Abstract
To identify, in high-resolution regions of DNA, the copy number changes associated with outcome in patients with diffuse large B-cell lymphoma (DLBCL), a disease with an approximately 50% mortality rate, we performed array comparative genomic hybridization (array-CGH) on specimens from 64 patients with newly diagnosed DLBCL treated with anthracycline-based chemotherapy. For the entire cohort, 55 commonly gained/lost regions, ranging in size from less than 1 Mbp to entire chromosomes, were identified using 1- to 2-Mbp and 2- to 4-Mbp resolution BAC arrays. Copy number changes of 9 minimal regions significantly correlated with overall survival, of which 6 were 10 Mbp or smaller. On multivariate analysis, loss of chromosomes 2 (2.4-4.1 Mbp) and 16 (33.8-35.6 Mbp) were found to be prognostic indicators of poor survival, independent of clinical features routinely used to predict outcome. Loss of chromosome 1 (78.2-79.1 Mbp) was predictive of good outcome. For a subset of 55 specimens classified according to cell-of-origin expression signature subtype, gain of chromosome 12 (45.4-53.8 Mbp) was found to be significantly associated with the germinal center B-cell-like DLBCL subtype. Overall, array-CGH identified relatively small genomic regions associated with outcome, which, along with follow-up expression studies, may reveal target genes important in DLBCL clinical behavior. (Blood. 2006;107:2477-2485)
Introduction
Diffuse large B-cell lymphoma (DLBCL) accounts for approximately one third of all non-Hodgkin lymphomas (NHLs).1 Collectively, these neoplasms are aggressive in clinical behavior but display great diversity in clinical, morphologic, and biologic characteristics, the underlying genetic and molecular etiology of which is not well understood. Genetically, DLBCLs are characterized by clonal chromosomal rearrangements often involving the immunoglobulin (IG) gene loci and somatic hypermutation at the IG heavy-chain locus, indicative of a germinal center (GC)-derived mature B-cell origin for these tumors.2 Sites of genomic gain and loss in DLBCL, as revealed by low-resolution genome scanning technologies such as chromosomal-comparative genomic hybridization (CGH), have variously been reported by us and others, and, more recently, by higher resolution array-CGH.3-7 In a few cases, a candidate target gene has been identified, but overall the role of genomic gain or loss in B-cell transformation remains unclear.8,9 Expression profiling of DLBCL has revealed 2 predominant subtypes, suggestive of distinct cells of origin.10-12 The GC B cell-like DLBCL (GCB-DLBCL) subtype expresses transcripts in common with normal GC B cells, whereas the activated B cell-like DLBCL (ABC-DLBCL) subtype expresses transcripts detected in normal activated peripheral B cells and plasma cells.10-12 However, approximately 10% to 35% of DLBCLs cannot be classified as either subtype using the present classification algorithm.12,13 IG-associated BCL2 translocations and amplification of the REL locus have been confirmed to be associated, though not exclusively, with the GCB-DLBCL subtype.11,14,15 Several additional genomic gains or losses have also been reported to correlate with subtype by array-CGH, though these have yet to be confirmed.16
DLBCL exhibits high response rates with standard anthracycline-based chemotherapy, but durable remission is achieved in only approximately 50% of patients.1 Adding the anti-CD20 antibody (rituximab) to accelerated treatment programs for patients with advanced-stage disease has improved overall survival (OS), though the impact on outcomes for patients with newly diagnosed disease remains under intense study. Prediction of outcome is routinely based on 5 presenting clinical features—patient age at diagnosis, tumor stage, serum lactate dehydrogenase (LDH) level, Eastern Cooperative Oncology Group (ECOG) performance status (PS), and extent of extranodal (EN) disease—all of which comprise the International Prognostic Index (IPI).17 To date, few molecular markers have been confirmed to be associated with outcome. GCB-DLBCL is thought to have a more favorable outcome than ABC-DLBCL, though this has not been observed in all cohorts studied.10-13,15,18 Conventional karyotype studies have identified breakpoints at 1q21 and deletions of 6q to be associated with poor outcome, and chromosomal-CGH has only variably confirmed an association between 17p loss and outcome.2,6 Of the few higher resolution array-CGHs performed to date, most have had a primary focus of target gene identification rather than clinical correlation.7-9 Thus, in the current study, we submitted a panel of specimens from 64 patients with newly diagnosed DLBCL treated with anthracycline-based frontline therapy to comprehensive array-CGH analysis to identify regions of DNA copy number changes associated with expression signature subtype and outcome.
