Abstract
In this study, we demonstrated that STAT3, a well-characterized transcription factor expressed in continuously activated oncogenic form in the large spectrum of cancer types, induces in malignant T lymphocytes the expression of DNMT1, the key effector of epigenetic gene silencing. STAT3 binds in vitro to 2 STAT3 SIE/GAS-binding sites identified in promoter 1 and enhancer 1 of the DNMT1 gene. STAT3 also binds to the promoter 1 region and induces its activity in vivo. Treatment of the malignant T lymphocytes with STAT3 siRNA abrogates expression of DNMT1, inhibits cell growth, and induces programmed cell death. In turn, inhibition of DNMT1 by a small molecule inhibitor, 5-aza-2-deoxy-cytidine, and 2 DNMT1 antisense DNA oligonucleotides inhibits the phosphorylation of STAT3. These data indicate that STAT3 may in part transform cells by fostering epigenetic silencing of tumor-suppressor genes. They also indicate that by inducing DNMT1, STAT3 facilitates its own persistent activation in malignant T cells. Finally, these data provide further rationale for therapeutically targeting STAT3 in T-cell lymphomas and, possibly, other malignancies.
Introduction
Aberrant DNA methylation plays a key role in carcinogenesis by frequently leading to epigenetic silencing of the expression of tumor-suppressor genes.1-3 DNA methylation typically affects areas rich in CpG sequences within the gene promoter region and is mediated by DNA methyltransferase 1 (DNMT1) and other members of the DNMT family.4 Despite the critical role of DNMT1 in carcinogenesis,5 regulation of its expression remains poorly understood.6,7
Cell signal transducer/transcription factors (STATs) are members of the ubiquitously expressed family of transcription factors activated in response to growth factors and cytokines.8 Activation of STATs requires their phosphorylation and translocation into the nucleus to initiate transcription of the target genes. Persistent activation of STATs, particularly STAT3, has been implicated in the pathogenesis of a whole spectrum of malignancies,9 including at least 2 types of T-cell lymphoma.10-13
Here we report that STAT3 directly induces the expression of DNMT1 in malignant T cells. Interestingly, DNMT1 expression is required for the maintenance of STAT3 activation. These findings indicate that STAT3 may in part mediate malignant cell transformation by facilitating DNA methylation and, consequently, epigenetic gene silencing of tumor-suppressor genes. In addition, by inducing DNMT1, STAT3 secures its own persistent activation by enabling epigenetic silencing of its negative regulator(s), such as SHP-1.14 These observations provide evidence for the direct link between aberrant STAT signaling and the promotion of epigenetic gene silencing and further support the notion of therapeutically targeting STAT3 in cancer cells.
Materials and methods
Cells and tissues
The T-cell lymphoma cell lines used in this study have been described in detail.13 In brief, PB-1, 2A, and 2B cell lines were established from a patient with a progressive T-cell lymphoma involving skin.11 SUDHL-1, JB6, and Karpas 299 cell lines were derived from an anaplastic lymphoma kinase–expressing T-cell lymphoma.12 The HUT102B line represents HTLV-I–related adult-type T-cell lymphoma/leukemia. RAJI and DAUDI cell lines were derived from the Burkitt lymphoma. Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors. Activated PBMCs (PBMC/phytohemagglutinin [PHA]) were obtained by 3-day stimulation with the mitogen (PHA; Sigma). Cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin/streptomycin mixture, and 2 mM l-glutamine. Tissue sections of lymph nodes and skin were obtained from 15 patients with advanced cutaneous T-cell lymphoma with histologic evidence of large-cell transformation.14
Western blot analysis
For DNMT1 and STAT3 expression analysis, 50 μg nuclear protein samples were electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels for 120 minutes at 80 V and were transferred onto membranes for 60 minutes at 100 V in 10% methanol transfer buffer. For caspase-3 analysis, 50 μg protein samples were separated on 15% SDS-PAGE and were transferred onto membranes at 80 V for 3 hours in 20% methanol transfer buffer. Membranes were then probed with the appropriate antibody. Immunoreactive proteins were visualized using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotec, Uppsala, Sweden) and were exposed to x-ray film. Antibodies against DNMT1, STAT1, STAT3, STAT4, STAT5, STAT6, DNMT3b, and actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and antibodies against STAT3 phosphorylated at Tyr705 and cleaved caspase 3 were obtained from Cell Signaling (Beverly, MA).
Immunohistochemical staining
Staining was performed as described12,13 with formalin-fixed tissue sections with the use of antigen retrieval and streptavidin-biotin complex (Research Genetics, Huntsville, AL) techniques and the antibodies against phospho-STAT3 (Cell Signaling Technology, Beverly, MA) and DNMT1 (Santa Cruz Technology, Santa Cruz, CA). Slides were counterstained with hematoxylin and examined with a Leica DMLB microscope (Leica Microsystems, Wetzlar, Germany) using dry lens and the following objectives: N Plan 40×/0.65 NA for 400× magnification and N plan 10×/0.25 NA for 100× magnification. Photomicrographs were taken with a Spot Insight color digital camera, version 3.2.0 (Diagnostic Instruments, Sterling Heights, MI). Spot software version 3 was used to acquire images, and image contrast was adjusted with Adobe Photoshop CS 8.0 software (Adobe Systems, San Jose, CA).
