-glycan attached to N72, both of which are involved in collagen and CRP binding.48,49 The GPVI structure shows that the side chain of V34 is buried and its mutation is likely to alter the conformation of the BC loop in D1, which includes L36 and is immediately adjacent to N72.
In our computational model of CRP docked to GPVI, the CRP binding groove is located within the primary cluster of basic residues (K41, K59, R60, and R166). K41 is centrally positioned within the floor of the putative binding groove and contacts CRP directly. Furthermore, the docked CRP interacts with the side chain of R166 and is within reach of the K59 and R60 side chains. The docking predictions were based strictly on geometric and energetic criteria and did not take into account any mutagenesis data. Therefore, the correlation between the docking prediction and mutagenesis results suggests that the predicted CRP binding mode is a reasonable approximation of the physiologic interaction of GPVI with CRP and collagen.
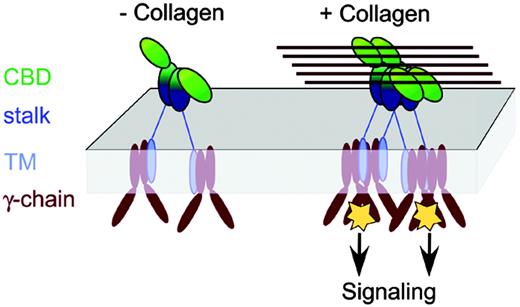
Implications for GPVI signaling. A model is shown for signaling by receptor clustering, in which clustering triggers a signaling cascade via the FcR γ-chain coreceptor. This would resemble TCR signaling via the related ζ-chain coreceptor, which occurs by clustering of at least 2 TCR complexes in an orientation-independent manner, with the proximity of the TCR complexes correlating to the strength of the signal.54 An individual GPVI dimer would not necessarily allow the γ-chains to approach closely enough to trigger signaling, due to the long GPVI stalk region. However, multiple GPVI molecules bound to a collagen fiber would juxtapose several γ-chains in a small volume, triggering activation. Although GPVI in the model is illustrated as a dimer, the proposed mechanism of GPVI clustering by fibrous collagen would also apply if GPVI were monomeric at the platelet surface.
Implications for GPVI signaling. A model is shown for signaling by receptor clustering, in which clustering triggers a signaling cascade via the FcR γ-chain coreceptor. This would resemble TCR signaling via the related ζ-chain coreceptor, which occurs by clustering of at least 2 TCR complexes in an orientation-independent manner, with the proximity of the TCR complexes correlating to the strength of the signal.54 An individual GPVI dimer would not necessarily allow the γ-chains to approach closely enough to trigger signaling, due to the long GPVI stalk region. However, multiple GPVI molecules bound to a collagen fiber would juxtapose several γ-chains in a small volume, triggering activation. Although GPVI in the model is illustrated as a dimer, the proposed mechanism of GPVI clustering by fibrous collagen would also apply if GPVI were monomeric at the platelet surface.
The cluster of residues including V34, L36, and the N-glycan at N72 may form a secondary binding site for CRP or collagen triple helices. Residue L36 is located on the surface of D1 approximately 1.4 nm (14 Å) from the putative binding groove, and these secondary residues are positioned such that they could interact with adjacent triple helices within an intact collagen fibril. The mutagenesis results are therefore consistent with the mode of interaction between GPVI and a collagen fibril illustrated in Figure 4B, in which a GPVI dimer binds simultaneously to several triple helices within the collagen fibril. This binding mode also suggests a model for the initiation of signal transduction triggered by receptor clustering that accompanies collagen binding (Figure 5).
These studies establish a structural basis for the ability of platelets to recognize and be activated by the vessel wall matrix protein collagen. Platelet activation is a critical step in the pathogenesis of human vascular diseases and new antiplatelet agents have revolutionized the immediate treatment of myocardial infarction. The early role of GPVI in arterial thrombus formation and the relative lack of bleeding associated with human GPVI-deficiency states suggest that new therapies aimed at inhibiting GPVI function might provide an ideal long-term treatment approach to these diseases. A structural understanding of collagen recognition by GPVI will provide a foundation for the development of such novel therapeutic agents.
Prepublished online as Blood First Edition Paper, April 6, 2006; DOI 10.1182/blood-2006-01-010215.
Supported in part by funds from the State of Ohio Eminent Scholar Program and an Award from the American Heart Association to A.B.H.
K.H. and A.B.H. designed research, performed research, analyzed data, and wrote the paper; and M.L.K. contributed vital reagents and wrote the paper.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Rhett Kovall, Tom Thompson, and Jeff Wilson for helpful advice, and Dr Pamela Bjorkman and Deb Conrady for comments on the article.
Coordinates have been deposited at the RCSB protein data bank (PBD ID 2GI7).

![Figure 4. Computational docking of CRP onto GPVI. (A) Side view of the surface of D1, illustrating the predicted binding of CRP within the binding groove. Two distinct docking algorithms placed CRP within the groove in D1 adjacent to the C′E region (shown in yellow). (B) Model of the GPVI-CRP complex predicted by the PatchDock and FTDock algorithms. The docking prediction for CRP determined by each program is shown in red with the C′E loop labeled for reference. Three crystal structures of CRP variants with their PDB IDs are illustrated above the predicted GPVI-CRP complex to show that the spacing and orientation of the docked CRP triple helices (red) match the conserved approximately 1.4 nm (14 Å) spacing observed in the CRP crystal structures, which is itself similar to that observed in intact collagen fibers. Of note, although collagen fibers have circular cross-sections, within a blood vessel the average fiber diameter is very large (∼ 30-40 nm [∼300-400 Å]) relative to the spacing between individual collagen triple helices (∼ 1.4 nm [14 Å]), such that the surface recognized by GPVI would be approximately planar. (C) Electrostatic potential of GPVI D1 mapped onto its surface, with the PatchDock CRP model superimposed. (D) Stereoview of the residues in D1 that contact CRP in one or both of the docked models. Side chains are colored according to chemical properties: yellow are hydrophobic, green are polar, and blue are positively charged. (E) Residues implicated in collagen or CRP binding are shown in color on the GPVI surface after adding modeled N-glycans. Purple residues (V34, K41, K59, and N-glycan) have been implicated in both collagen and CRP binding,25,26,48,49 green residues (L36, R60, R166) have been implicated in collagen binding only,25,48 and light-blue residues (F91, R117, Y118, F120, R139, S164) were mutated with no effect on either collagen or CRP binding.25](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/3/10.1182_blood-2006-01-010215/4/m_zh80150699210004.jpeg?Expires=1765917164&Signature=IM~zCAfLNnRBFEMpJKbXjeKAY5qJBk6iSzCF7~4CHc2fo0uIFrTWYShHF~l9fJWoIxabnVpDvmFmM7RpnV1lF-lTdnDrDG7~SrZsC0BvFlSFrRrdkCZs71F~m7JyFwq2773oXmCaay60cnwb5NmT1r7QbQCD5BuXagueb0p5kK869~ojRVUHEScw6Nq5OCpNxE4dPtOOSIbLYzmY7lRSV16rhEY2kii2ZOaV5mkuQOWdpWIAedtep91GR1mCGd5bTvpKpGDRh33lc9GPAsSeCTuMRXi9ht~DtrQ0gxDC8SXI68pizi4R2ABwB7h1--7wpvJN2jJMK9M28LdFeMNUOA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)