Patients, materials, and methods
Tumor specimens and DLBCL cell lines
The study cohort consisted of 64 patients at the Memorial Sloan-Kettering Cancer Center (MSKCC) with DLBCL newly diagnosed between March 1984 and October 1998 and treated at MSKCC with anthracycline-based therapy. For each patient, a fresh-frozen or OCT-embedded specimen was available for study. The study was approved by the institutional review board. Original diagnostic material of all patients was reviewed to confirm histology according to the World Health Organization classification.19 Patients excluded from the study were those with primary mediastinal DLBCL, evidence of histologic transformation from low-grade lymphoma, human immunodeficiency virus-associated lymphoma, and posttransplantation lymphoproliferative disorder. Medical records of all patients were reviewed to obtain relevant clinical information: patient age at diagnosis, LDH level, disease stage, PS, extent of EN disease, IPI score, treatment, response, time from start of treatment to last follow-up date or time-to-treatment failure (refractory, relapse, death from lymphoma or unknown cause) (TTF), and OS from start of treatment to last follow-up or death (death caused by lymphoma or of unknown cause) (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).17 Median follow-up time for OS among those alive was 90 months. OS significantly correlated with IPI score (P = .007) by log-rank test. DLBCL cell lines used in the study comprised the B-cell OCI-Ly NHL series (Ly1, Ly2, Ly4, Ly7, Ly8) (gift of Mark Minden, Ontario Cancer Institute, Toronto, ON, Canada), which were maintained as previously described by us.20
Array-CGH
Genomic (test) DNA was extracted from fresh-frozen or OCT-embedded specimens or from pellets of cell lines, as previously described.21 Karyotypically normal male DNA (Promega, Madison, WI) was used as reference DNA. Restriction and labeling of DNAs was performed as recommended by the manufacturer of the arrays (Spectral Genomics, Houston, TX). Briefly, 2 μg each DNA was digested with EcoRI or DpnII (10 U/μg) at 37°C for 16 hours. DNA was purified with the use of the Zymo Clean-up Kit (Zymo Research, Orange, CA) or the PCR Purification Kit (Qiagen, Valencia, CA), and each DNA was separately labeled with cyanine-5 (Cy5) and cyanine-3 (Cy3) dCTPs using a random labeling kit (Invitrogen, Carlsbad, CA). Labeled test and reference DNAs (forward hybridization, Cy5-labeled test and Cy3-labeled reference; reverse hybridization, Cy3-labeled test and Cy5-labeled reference) were mixed, coprecipitated with isopropanol, washed, and resuspended in hybridization solution (Spectral Genomics). DNA mixtures were denatured at 72°C for 10 minutes, prehybridized at 37°C for 30 minutes, and cohybridized to the arrays with coverslips for 16 or more hours at 37°C according to the manufacturer's recommended protocol. For 26 specimens, forward and reverse hybridizations were performed using an array consisting of 1552 target clones (BAC and P1) at an average resolution of 2 to 4 Mbp across the genome, whereas for the remaining 38 specimens, a second array was used consisting of 2621 BAC clones at an average of 1-Mbp resolution (SpectralChip 2600; Spectral Genomics). All clones were represented on the respective array in duplicate, with the genomic location of the clones provided by Spectral Genomics. After hybridization, the arrays were washed in 50% formamide/2 × SSC, 2 × SSC/0.1% NP-40, and 0.2 × SSC according to the manufacturer's recommendations. Images and signal intensities were acquired using a GenePix4000A (Axon Instruments, Burlingame, CA) dual-laser scanner in combination with GenePixPro 3.0 (Axon Instruments) imaging software.
Array data processing, normalization, and change-point analysis
Differences between the log base 2 median minus background values for Cy5 and Cy3 were used as the raw data for analyses (Cy5 minus Cy3 for forward hybridization and Cy3 minus Cy5 for reverse hybridization). Spots were excluded from analyses according to the following criteria: very low Cy3 or Cy5 (less than 6 on the log scale) or flagged by GenePixPro 3.0 software as irregular. Additional clones excluded from analyses were those with 10% or more polymorphism within a normal population (16 and 24 clones in the 2- to 4-Mbp and the 1-Mbp resolution arrays, respectively), mapped to the Y chromosome (4 clones for each array type), represented twice in the clone list (11 and 1 for the 2- to 4-Mbp and the 1-Mbp resolution arrays, respectively) and those of unknown map position (3 for the 2- to 4-Mbp resolution array).22 Thus, for the 2- to 4-Mbp resolution array, 1518 clones were used for normalization. All these clones were represented on the 1-Mbp resolution array, which included an additional 1074 clones (total, 2592 clones). Raw data were normalized for each specimen on a block-by-block basis because of spatial artifacts in images. The normalized value for each spot was calculated as the raw value minus the trimmed means for the row and column of the block plus the overall block-trimmed mean. The amount of trimming was 10%. After normalization, both values for each spot representing one clone were averaged or the value of one spot was used if the other had been excluded from analysis for the reasons described. Average values for forward hybridization were averaged with those of the matching reverse hybridization, giving a single final value for each clone.
A new modification of the circular binary segmentation (CBS) algorithm that we have described previously23 was used to identify segmental gains and losses along autosomes. Change-points (thus, defining segments) were found corresponding to P values below .05. Full details of the new algorithm are provided in Document S1. In addition, single clones that were in normal segments but that were 3 standard deviations (SDs) above or below the expected normal value were called gains or losses, respectively.