Isolation of total RNA, reverse transcription–polymerase chain reaction, TA cloning, and DNA sequencing
Total RNA was prepared from T-cell lymphoma cell lines and PBMCs using Trizol reagent (Life Technologies, Bethesda, MD), and cDNAs were made from 5 μg total RNAs using the Superscript Preamplification System (Life Technologies) according to the manufacturer's protocol. Oligo-dT primers were used to prime first-strand synthesis for all reactions. For polymerase chain reaction (PCR), 2 μL transcriptase reaction mixture was diluted with 50 μL PCR buffer containing 0.2 mM each deoxynucleotide triphosphate, 1.5 mM MgCl2, 1.25 U Taq DNA polymerase (Life Technologies), and 20 pmol each of primer. Primers specific for the DNMT1 gene were 5′-GTATCGCCTCTCTCCGTTTG -3′ (sense) and 5′-TACGGGCTTCACTTCTTGCT -3′ (antisense); primers for the GAPDH gene used as a control were 5′-ATGGGGAAGGTGAAGGTCGGAGTC-3′ (sense) and 5′-CCATGCCAGTGAGCTTCCCGTTC-3′ (antisense). PCR was performed for 30 cycles under the following conditions: 3 minutes at 94°C, 30 seconds at 94°C, 45 seconds at 60°C, 1 minute 30 seconds at 72°C, and 10 minutes at 72°C. PCR products were electrophoresed in 1.5% agarose gel. The specific band was excised and purified by the Qiaex II gel extraction kit (QIAGEN, Valencia, CA). Purified PCR products were subcloned into pCR2.1 using the TA Cloning Kit (Invitrogen, Carlsbad, CA). T7 primer or M13 reverse primer was used for sequencing. Each sequencing reaction contained 0.5 μg DNA, 3.2 pmol primer, and 4 μL sequencing mix in a final volume of 10 μL. Sequencing products were analyzed by an automated DNA sequencer.
Electrophoretic mobility shift assay
The following oligonucleotide duplexes containing STAT3 candidate binding sites (TT N5-6 AA) were used: for promoter 1, 5′-ATTGGAAACTGAGGACTTTACTCAAG-3′ (sense) and 5′-CTTGAGTAAAGTCCTCAGTTTCCAAT-3′ (antisense); for enhancer 1, 5′-AGCCACGTGGTGCTTAAAAAA-3′ (sense) and 5′-TTTTTTAAGCACCACGTGCGT-3′ (antisense). Oligonucleotides in mutated form had the following sequences: for promoter 1, 5′-ATTGGAAACTGAGGACCCTACTCGGG-3′ (sense) and 5′-CCCGAGTAGGGTCCT CAGTTTCCAAT-3′ (antisense); for enhancer 1, 5′-AGCCACGTGGTGCGCAAAACC-3′ (sense) and 5′-GGTTTTGCGCACCACGTGGCT-3′ (antisense). Nuclear proteins were extracted with NE-PERTM Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). Protein–DNA complexes were detected using biotin end-labeled double-stranded DNA probes prepared with the Biotin 3′ End DNA Labeling Kit (Pierce). Electrophoretic mobility shift assay (EMSA) was performed with a LightShift Chemiluminescent EMSA Kit (Pierce). Briefly, nuclear extracts (6 μg protein) and the 1 × binding buffer with 2.5% glycerol, 5 mM MgCl2, 50 ng/μL poly (dI-dC), 0.05% NP-40, 1 mM DTT, and 20 fmol biotin 3′-end labeled double-stranded oligonucleotide were incubated at room temperature for 15 minutes in a volume of 20 μL. For STAT3 supershift analysis, an anti–STAT3 polyclonal antibody (Santa Cruz; 1 μg per reaction) was incubated with the nuclear proteins on ice for 20 minutes before labeled oligonucleotide was added. Reaction products were separated by electrophoresis (5% acrylamide [29:1 acryl/bis]) in 0.5 × TBE. After electrophoresis, the protein–DNA complexes were transferred onto nylon membranes and detected using chemiluminescence (LightShift kit; Pierce).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed as described previously.13 In brief, cells were cross-linked with 1% formaldehyde at 37°C for 10 minutes. The cross-linking reaction was quenched by washing the cells several times with cold PBS, buffer I (0.25% Triton X 100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5), and buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Cells were centrifuged, resuspended in cold IP buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1, and protease inhibitors), and sonicated to an average fragment size of 300 to 600 bp. Solubilized chromatin was clarified by centrifugation at 21 000g, and the supernatant was preincubated for 2 hours with protein A/G agarose beads. Precleared chromatin was incubated overnight at 4°C with 2 μg anti-STAT3. Immune complexes were bound to protein A agarose beads at 4°C for an additional 2 to 3 hours. Beads were washed with TSE + 150 mM NaCl buffer (0.1% SDS, 1% Triton X 100, 2 mM EDTA, 20 mM Tris, pH 8.1, 150 mM NaCl), TSE + 500 mM NaCl buffer (0.1% SDS, 1% Triton X 100, 2 mM EDTA, 20 mM Tris, pH 8.1, 500 mM NaCl), Buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate 1 mM EDTA, 10 mM Tris, pH 8.1), and TE. Protein–DNA complexes were eluted from the protein A agarose beads with 1% SDS and 0.1 M NaHCO3 at room temperature for 10 minutes with gentle agitation. The supernatant was transferred to a fresh tube, and cross-links were reversed overnight at 65°C. Samples were treated with Proteinase K (Roche Biochemicals, Mannheim, Germany) at 37°C for 2 hours and were extracted once with phenol, and the DNA was precipitated with 2.5 vol ethanol plus 20 μg glycogen as carrier. Precipitated DNA was pelleted, washed once with 70% ethanol, dried, and resuspended in 25 μL water. DNA was analyzed by PCR using specific primer pairs: for promoter 1, 5′-CCCACCTAAGGTCGTATAGCC-3′ (sense), 5′-GTCCAGCTCCACGTTTCCT-3′ (sense); for enhancer 1, 5′-TCTAAAACTCCTGGGCTCAA-3′ (sense), 5′-CCCATGCCACTTCTATAC CC-3′ (antisense); for the control, 5′-AGGTGGGTGGATCACTTGAG-3′ (sense), 5′-GTGGAACAGCCA ACAATCCT-3′ (antisense).