Clinical and expression subtype correlations
Only autosomal gains or losses were considered in clinical and expression subtype correlations. To use the data derived from the 2 array types, clones not represented on the smaller array were assigned as gained, normal, or lost, using the corresponding segmental results defined by the constrained CBS method for each specimen. Clones not represented on the smaller array that fell outside CBS-defined segments were treated as missing. Common regions of gain or loss for the entire cohort were identified as described in Document S1. Associations with gain or loss of a region with OS and TTF were quantitated using the log-rank test. Multivariate analysis was performed using the Cox regression model.24 All other comparisons between clinical variables or expression subtype and regional gain/loss were performed using the Fisher exact test.25
Immunohistochemistry
Immunohistochemical analysis was performed on consecutive tissue microarray sections or whole sections of 55 available biopsy specimens, as previously described.26 Pretreatment protocols were used according to the primary antibody, as follows: MUM1 (monoclonal; Dako Cytomation, Carpenteria, CA; EDTA, pH 8.0, 1:100); CD10 and BCL6 (monoclonal; Novocastro Laboratories, Newcastle, United Kingdom; citric acid, pH 6.0 and pH 8.0, respectively, 1:100); and CD5 (monoclonal; Dako Cytomation; EDTA, pH 8.0, 1:200). For CD5, a specimen was considered positive if greater than 50% of tumor cells exhibited staining. If greater than 30% of tumor cells exhibited staining for CD10, BCL6, or MUM1, the specimen was called positive for the respective antibody. Specimens were classified as GCB-DLBCL or non-GCB-DLBCL using the published algorithm.27
Expression array signature subgroup classification
Of the entire cohort, 38 specimens had previously been submitted to expression profiling as part of a larger subset using the Affymetrix HG-U95A oligonucleotide arrays.15,28 The expression signature subgroup of each tumor was redetermined according to the method of Wright et al,12 using 18 (represented by 34 probe sets) of the reported 27 genes for classification, as detailed in the online supplement.
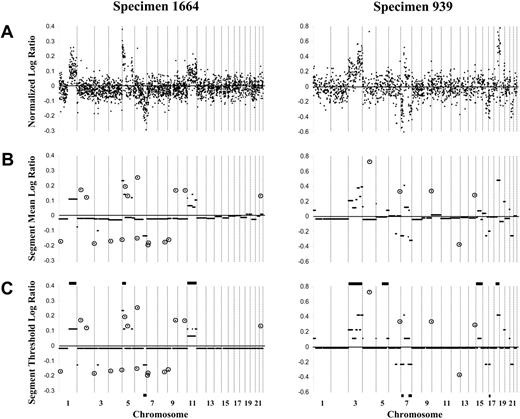
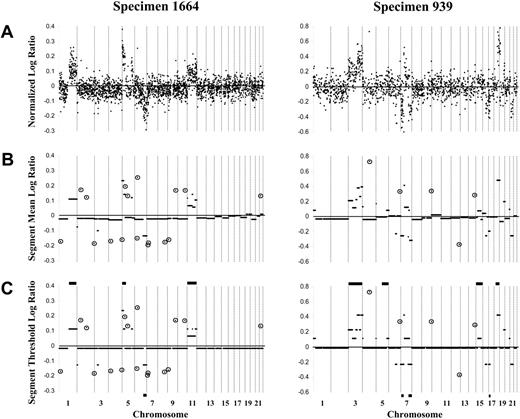
Representative array-CGH log ratio plots of 2 DLBCL specimens (1664 and 939) using the 1-Mbp and 2- to 4-Mbp resolution arrays, respectively. (A) Custom normalization was performed, and an averaged log ratio for each clone was calculated for the duplicate forward and reverse hybridizations for each clone. Clones are organized according to linear chromosome position. For the 1-Mbp resolution array exemplified by specimen 1664, normalized log ratios are shown for 2436 autosomal clones. For the 2- to 4-Mbp resolution array exemplified by specimen 939, normalized log ratios are shown for 1421 autosomal clones. (B) CBS analysis identified segments (minimum of 2 contiguous clones) with the same distribution of ratios. (C) Constrained CBS analysis then defined segments of normal copy number (log ratio, approximately 0) compared with those of gain (log ratio, greater than 0) or loss (log ratio, less than 0) of copy number. Horizontal bars on the top and bottom of panel C show the gains and losses, respectively, of the specimen determined by chromosomal-CGH. Circled clones represent singleton clones (within CBS-defined segments of normal copy number) with a log ratio more than 3 SDs from the expected normal value and considered singleton genomic gain or loss.
Representative array-CGH log ratio plots of 2 DLBCL specimens (1664 and 939) using the 1-Mbp and 2- to 4-Mbp resolution arrays, respectively. (A) Custom normalization was performed, and an averaged log ratio for each clone was calculated for the duplicate forward and reverse hybridizations for each clone. Clones are organized according to linear chromosome position. For the 1-Mbp resolution array exemplified by specimen 1664, normalized log ratios are shown for 2436 autosomal clones. For the 2- to 4-Mbp resolution array exemplified by specimen 939, normalized log ratios are shown for 1421 autosomal clones. (B) CBS analysis identified segments (minimum of 2 contiguous clones) with the same distribution of ratios. (C) Constrained CBS analysis then defined segments of normal copy number (log ratio, approximately 0) compared with those of gain (log ratio, greater than 0) or loss (log ratio, less than 0) of copy number. Horizontal bars on the top and bottom of panel C show the gains and losses, respectively, of the specimen determined by chromosomal-CGH. Circled clones represent singleton clones (within CBS-defined segments of normal copy number) with a log ratio more than 3 SDs from the expected normal value and considered singleton genomic gain or loss.
FISH analysis
Fluorescence in situ hybridization (FISH) analysis was performed using the BAC clones RP11-79H7 and RP11-121J7, mapped to 13q31.3, and RP11-25E13 and RP11-207D10, mapped to 13q33.2-q33.3 (Invitrogen). DNA from the BAC clones was isolated and directly labeled with either Spectrum Orange-dUTP or Spectrum Green-dUTP (Vysis, Downers Grove, IL) by nick translation and hybridized according to manufacturer-supplied protocols and as described by us.29 All BAC clones were first hybridized to normal human metaphase chromosomes to verify location and to exclude chimerism. In the 5 B-cell lymphoma cell lines, a minimum of 10 metaphases and 200 intact, nonoverlapping interphase nuclei were scored using a E800 microscope (Nikon, Tokyo, Japan) equipped with the Quips PathVysion system (Applied Imaging, Santa Clara, CA).
Results
Array-CGH analysis of 64 DLBCL specimens
Autosomal DNA copy number changes in specimens from 64 patients with newly diagnosed DLBCL were determined by array-CGH. In 26 specimens, forward and reverse hybridization was performed using Spectral Genomics arrays of BAC/P1 clones of 2- to 4-Mbp resolution across the genome, and for the remaining 38 specimens, an array of BAC clones of 1-Mbp resolution was used. Figure 1A shows averaged, normalized log ratio plots for the autosomal genome obtained for specimens 1664 and 939, which are representative of the data derived from the 2 array types. CBS analysis was performed for each tumor specimen to identify segments (minimum of 2 contiguous clones) with the same distribution of ratios (Figure 1B), followed by a constrained version of CBS to define segments of normal copy number compared with those with gain or loss of copy number (Figure 1C). Segmental gains and losses ranged in size from 1 Mbp to entire chromosomes and were detected in all but 3 specimens. On average, 7.0 ± 5.7 segmental gains per specimen were detected (range, 0-33 segmental gains), with slightly fewer segmental losses per specimen (5.7 ± 4.9; range, 0-21 segmental losses) (Table 1). Single clones within CBS-defined segments of normal copy number and with a log ratio more than 3 SDs from the expected normal value were considered singleton genomic gains/losses (Figure 1B-C). Specimens exhibited on average 8.6 ± 6.9 single clone gains (range, 0-40 gains) and 9.1 ± 6.3 single clone losses (range, 0-26 losses) (Table 1).
Chromosomal-CGH had previously been performed on 45 of the specimens in the current cohort.4,5 As expected, array-CGH generally detected a larger number of genomic copy number changes than chromosomal-CGH per specimen, as exemplified by specimens 1664 and 939 in Figure 1C. In 27 specimens, all changes detected by chromosomal-CGH were also detected by array-CGH (including specimens 1664 and 939). For 6 specimens in which no autosomal changes were found by chromosomal-CGH, changes were now detected by array-CGH. For the remaining 12 specimens, 9 showed partial overlap in the changes detected by the 2 techniques, 2 showed single alterations by chromosomal-CGH not detected by array-CGH, and 1 showed discrepant changes. Thus, overall, array-CGH confirmed genomic copy number changes revealed by chromosomal-CGH and identified changes that were previously undetected.
Common regions of genomic copy number changes in DLBCL
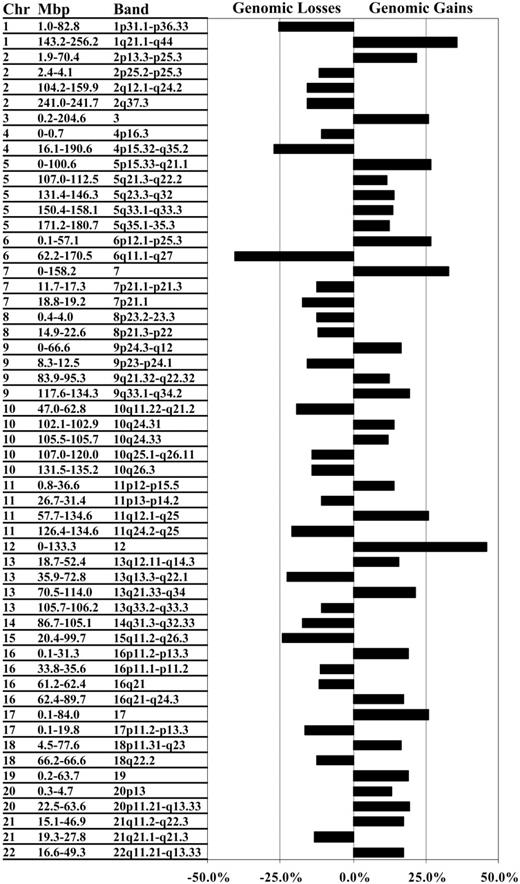
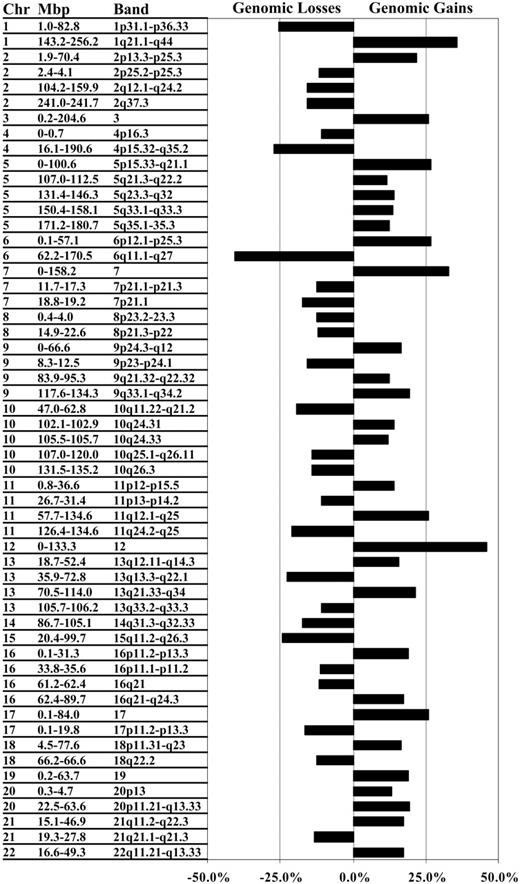
Data for the 2 array types were used to define autosomal regions frequently gained or lost in the entire cohort. To do this, clones not represented on the smaller 2- to 4-Mbp resolution array were assigned as gained, normal, or lost using the corresponding segmental results defined by the constrained CBS method, or they were defined as missing if outside the CBS-defined segments. Table S2 provides the assignment for each specimen for each autosomal clone identified either as a segmental or singleton copy number change. Common regions of gain or loss were identified where at least 2 contiguous clones exhibited a gain or a loss in at least 10% of all specimens. Using these criteria, common regions of copy number changes were detected on all autosomes, ranging in size from less than 1 Mbp to entire chromosomes (Table 2). In total, the 64 DLBCL specimens exhibited 29 common regional gains and 26 common regional losses, the physically largest of