Cell transfection
T-cell lymphoma cell lines 2A and 2B were transfected using the Lipofectamine 2000 (Gibco/BRL) or Cell Line Nucleofector Kit T (Amaxa Biosystems, Cologne, Germany). STAT3 and control non–sense siRNA mixtures were purchased from Upstate Biotechnology (Lake Placid, NY). AS-ON(1) (5′-AACCATGAGCACCGTTCTCC-3′) and DNMT1 AS-ON (2) (5′-TTCATGTCAGCCAAGGCCAC-3′), corresponding to the exon 1 and 3′ untranslated region, respectively, were synthesized according to the published sequence,15 and control, scrambled oligonucleotides were high-performance liquid chromatography (HPLC)–purified and FITC-labeled.
Cell proliferation and survival (MTT enzyme conversion) assay
Cells transfected with STAT3 or control non–sense siRNA were examined in quadruplicate in 96-well plates. After 72 hours, the relative number of cells was determined with MTT reduction colorimetric assay (Promega, Madison, WI) and absorbance was measured at 570 nm with a 96-well plate reader.
Flow cytometry assay
Transfected cells were quantified by measuring green fluorescence in FL1 on the flow cytometer for transfection efficiency. Annexin-V staining was performed with the annexin-V–FLUOS Staining Kit (Roche) according to the manufacturer's protocol. In brief, the cells were collected, washed, and resuspended in 100 μL binding buffer containing annexin-V–FLUOS and/or propidium iodide. Samples were incubated for 15 minutes at room temperature and were analyzed with a Becton Dickinson FACScan flow cytometer and CellQuest software to acquire and analyze the data.
Luciferase reporter assay
DNMT1 promoter and enhancer regions were amplified from genomic DNA and were isolated from 2A cells and PHA blasts with primers designed from GenBank sequences (AC010077) DNMT1 forward (5′-TAGGTACCTGAGATGCAGTTTCTCTATGTT ACC-3′) and DNMT1 reverse (5′-TAACTCGAGATCTCGGAGGCTTCAGCA-3′). Italicized sequences represent nucleotides added to the complementary sequences to generate Kpn1 and Xho1 unique restriction digest sites. The resultant 718-bp PCR product was gel purified and cloned into the pGL3-basic luciferase reporter construct (Promega) to generate the DNMT1-promoter/enhancer-pGL3 construct. Constructs were validated by sequence analysis. Site-directed mutagenesis was used to mutate the STAT3-binding sites with the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Briefly, primers containing the desired mutations of STAT3-binding sites were generated as follows: the mutant within DNMT1 promoter 5′-GGTTGGATTGGAACTGAGGACTGGACTCCCGGGCTCTCAC-3′ and the mutants within DNMT1 enhancer 5′-CAGGCGTGAGCCACGTGGTGCAAAAAATTGGCAACAAAAAAC-3′. It was confirmed that all plasmids contained the desired mutations by DNA sequencing. They were introduced to 1 × 105 SUDHL-1 cells per well and were transiently transfected in duplicate with Lipofectamine 2000 (Invitrogen) according to manufacturer's directions, with the designated combination of DNMT1/luciferase constructs at 1.0 μg and 100 ng phRL (renilla luciferase) TK plasmids (Promega) for transfection efficiency. Twenty-four hours after transfection, the cells were washed, lysed, and sequentially evaluated for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured for 10 seconds after a 2-second delay using a BD Monolight 3010 luminometer (BD Biosciences, San Jose, CA). Variation in transfection efficiency was normalized by dividing the construct luciferase activity by the corresponding renilla luciferase activity. Promoter with enhancer activity is reported as the mean ± SD.
Results
Coexpression of phosphorylated STAT3 and DNMT1 in malignant T-cell lymphoma cells
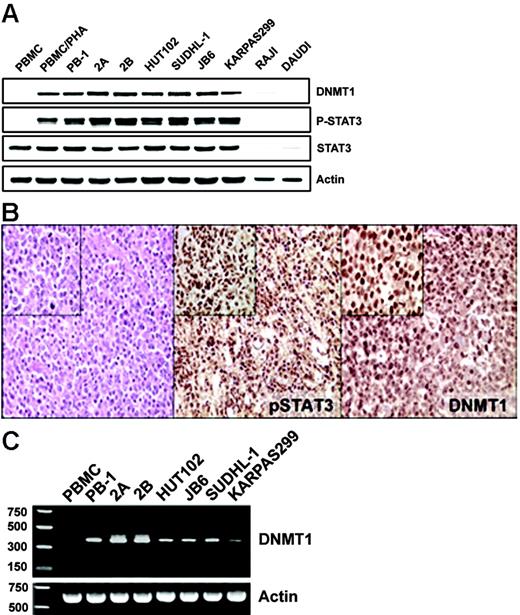
Our previous studies documented the expression of STAT3 in activated form in several populations of malignant T cells.10-13 More recently, we also identified the expression of DNMT1 in some of these populations.13 To examine this apparent coexpression in greater detail, we directly compared the expression of DNMT1 and phospho-STAT3 in several cell lines from different types of T-cell lymphoma. Although the control, normal resting T-cell–rich peripheral blood mononuclear cells (PBMCs) and Burkitt lymphoma lines (RAJI and DAUDI) failed to express both these proteins, mitogen (PHA)–activated PBMCs and all malignant T-cell lines examined expressed both phospho-STAT3 and DNMT1 (Figure 1A). To determine whether the expression of phospho-STAT3 was associated with the expression of DNMT1 protein in native T-cell lymphoma cells, we performed immunohistochemical staining of tissue samples derived from 15 patients with cutaneous T-cell lymphoma. Both proteins were expressed in essentially all malignant cells in all the patients examined (Figure 1B; a representative image). The scattered negative cells, consistent with resting lymphocytes, served as an internal negative control. Similar results were obtained when phospho-STAT312 and DNMT113 expression were examined previously in separate studies, in the same set of 10 patients with T-cell lymphoma expressing anaplastic lymphoma kinase (ALK). These results suggested that STAT3 might, indeed, be involved in the regulation of DNMT1 expression.