which were consistent with karyotypic and chromosomal-CGH findings (Figure 2).2-6 Improved resolution afforded by array-CGH led in some instances to refinement of the smallest gained or lost region within a previously reported larger region, but at higher frequency. This is exemplified by chromosome 20, in which gains of 20p (approximately 25 Mbp) had been reported by us to occur at a frequency of 5% to 10% using chromosomal-CGH. In the current study, array-CGH narrowed the frequently gained region to 4.4 Mbp now evident in 13.3% of specimens (Table 2).5 In other instances, subregions within regions could be defined based on changes in the frequencies of copy number change across regions, leading to narrowing of a region or to identification of 2 or 3 peaks of gain or loss within a region (Table 2). Region narrowing is exemplified by the loss of chromosome 10 (47.0-62.8 Mbp) comprising approximately 15 Mbp (10q21.1), which displayed a 3.9-Mbp subregion of loss observed in approximately 20% of specimens. Identification of multiple peaks of gain or loss within a region is exemplified by the loss of chromosome 1 of approximately 80 Mbp (1p) in which 2 subregions of loss could be discerned: 1.5 to 9.0 Mbp, which includes the TP73 locus recently shown to display loss in DLBCL by FISH, and 78.2 to 79.1 Mbp, which includes 5 known genes that may represent putative targets of loss.30 For the regional gain of chromosome 2 (1.9-70.4 Mbp), 2 subregions of gain were found: 9.0 to 56.0 Mbp and 59.3 to 63.9 Mbp, with the latter subregion including REL and BCL11A. These loci have previously been reported by us and others3-6,15,21,31 to occur frequently in higher copy numbers in DLBCL. Thus, array-CGH confirmed previously identified common regions of genomic gain or loss and, importantly, provided further definition of the size and genomic location of the most commonly gained and lost regions.
DNA copy number changes associated with expression signature subgroup
Only 38 specimens of the entire cohort had been submitted for expression profiling, of which 26 could be classified into an expression signature subgroup with greater than 90% probability according to the method of Wright et al12 (Table S1). Thus, to increase the analyzable cohort size for correlation of genomic copy number changes with expression signature subgroup, 55 available specimens were classified based on the immunostaining patterns of CD10, BCL6, and MUM1, as described.27 CD10 expression was observed in 42% of specimens, BCL6 in 55%, and MUM1 in 28% (Table S1). According to the staining algorithm, 34 (62%) specimens were classified as GCB-DLBCL and 21 (38%) as non-GCB-DLBCL. Only 9 (16%) specimens were negative for all 3 antibodies, which could account for the lower percentage of non-GCB-DLBCL determined in this study compared with 58% reported by Hans et al,27 in which 33% of specimens were negative for all antibodies. The 55 specimens included 22 submitted to expression profiling that could be classified with greater than 90% probability. Of these 22, 19 (86%) exhibited agreement between subgroup classification by the 2 methods. Consistent with our previous findings, non-GCB-DLBCL did not exhibit poorer OS than GCB-DLBCL in this cohort when classified by the immunohistochemical method (P = .62).15
Because there was good agreement between the 2 methods for subtype classification, correlations of genomic gains and losses with expression signature subgroup were performed using a 2-tailed Fisher exact test on the 55 specimens classified by the immunohistochemical method. Gain of a narrow region on chromosome 12 (45.4-53.8 Mbp) showed a significant association with the GCB-DLBCL subtype (P = .026), whereas gain of the entire chromosome 12 showed a trend toward significance (P = .057) (Table 3). Consistent with previous findings, gain of chromosome 2 (1.9-70.4 Mbp) exhibited a trend toward a significant association (P = .054) with the GCB-DLBCL subtype.11,15,16 Specimens of the GCB-DLBCL subtype also exhibited higher frequencies of gain of chromosome 9 (83.9-95.3 Mbp), loss of chromosome 11 (130.6-134.6 Mbp), and loss of chromosome 13 (105.7-106.2 Mbp) than the non-GCB-DLBCL subtype (Table 3), though not with statistical significance. None of the genomic copy number changes associated significantly with the non-GCB-DLBCL subtype.