Coexpression of activated STAT3 and DNMT1 in malignant T-cell lymphoma cells. (A) Western blot analysis with the depicted antibodies of the nuclear protein extracts from cell lines derived from T-cell lymphomas involving skin (PB-1, 2A, 2B), HTLV-I–associated (HUT102B), and anaplastic lymphoma kinase–positive (SUDHL-1, JB6, Karpas 299) malignancies. Normal T-cell–rich PBMCs, both resting and PHA activated, and B cells (RAJI and DAUDI) served as controls. (B) Immunohistochemical analysis of the cutaneous T-cell lymphoma tissues. (left) Hematoxylin-eosin staining was used to visualize malignant T cells. (middle) Anti–phospho(p)-Y705STAT3 staining. (right) Anti-DNMT1 staining. Original magnification: larger images, 100 ×; insets, 400 ×. Depicted results that show coexpression of p-STAT3 and DNMT1 by malignant T cells are representative for all biopsy tissue samples of the 15 examined patients. (C) RT-PCR analysis of DNMT1 mRNA expression in the same diverse T-cell lymphoma cell line depicted in Figure 1A, with PBMCs serving as control.
Coexpression of activated STAT3 and DNMT1 in malignant T-cell lymphoma cells. (A) Western blot analysis with the depicted antibodies of the nuclear protein extracts from cell lines derived from T-cell lymphomas involving skin (PB-1, 2A, 2B), HTLV-I–associated (HUT102B), and anaplastic lymphoma kinase–positive (SUDHL-1, JB6, Karpas 299) malignancies. Normal T-cell–rich PBMCs, both resting and PHA activated, and B cells (RAJI and DAUDI) served as controls. (B) Immunohistochemical analysis of the cutaneous T-cell lymphoma tissues. (left) Hematoxylin-eosin staining was used to visualize malignant T cells. (middle) Anti–phospho(p)-Y705STAT3 staining. (right) Anti-DNMT1 staining. Original magnification: larger images, 100 ×; insets, 400 ×. Depicted results that show coexpression of p-STAT3 and DNMT1 by malignant T cells are representative for all biopsy tissue samples of the 15 examined patients. (C) RT-PCR analysis of DNMT1 mRNA expression in the same diverse T-cell lymphoma cell line depicted in Figure 1A, with PBMCs serving as control.
Transcriptional activity of promoter 1 of the DNMT1 gene in malignant T cells
Four different transcription initiation sites, 1 major and 3 minor, have been identified in the DNMT1 gene.6 Gene transcription from these sites is regulated by independent enhancers and promoters, with the latter located directly upstream of the sites. The major initiation site located upstream of exon 1 is driven by promoter 1 and displays a certain degree of basal activity, whereas the 3 minor transcription initiation sites have minimal basal activity.6 Because the anti-DNMT1 antibody that detected DNMT1 in the malignant T cells yielded a single band (Figure 1A) of approximately 200 kDa, consistent with a full-length isoform, and because the antibody was developed to recognize the N-terminus of the protein shared by the different isoforms, we reasoned that transcription of the DNMT1 gene is driven in the malignant T cells by promoter 1. To confirm transcriptional activation of promoter 1, we performed reverse transcription (RT)–PCR using a primer pair that covered cDNA fragment from the + 60 site (encoded by exon 1) to the + 568 site (encoded by exon 4). Surprisingly, the obtained RT-PCR product (Figure 1C) of 380 bp was smaller than predicted because of the splicing of the 130-bp internal fragment, as confirmed by sequence analysis (data not presented). Regardless of this shortening, the very presence of the mRNA documented functional activity of promoter 1 in malignant T cells.
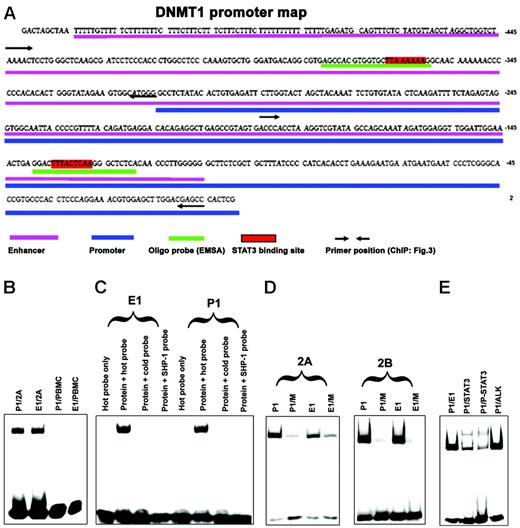
Binding of STAT3 to promoter 1 and enhancer 1 SIE/GAS sites in vitro
We next analyzed the DNA sequence of the promoter 1 region for the presence of potential STAT3-binding sites. All STATs bind to similar SIE/GAS DNA sequences that contain the TT and AA base tandems, separated in STAT3 by 4 to 6 bases (5′-TTN4-6AA-3′), where the identity of N may be diverse.13,16 Our analysis identified 2 potential STAT3 SIE/GAS-binding sites, one within promoter 1 and the other within enhancer 1 (positions – 127 to –135 and – 360 to – 367, respectively) as depicted in Figure 2A. To determine whether STAT3 binds to these 2 identified SIE/GAS sequences, we performed EMSA with DNA oligonucleotide probes that contained the putative STAT3-binding sites. As shown in Figure 2B, nuclear protein extracts from malignant T cells that contained activated STAT3, but not from normal PBMCs in which STAT3 was quiescent, displayed strong binding to both probes. Binding was completely abrogated when the same, yet unlabeled, DNA probe or an unlabeled probe that contained a STAT3-binding site from the SHP-1 gene promoter13 was added at 40-fold excess to the samples before the labeled probe was added (Figure 2C). To document the binding specificity even further, 2 probes were prepared in which the TT and AA pairs within the STAT-binding site were substituted with CC tandems, with the remainder of the 26-base probes unchanged. As shown in Figure 2D, the substitutions greatly diminished to essentially abrogate the binding. Finally, to document that it was indeed STAT3 and not another STAT that bound to the probes, we performed EMSA supershift analysis using an anti-STAT3 antibody. As shown in Figure 2E, anti-STAT3, but not an irrelevant anti-ALK, antibody markedly diminished density of the band corresponding to the protein–oligonucleotide complex and led to formation of the slower migrating “supershifted” band. In combination, these results provided strong evidence that activated STAT3 directly binds to promoter 1 and enhancer 1 of the DNMT1 gene.
Association of STAT3 with SIE/GAS sites identified inDNMT1gene promoter 1 and enhancer 1 in vitro. (A) Map of the DNMT1 gene promoter and enhancer region with highlighted STAT3-binding sites and corresponding DNA oligonucleotide probes used in EMSA and primer sets used in ChIP (for comparison, see Figure 4). (B) EMSA with DNA oligonucleotides that corresponded to promoter 1 (P1) and enhancer 1 (E1) STAT3-binding sites with nuclear protein extracts from malignant T cells 2A and normal PBMCs as control. (C) Binding of the 2A T-cell line protein extracts (protein) to the enhancer 1 and promoter 1 STAT3-binding sites in the presence and absence of the unlabeled (cold and SHP-1) probes. (D) Binding of the 2A and 2B T-cell line protein extracts to promoter 1 and enhancer 1 STAT-binding sites either wild-type (P1 and E1, respectively) or with mutated (M) TT and AA pairs by substitution with the CC pairs. (E) Supershift EMSA with the anti-STAT3 and control anti–anaplastic lymphoma kinase (ALK) antibody.
Association of STAT3 with SIE/GAS sites identified inDNMT1gene promoter 1 and enhancer 1 in vitro. (A) Map of the DNMT1 gene promoter and enhancer region with highlighted STAT3-binding sites and corresponding DNA oligonucleotide probes used in EMSA and primer sets used in ChIP (for comparison, see Figure 4). (B) EMSA with DNA oligonucleotides that corresponded to promoter 1 (P1) and enhancer 1 (E1) STAT3-binding sites with nuclear protein extracts from malignant T cells 2A and normal PBMCs as control. (C) Binding of the 2A T-cell line protein extracts (protein) to the enhancer 1 and promoter 1 STAT3-binding sites in the presence and absence of the unlabeled (cold and SHP-1) probes. (D) Binding of the 2A and 2B T-cell line protein extracts to promoter 1 and enhancer 1 STAT-binding sites either wild-type (P1 and E1, respectively) or with mutated (M) TT and AA pairs by substitution with the CC pairs. (E) Supershift EMSA with the anti-STAT3 and control anti–anaplastic lymphoma kinase (ALK) antibody.
Association of STAT3 with promoter 1 and enhancer 1 of DNMT1 gene in vivo
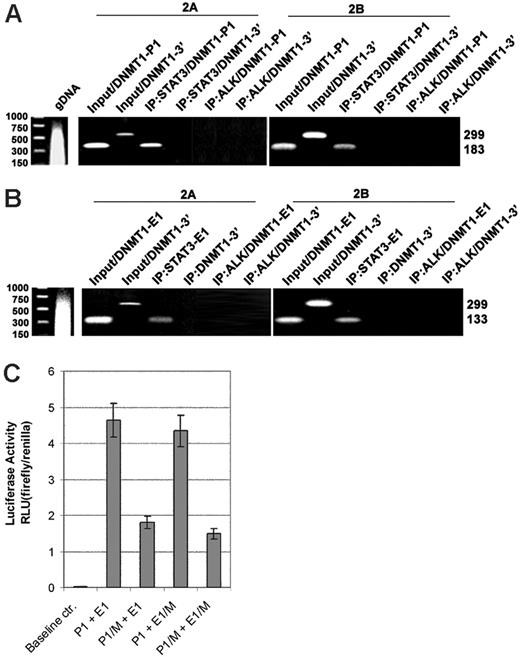
To demonstrate the direct interaction between STAT3 and DNMT1 promoter 1 and enhancer 1 in vivo, we carried out the ChIP assays using an anti-STAT3 antibody, a control anti-ALK antibody, and 3 sets of PCR primers that covered promoter 1, enhancer 1 (both depicted in Figure 2A), and, as an additional control, the 3′-end of the DNMT1 gene located approximately 10 kb downstream. The exact positions of the former 2 primer pairs are depicted in Figure 2A. Although the promoter 1 and 3′-end–specific primers efficiently generated PCR products from the input (ie, the intact cell lysate), only the promoter 1–specific primers yielded a strong band from the STAT3 antibody immunoprecipitate (Figure 3A). The 3′-end–recognizing primers failed to produce a detectable band. Similarly, only the enhancer 1–specific primer, not the 3′-end–specific primers, generated a band from the STAT3 antibody precipitate (Figure 3B). The anti-ALK antibody precipitate did not generate any bands when either promoter 1–specific (Figure 3A) or enhancer 1–specific primers were used. These results demonstrate in vivo binding of STAT3 to promoter 1 and enhancer 1.
To demonstrate that STAT3 binding to the DNMT1 promoter/enhancer region activates the promoter, we performed a luciferase reporter assay using constructs containing the promoter and enhancer in the wild-type or mutated form, or both (Figure 3C). On transfection into SUDHL1 cells that constitutively expressed activated STAT3, the wild-type promoter/enhancer construct displayed high degree of activity (more than 450 times) over the baseline control construct containing only the luciferase gene. Mutation of the STAT3-binding site by replacement of the AA/TT pairs with CC tandems resulted in the profoundly (greater than 50%) diminished activity of the promoter. Similar mutation of the STAT3-binding site within the enhancer had a visible but overall minor effect, either alone or in combination with mutation of the promoter. These observations suggest that transcriptional activation by STAT3 of the promoter, rather than the enhancer, may be the critical aspect of STAT3-induced expression of the DNMT1 gene.