DNA copy number changes associated with clinical features
Table 4 lists common regions and subregions of DNA copy number changes that associated significantly with a clinical feature (P < .05). By Fisher exact test, several regions of genomic gain or loss were associated with 1 or more of the 5 features used to determine the IPI score. In this panel of newly diagnosed DLBCLs, 4 regions of gain (excluding 2 larger regions that were narrowed) and 2 regions of loss were significantly associated with longer OS by the log-rank test (Table 4). Of these, gains of chromosome 3 and 19 also correlated with clinical features known to be associated with good outcome. Loss of chromosomes 2 (2.4-4.1 Mbp) and 16 (33.8-35.6 Mbp) and gain of chromosome 13 (85-91.9 Mbp) were significantly associated with inferior survival (Table 4). Using a 5% false discovery rate cutoff to correct for multiple comparisons, only the loss of chromosome 16 (33.8-35.6 Mbp) remained significant.32 Of the 9 minimal regions associated with OS, 6 were also significantly associated with TTF in a consistent manner (Table 4). The region of gain of chromosome 13, identified in the current study to be associated with poor outcome, overlapped with a region previously reported to associate with poor outcome in a subset of DLBCLs that expressed CD5.7 Thus, to determine whether in the present cohort this association was restricted to CD5+ DLBCL, sections for 41 available specimens were submitted to immunohistochemical analysis for CD5 staining (Table S1). For this smaller group, gain of the 13q region had a trend toward significance with poor outcome (P = .055), but none of the specimens exhibited CD5+ staining, indicating in the present cohort the association is also evident in CD5- DLBCL.
Common regions of genomic gain and loss in 64 DLBCL specimens. The tree shows the 55 genomic regions that exhibited gain or loss in at least 10% of the specimens. For each region, a corresponding cytogenetic location and the respective frequency of change within the cohort are given.
Common regions of genomic gain and loss in 64 DLBCL specimens. The tree shows the 55 genomic regions that exhibited gain or loss in at least 10% of the specimens. For each region, a corresponding cytogenetic location and the respective frequency of change within the cohort are given.
To determine whether the gains and losses associated with OS were independent of IPI in predicting outcome, each of the 9 minimal regions was entered separately, along with IPI score, into multivariate analysis using the Cox regression model. Loss of chromosomes 2 (2.4-4.1 Mbp) and 16 (33.8-35.6 Mbp) associated independently with inferior survival, with hazards ratios of 3.32 (P = .033; 95% confidence interval [CI], 1.10-6.51) and 6.86 (P = .0003; 95% CI, 2.41-10.08), respectively. Loss of chromosome 1 (78.2-79.1 Mbp) was an independent predictor of better outcome, with a hazard ratio of 0.23 (P = .019; 95% CI, 0.07-0.91). Again, only the loss of chromosome 16 (33.8-35.6 Mbp) remained significant after adjustment for multiple comparisons. Overall, genomic copy number changes of 9 previously unidentified minimal regions were associated with OS and consisted of 3 that were additionally found to be independent of IPI score in the prediction of outcome in DLBCL.