Binding and activation of STAT3 toDNMT1gene promoter/enhancer region in vivo. Cell lysates from T-cell lymphoma cell lines 2A and 2B were examined in the chromatin immunoprecipitation (ChIP) assay before (input) and after immunoprecipitation using anti-STAT3 antibody with an unrelated (ALK) antibody serving as the negative control. DNA binding was detected in the ChIP assay using PCR primer pairs corresponding to promoter 1 (A, P1) and enhancer 1 (B, E1) regions. Binding to the 3′-end of the DNMT1 gene served as an additional negative control. (C) STAT3 effect on activity of the DNMT1 promoter and enhancer. SUDHL-1 cells that constitutively expressed activated STAT3 were transfected with firefly luciferase constructs containing promoter 1 and enhancer 1 of the DNMT1 gene in either wild-type form (P1 and E1, respectively) or with mutated (M) TT and AA pairs that were substituted with the CC pairs. Results are presented in relative luciferase unit (RLU) after normalization based on expression of the cotransfected renilla luciferase gene.
Binding and activation of STAT3 toDNMT1gene promoter/enhancer region in vivo. Cell lysates from T-cell lymphoma cell lines 2A and 2B were examined in the chromatin immunoprecipitation (ChIP) assay before (input) and after immunoprecipitation using anti-STAT3 antibody with an unrelated (ALK) antibody serving as the negative control. DNA binding was detected in the ChIP assay using PCR primer pairs corresponding to promoter 1 (A, P1) and enhancer 1 (B, E1) regions. Binding to the 3′-end of the DNMT1 gene served as an additional negative control. (C) STAT3 effect on activity of the DNMT1 promoter and enhancer. SUDHL-1 cells that constitutively expressed activated STAT3 were transfected with firefly luciferase constructs containing promoter 1 and enhancer 1 of the DNMT1 gene in either wild-type form (P1 and E1, respectively) or with mutated (M) TT and AA pairs that were substituted with the CC pairs. Results are presented in relative luciferase unit (RLU) after normalization based on expression of the cotransfected renilla luciferase gene.
STAT3 depletion inhibits DNMT1 expression
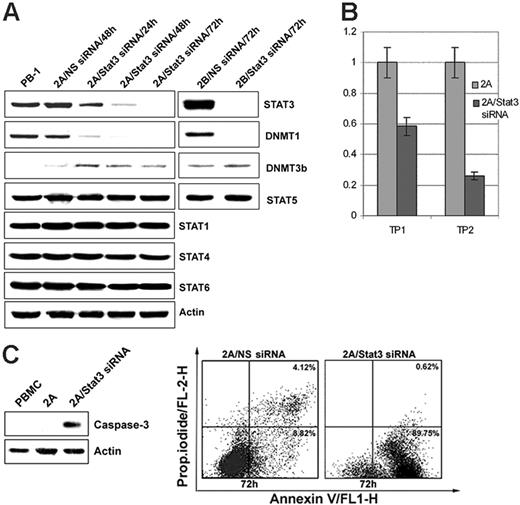
To demonstrate directly that STAT3 is involved in the induction of DNMT1 expression, malignant T cells were treated with a STAT3 siRNA mix. As shown in Figure 4A, STAT3 expression was significantly diminished by 24 hours from the treatment initiation and was essentially abrogated by 48 to 72 hours. STAT3 depletion also resulted in the gradual inhibition of DNMT1 expression with similar kinetics, including lack of detectable DNMT1 at the 72-hour time point. Expression of another member of the DNMT family, DNMT3b, and of other depicted control proteins remained unaffected, indicating that STAT3 activity is specifically directed toward DNMT1. STAT3 depletion was also associated with the inhibition of cell growth, as determined by MTT conversion (Figure 4B), and the triggering of apoptotic cell death, as determined by activated caspase 3 expression (Figure 4C, left panel) and cell-surface binding of annexin V (Figure 4C, right panel).
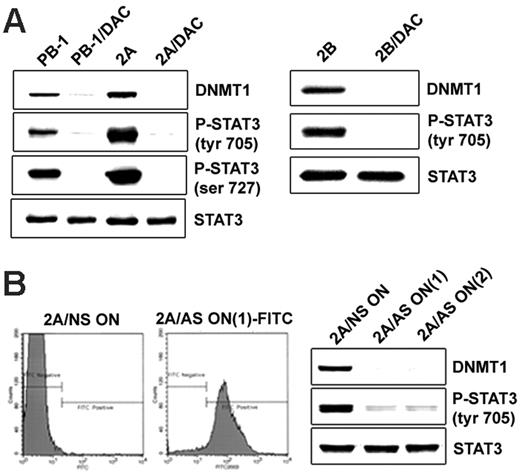
DNMT1 inhibition results in the suppression of STAT3 activation
Our previous studies have demonstrated that in malignant T cells from lymphoma involving skin, STAT3 activation is mediated by the Jak1/Jak3 complex linked to the cytokine receptor common gamma chain.10,11,13 SHP-1 phosphatase, the negative regulator of Jaks,17 is epigenetically silenced in malignant T cells.13,14 Inhibition of DNMT1 activity leads to the re-expression of SHP-113,14 and the dephosphorylation of Jak3.14 To determine whether DNMT1 inhibition leads to impaired activation of STAT3, we treated malignant T cells with a small molecule inhibitor, 5-aza-2-deoxycytidine (DAC), which affects DNMT1 primarily and other members of the DNMT family to a lesser degree.18 As shown in Figure 5A, DAC treatment resulted in malignant T cells in the essentially complete dephosphorylation of STAT3 at the key Tyr705 and Ser727 without affecting STAT3 expression. This finding indicates that the activity of DNMTs, specifically of DNMT1, is indeed critical for sustained STAT3 activation in malignant T cells. To provide direct evidence that STAT3 dephosphorylation is caused by the inhibition of DNMT1 rather than of another DNMT, we treated malignant T cells with 2 DNMT1-specific antisense oligonucleotides that have been extensively characterized previously.5,15,19 These oligonucleotides, designated DNMT1 AS-ON(1) and DNMT1 AS-ON(2), were labeled with fluorescein to permit evaluation of their uptake by target cells. To determine the degree of such uptake by malignant T cells, we examined by flow cytometry the 2A and 2B cells treated with these DNMT1 oligonucleotides. As shown in Figure 5B (left panel), essentially all 2A malignant T cells incorporated DNMT1 AS-ON(1). The same result was obtained when the 2B T cells were transfected with the DNMT1 AS-ON(1) and when either 2A or 2B cells were treated with the DNMT1 AS-ON(2) (data not shown). Both oligonucleotides profoundly inhibited STAT3 phosphorylation in 2A cells (Figure 5B; right panel) and 2B cells (not shown). These results indicate that by inducing DNMT1 expression, STAT3 secures its own persistent activation, likely by facilitating DNMT1-mediated silencing of SHP-113 and possibly other negative regulators of its activation.
STAT3 depletion by siRNA inhibits expression of DNMT1. (A) Expression of STAT3, DNMT1, and the other listed proteins after treatment of the T-cell lymphoma cell lines 2A and 2B for the indicated times with STAT3 siRNA or control, non–sense (NS) siRNA. Untreated PB-1 line served as an additional control. (B) STAT3 siRNA–mediated inhibition of cell growth of 2A cells as determined in the MTT enzymatic conversion assay. Time point (TP) 1 designates a single siRNA treatment at the experiment inception (0 hour), and TP2 designates 2 treatments at 0 hour and at 48 hours. The assay readout was performed at 72 hours. (C) STAT3 siRNA–mediated induction of apoptotic cell death as determined in 2A cells by expression of cleaved caspase 3 (left panel) and annexin V binding (right panel).
STAT3 depletion by siRNA inhibits expression of DNMT1. (A) Expression of STAT3, DNMT1, and the other listed proteins after treatment of the T-cell lymphoma cell lines 2A and 2B for the indicated times with STAT3 siRNA or control, non–sense (NS) siRNA. Untreated PB-1 line served as an additional control. (B) STAT3 siRNA–mediated inhibition of cell growth of 2A cells as determined in the MTT enzymatic conversion assay. Time point (TP) 1 designates a single siRNA treatment at the experiment inception (0 hour), and TP2 designates 2 treatments at 0 hour and at 48 hours. The assay readout was performed at 72 hours. (C) STAT3 siRNA–mediated induction of apoptotic cell death as determined in 2A cells by expression of cleaved caspase 3 (left panel) and annexin V binding (right panel).
DNMT1 inhibition results in the suppression of STAT3 activation. (A, left) Two T-cell lymphoma cell lines (PB-1 and 2A) treated with the DNMT inhibitor DAC were examined for expression of the noninhibited DNMT1 protein (upper lane) and STAT3 phosphorylation at Tyr705 (upper middle lane) and Ser727 (lower middle lane) and for expression of total STAT3 (lower lane). Untreated cells served as a control. (A, right) Expression of noninhibited DNMT1, p-Y705STAT3, and total STAT3 in the additional T-cell lymphoma cell line, 2B, before and after treatment with DAC. (B, left) 2A cells were treated for 72 hours with 2 fluorescein-labeled DNMT1 antisense DNA oligonucleotides, designated DNMT1 AS-ON(1) and DNMT1 AS-ON(2), and non–sense DNA oligonucleotide (NS-ON) as control. Flow cytometry analysis showed efficiency of the uptake of FITC-labeled DNMT1 AS-ON(1). (B, right) Western blot expression of DNMT1, phospho-STAT3, and total STAT3 after treatment with control NS-ON, DNMT1 AS-ON(1), and AS-ON(2).
DNMT1 inhibition results in the suppression of STAT3 activation. (A, left) Two T-cell lymphoma cell lines (PB-1 and 2A) treated with the DNMT inhibitor DAC were examined for expression of the noninhibited DNMT1 protein (upper lane) and STAT3 phosphorylation at Tyr705 (upper middle lane) and Ser727 (lower middle lane) and for expression of total STAT3 (lower lane). Untreated cells served as a control. (A, right) Expression of noninhibited DNMT1, p-Y705STAT3, and total STAT3 in the additional T-cell lymphoma cell line, 2B, before and after treatment with DAC. (B, left) 2A cells were treated for 72 hours with 2 fluorescein-labeled DNMT1 antisense DNA oligonucleotides, designated DNMT1 AS-ON(1) and DNMT1 AS-ON(2), and non–sense DNA oligonucleotide (NS-ON) as control. Flow cytometry analysis showed efficiency of the uptake of FITC-labeled DNMT1 AS-ON(1). (B, right) Western blot expression of DNMT1, phospho-STAT3, and total STAT3 after treatment with control NS-ON, DNMT1 AS-ON(1), and AS-ON(2).