FISH validation of gains and losses identified by array-CGH
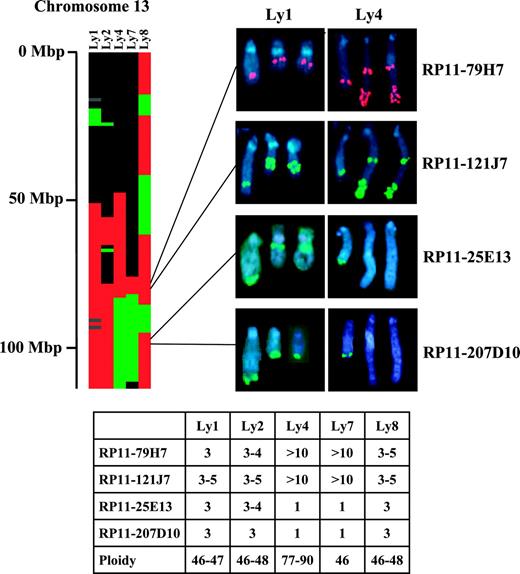
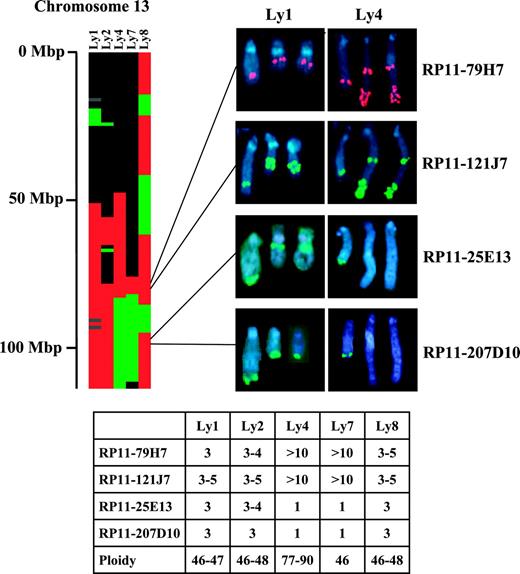
Genomic copy number changes identified by array-CGH were validated by FISH in 5 B-cell lymphoma cell lines. Array-CGH was performed and analyzed as described. Table S2 provides the assignment for each cell line for each autosomal clone, either as a segmental or as a singleton copy number gain or loss. By array-CGH, all 5 cell lines exhibited a gain within a region of chromosome 13 (85-91.9 Mbp) that was commonly gained in DLBCL specimens and was associated with poor outcome (Table 2; Figure 3). For validation of this gain, FISH analysis of the cell lines was performed using 2 BAC clones (RP11-79H7, RP11-121J) contained within the region. Figure 3 shows the partial karyotypes confirming an increase in copy number from 3 to more than 10 of the 2 BACs in all 5 cell lines. For validation of loss, FISH analysis was performed with 2 BAC clones (RP11-25E13, RP11-207D10) that were mapped within a common region of loss on chromosome 13 (105.7-106.2 Mbp) in the DLBCL specimens and by array-CGH exhibited loss in the cell lines Ly4 and Ly7. Cell lines Ly1, Ly2, and Ly8 exhibited a gain of this region by array-CGH. Figure 3 shows the FISH hybridization patterns obtained for these 2 BACs, by which 3 to 5 copies per cell were evident in Ly1, Ly2, and Ly8 and only 1 copy per cell was evident in Ly4 and Ly7, thus validating the array-CGH findings.
Discussion
Application of low-resolution genome scanning technologies has identified few genomic alterations at the chromosome band level associated with outcome in DLBCL. In the current study, we describe, for the first time, results of comprehensive array-CGH analysis of a large panel of specimens from patients with newly diagnosed DLBCL that have led to the identification of regions of genome copy number changes predictive of outcome and independent of IPI. All 3 regional prognosticators were smaller than 5 Mbp, which is far below the resolution of chromosomal CGH; therefore, it was not unexpected that such copy number changes would go undetected using the lower resolution technologies. In addition, applying an advanced algorithm (constrained CBS) to define segments of copy number change within specimens provided greater sensitivity in the detection of changes than had previously been possible. In all, 55 common regions of copy number gain or loss were identified, ranging in size from less than 1 Mbp to entire chromosomes. For the larger regions, there was good correlation with previous chromosomal CGH studies performed on DLBCL and for which array-CGH afforded further delineation into smaller subregions.
It is unclear at present whether the 2 broad expression signature subtypes of DLBCL truly represent DLBCL clonally derived from the 2 different stages of B-cell differentiation: GCB-DLBCL from GC B-cells and non-GCB- or ABC-DLBCL from a B-cell undergoing plasmacytic differentiation through the GC.10,12 In this model, it was expected that some genetic abnormalities would be shared by the 2 groups associated with GC passage, whereas others would be specific to each subtype. Another, less likely, model is that in which the non-GCB- or the ABC-DLBCL subtype arose from a B-cell undergoing plasmacytic differentiation outside the GC, in which few genetic abnormalities would be expected to be shared by the 2 broad subtypes.12 Our study supports the former model in that few regions displayed different frequencies of gains or losses between the 2 subtypes. Gain of the REL locus and t(14;18)(q32;q21) have been reported to occur predominantly, though not exclusively, within GCB-DLBCL.11,14,15 In the current study, we confirmed the former association, whereas the latter abnormality was not detected by CGH. Additionally, we report gain of an approximately 9 Mbp region on chromosome 12 (45.4-53.8 Mbp) to be significantly associated with the GCB-DLBCL subtype. This region was part of a larger region on 12q recently documented to be associated with the GCB-DLBCL subtype.16 In that report, changes in the copy number of several genomic regions by array-CGH were found to correlate with both expression signature subtypes. Discrepancies between the studies could be accounted for by different methodologies used to define regional copy number changes and determining subtype classification, as well as by the high proportion of CD5+ DLBCL within that study cohort.
Genomic copy number changes of chromosome 13 detected by array-CGH and validated by FISH in lymphoma cell lines. The heat map shows the genomic copy number changes of chromosome 13 in 5 lymphoma cell lines. Red indicates gain; green, loss; black, normal; and gray, unknown. Scale on the left shows physical distance from the telomere of the q arm. FISH analysis of metaphase chromosomes of 2 representative cell lines (Ly1 and Ly4) are shown with 2 BACs (RP11-79H7 and RP11-121J7) exhibiting gain by array-CGH, and with 2 BACs (RP-25E13 and RP11-207D10) showing gain by array-CGH in Ly1 and loss in Ly4. The average number of copies per cell, as detected by FISH analysis and the ploidy of each cell line, are tabulated.
Genomic copy number changes of chromosome 13 detected by array-CGH and validated by FISH in lymphoma cell lines. The heat map shows the genomic copy number changes of chromosome 13 in 5 lymphoma cell lines. Red indicates gain; green, loss; black, normal; and gray, unknown. Scale on the left shows physical distance from the telomere of the q arm. FISH analysis of metaphase chromosomes of 2 representative cell lines (Ly1 and Ly4) are shown with 2 BACs (RP11-79H7 and RP11-121J7) exhibiting gain by array-CGH, and with 2 BACs (RP-25E13 and RP11-207D10) showing gain by array-CGH in Ly1 and loss in Ly4. The average number of copies per cell, as detected by FISH analysis and the ploidy of each cell line, are tabulated.
Deletions of 6q and breakpoints at 1q21 have previously been reported by karyotype analysis as chromosomal abnormalities associated with inferior survival in DLBCL.2 Although breakpoints are not detected by CGH unless accompanied by genomic gain or loss, 6q deletions are detected, but no significant association with OS has been confirmed for this abnormality by chromosomal- or array-CGH studies.4-7,16 In the current study, loss of chromosome 6 (62.2-170.5 Mbp) was associated with greater TTF, which perhaps can be explained by small deletions detected by array-CGH in specimens previously thought not to carry deletions in this region and by the heterogenous location of such deletions. By chromosomal-CGH, loss of 17p harboring the TP53 gene has variously been reported to portend a poor outcome, though not in our reported cohorts.4-6 In the current array-CGH study, loss of this region was detected in 16% of specimens and only showed a trend toward significance with poor outcome (P = .057). Recent array-CGH studies by one group have inconsistently reported gain of several clones on 5p to be associated with good outcome in CD5- DLBCL specimens, loss of clones at 1p34.21-36.3 and gain of clones at 13q21.1-q31.3 and at 13q31.3-q34 to be associated with worse survival in CD5+ DLBCL, and loss of the CDKN2A locus mapped to 9p21 occurring with greater frequency in more aggressive DLBCL, irrespective of CD5 status.7,16 In comparison, in the current cohort studied, gain of a corresponding 13q21.1-q31.3 region (narrowed to 85.0-91.9 Mbp) was found to portend inferior survival in patients with CD5- DLBCL but was not found to be independent of IPI. In contrast, loss or gain of the other regions was not commonly detected in 10% or more of specimens (for example, 9p21, where deletion of the respective region was only detected in 4 of the 64 specimens) or did not significantly associate with outcome in the present cohort. This could be accounted for by use in the current study of advanced algorithms to define segments of copy number change, consideration of regional gains or losses in correlative analyses rather than single BAC clones, and differences in the patient cohorts. Other array-CGH studies on DLBCL have focused on copy number changes of specific regions, oncogenes, and tumor-suppressor genes.8,9,33 In the case of the amplicon covering 13q31-q32, follow-up expression studies have revealed that C13orf25 rather than GPC5, as previously suggested, is a good candidate for the target gene of the copy number change.9,34 A recent study has indicated that the C13orf25 transcript is a functional precursor of a series of micro-RNAs belonging to the mir-17-92 polycistron, which, when overexpressed with myc, accelerated tumor development in a murine B-cell lymphoma model.35 This locus lies within the region represented by the BACs on the array and those used for FISH in the current study to confirm genomic duplications in the lymphoma cell lines. Furthermore, in 6 of 10 independent DLBCL specimens with karyotypic 13q abnormalities, extra copies were detected by FISH, usually through duplication (data not shown). The functional significance of gain of this region and overexpression of the mir-17-92 polycistron in relation to outcome remain to be fully determined. For the 3 novel regions identified to associate with outcome independent of IPI, few Refseq genes have been mapped that represent candidate target genes. For chromosome 1 (78.2-79.1 Mbp), the genes mapped are PTGFR, IFI44L, and IFI44, and for chromosome 2 (2.4-4.1 Mbp), they are RNASEH1, RPS7, COLEC11, and ALLC. For the chromosome 16 (33.8-35-6 Mbp) loss that remained significantly associated with inferior survival after correction for multiple comparisons, there were 2 mapped genes. One was FLJ43855, which exhibited similarity with a sodium- and chloride-dependent creatine transporter, and the other was a p53-inducible gene, TP53TG3, whose expression has also been noted to increase in several breast cancer cell lines after doxorubicin treatment.36,37
Overall, array-CGH of a panel of DLBCL specimens provided comprehensive high-resolution scanning of the DLBCL genome and identified multiple regions of common copy number changes, several of which significantly correlated with outcome. These clinical correlations now must be validated in a prospectively ascertained, uniformly treated cohort of patients, and follow-up expression studies of the novel prognostic regions are required to identify target genes.
Prepublished online as Blood First Edition Paper, November 29, 2005; DOI 10.1182/blood-2005-07-2950.
Supported in part by research grants from the National Institutes of Health.
W.C. and J. Houldsworth contributed equally to the study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Marina Ashinov and Irina Linkov in the MSKCC Immunohistochemistry Core Facility for expert technical assistance.