Discussion
STAT3 promotes malignant cell transformation in various cells by impacting on a whole spectrum of their functions.9 Depending on the cell type, STAT3 mediates cell proliferation, inhibits programmed cell death, and promotes angiogenesis. It accomplishes these diverse tasks primarily by inducing the expression of a number of genes that code for c-myc, cyclin D1, Bcl-xL, survivin, VEGF-I, and other proteins. More recent studies indicate that STAT3 also plays a role in suppressing the anticancer immune response by indirectly affecting tumor-infiltrating antigen-presenting cells.20 Here we report that in malignant T cells, STAT3 induces the expression of DNMT1. This finding not only describes a new functional effector of STAT3 but also reveals a novel mechanism of cell transformation by this oncogenic factor that is ubiquitously expressed in various types of malignancy.9 Furthermore, it provides evidence of interdependence between the aberrant cell signaling that is usually, if not always, initiated in cancer cells by a genetic abnormality such as DNA mutation or chromosomal translocation, with an inhibition of tumor-suppressor gene expression that is frequently mediated by epigenetic silencing of the affected genes. Numerous studies have documented the central role of DNMT1 in the epigenetic gene silencing in cancer.1,3,21 Its main function appears to be the maintenance of promoter DNA methylation of the affected genes during cell division and, hence, preservation of inactivation of the tumor-suppressor genes in the malignant cells. Although many genes have been identified as epigenetically silenced in cancer cells, recent studies indicate that the scope of gene silencing by this mechanism, and hence the role of DNMT1 in oncogenesis, may be even bigger than realized.22 Accordingly, CpG-enriched areas, or CpG islands, have been identified in the promoter regions of approximately 28 000 genes. Of these, at least 8% seem aberrantly methylated in any given tumor type. With regard to the cutaneous T-cell lymphoma examined here, we have previously identified epigenetic gene silencing as the mechanism underlying inhibition of the SHP-1 tyrosine phosphatase gene14 ; the observation was subsequently extended by several groups23-25 to other types of hematologic malignancies. Negative regulator of cell-cycle progression INK4a/p16 and the related genes26 are also frequently epigenetically silenced in this type of T-cell lymphoma. Furthermore, a recent study27 identified more than 30 additional genes, including BCL7a, PTPRG, and p73, as inactivated by DNA methylation of their promoters in this type of malignancy.
Inhibiting DNMT1 function and expression results in T-cell malignancy in STAT3 dephosphorylation (Figure 5). This finding indicates that the STAT3-mediated induction of DNMT1 expression leads, among other outcomes, to the securing by STAT3 of its own perpetual activation. This STAT3 self-promoting effect presumably occurs through the DNMT1-mediated epigenetic silencing of inhibitors of signaling pathway(s) leading to STAT3 activation. Consequently, the persistently active STAT3 permits maintenance of the neoplastic phenotype of the affected cells. Our previous studies have documented that STAT3 is activated in such cells by the cytokine common gamma chain–associated Jak1/Jak3 tyrosine kinase complex,10,11,13 which remains constantly activated in part because of the loss of expression of its inhibitor, SHP-1 tyrosine phosphatase, and28 in turn because of the epigenetic silencing of the SHP-1 gene.14 Our recent report13 demonstrates that STAT3 itself is responsible for SHP-1 gene silencing by attracting to the gene promoter at least 2 members of the epigenetic gene silencing machinery, DNMT1 and HDAC1. This observation, combined with the current findings (Figure 5), indicates that in malignant T cells, STAT3 uses a 2-step approach to ensure its own perpetual activation by first inducting the expression of DNMT1 and then mobilizing the methyltransferase to silence the SHP-1 gene.
Several other diverse transcription factors, such as c-fos, Fli-1, Sp1, SP3, and E2F, have been reported recently as involved in the regulation of DNMT1 expression.6,7,29,30 Whether these factors, including STAT3, regulate the DNMT1 gene together or separately, depending on the cell type, is unknown. One of the factors, c-fos, is unable to up-regulate the function of promoter 1, identified by us as the target of STAT3, whereas it does augment activity of the 3 minor transcription initiation sites.6 Furthermore, E2F seems to inhibit rather than activate DNMT1 transcription unless it cooperates with another, unidentified, transcription factor.7 Finally, it may be relevant to our findings—considering STAT3 is the key effector of signaling by this cytokine—that IL-6 has recently been identified in a multiple myeloma cell as the major inducer of the DNMT1 line.27 Nevertheless, our results reported here identify STAT3 as the major inducer of DNMT1 transcription, at least in malignant T lymphocytes.
Our findings also have potential therapeutic implications for T-cell lymphoma and likely other malignancies. The DNMT inhibitor DAC and the closely related 5-aza-cytidine have been successfully applied clinically, mostly in patients with myelodysplastic syndrome and other myeloproliferative disorders.31,32 However, these drugs entail considerable toxicity, in part because of their targeting not only DNMT1 but also other members of the family.18 DNMT1 antisense oligonucleotides,5,15 which we used in this study (Figure 5), are also undergoing clinical evaluation.27 However, the clinical efficacy of antisense oligonucleotides in general, and of those targeting DNMT1 in particular, remains to be proven. Our finding that STAT3 induces DNMT1 expression provides further rationale for directly targeting STAT3 in malignant cells.33,34 The critical role of STAT3 in oncogenesis has recently been documented in vivo in the mouse models of skin17 and breast35 cancers and in anaplastic lymphoma kinase–expressing T-cell lymphoma.36 Accordingly, peptides that interfere with STAT3 function37 or that induce STAT3 ubiquitination38 have been constructed. Furthermore, peptidomimetic small molecule STAT3 inhibitors that impair STAT3 dimerization have also been synthesized,39 suggesting that the generation of clinically useful anti-STAT3 compound(s) might follow. Inhibiting STAT3 activation indirectly by targeting its activators, such as Jaks and Src,9 in malignant cells represents an attractive alternative. Accordingly, pharmacologic grade inhibitors of Jaks38 and Src39 are available; the latter are already undergoing evaluation in clinical trials.
In summary, we demonstrated that STAT3 induces DNMT1 expression in malignant T cells by binding to promoter 1 and enhancer 1 of the DNMT1 gene. Preservation of the continuous activation of STAT3 is one of the consequences of DNMT1 expression in such cells. This study describes a new mechanism by which STAT3 mediates malignant cell transformation and, in more general terms, provides evidence of the direct link between oncogenic, aberrant cell signaling and the epigenetic gene silencing that typically affects tumor-suppressor genes. Finally, results of this study provide further rationale for targeting STAT3 therapeutically.
Prepublished online as Blood First Edition Paper, April 18, 2006; DOI 10.1182/blood-2005-08-007377.
Supported in part by the National Cancer Institute (grants R01-CA89194 and R01-CA96856) and the Danish Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.