Our previous studies have identified mechanisms by which cytokine production, blocked by Ly49G2 receptor cross-linking, can be overridden. In this study we analyzed the regulation of other ITAM-positive receptor signaling on NK, NKT, and T cells and characterized the biochemical pathways involved in this signaling. Our studies demonstrate that cross-linking of NKG2D and NK1.1 results in a synergistic NK IFN-γ response when combined with IL-12 or IL-18. Examination of NKT- and T-cell responses demonstrated that cross-linking of NKG2D and CD3 resulted in potent synergy when combined with IL-12 and, to a lesser degree, with IL-18. We have now found that both the p38 MAP kinase and the ERK-dependent signal transduction pathways are required for the synergistic response. Further mechanistic examination of the synergy indicated a potent up-regulation of total IFN-γ mRNA in the nuclear and the cytoplasmic compartment, but mRNA half-life was not affected. Fifteen minutes of IL-12 pretreatment was sufficient to result in maximal synergistic activation, indicating that the response of the cells to the IL-12 signal was rapid and immediate. Thus, our data demonstrate that multiple convergent signals maximize the innate immune response by triggering complementary biochemical signaling pathways.
Introduction
Murine natural killer (NK) cells express multiple Ly49 receptors1-5 that either inhibit or activate NK cell functions, including cytolysis and cytokine secretion. A functionally similar family of molecules exists on human NK cells—the killer cell immunoglobulin-like receptors (KIRs). The inhibitory Ly49 receptors (Ly49A, C, G and I) inhibit NK cell function on binding of class 1 ligands on target cells.6-8 These Ly49 inhibitory receptors—and inhibitory KIRs—contain cytoplasmic immune receptor tyrosine-based inhibitory motifs (ITIMs) that are phosphorylated on stimulation, leading to the recruitment of SHP-1 phosphatase and attenuation of intracellular signals. In contrast, the ITAM-associated activating receptors (eg, Ly49D and Ly49H) mobilize intracellular Ca2+, induce cytokine mRNA and protein production, and mediate reverse antibody-dependent cellular cytotoxicity (ADCC) in the presence of specific mAbs.9-12
Circulating NK cells expressing activating Ly49 also express coreceptor paired inhibitory Ly49. Thus, effector cells that express the activating Ly49D receptor that binds H2-Dd as a ligand also coexpress, at very high levels, inhibitory Ly49G2 or Ly49A13-15 receptors that also bind H2-Dd and inhibit the activating function. Based on this coexpression, engagement of activating Ly49 NK receptors in vivo appears constantly at odds with inhibitory forces. Our previous studies demonstrated that cross-linking of activating Ly49D murine NK cell receptors can potently synergize with IL-12 for selective and synergistic production of IFN-γ, both in vitro and in vivo. Importantly, IL-12 was the key signal needed for overriding the inhibitory receptor blockade for cytokine production.
Given that there are numerous coreceptor systems in the T-cell system that require 2 signals to induce sufficient cellular activation, we postulated that other NK cell receptors may require 2 positive signals to override the ever-vigilant inhibitory receptor blockade. Thus, we sought to examine a model in which the secretory function of activating receptors, in addition to the Ly49 family, might be triggered by coreceptor function. Furthermore, as reported here, we have now characterized the biochemical pathways required for the expression of IFN-γ in response to multiple, yet distinct, extracellular signals.
Materials and methods
Reagents
Alpha (α) GalCer (KRN7000) was graciously provided by Kirin Brewery (Tokyo, Japan). The ceramide reagents were first dissolved in DMSO, then diluted in phosphate-buffered saline (PBS) containing 0.5% Tween 20. Control diluent or PBS was used as a control for all studies. MAP kinase inhibitors SB203580 (source) and U0126 (source) were used at a final concentration of 1 μM.
Cell lines
B-cell lines (A20 and A20/CD1d, generously provided by M. Kronenberg, La Jolla Institute, San Diego, CA) were pretreated with various reagents for 30 minutes at 37°C, washed, and mixed with sorted populations, and supernatants were collected for analysis after specified culture time.
NK cell isolation
Liver NK cells were isolated from C57BL/6 (B6) mice, as previously described.14 Animal care was provided in accordance with the procedures outlined in the “Guide for the Care and Use of Laboratory Animals” (National Institutes of Health Publication No. 86-23, 1985). Liver mononuclear cells were used either untreated (15%-25% CD3-, NK1.1+) or after in vivo IL-2 treatment (35%-70% CD3--, NK1.1+), followed by lineage depletion (with CD3, CD19, and CD24) (greater than 90% CD3-, NK1.1+) or after in vitro expansion with IL-2 (6000 IU/mL recombinant IL-2) (Chiron, Emeryville, CA), as previously described.16 In vivo IL-2 treatment was conducted as previously described using a plasmid containing the murine Il2 gene.17
Antibodies used
The monoclonal antibodies 4E5 (Ly49D), 3D10 (Ly49H), and 3A10 (NKG2D) were previously described11 or were provided by Dr Wayne Yokoyama (Washington University, St Louis, MO). Rat IgG (Becton Dickinson/PharMingen, San Jose, CA) was used as a control for flow cytometry and functional studies. Rabbit F(ab')2 anti-rat IgG was used as a cross-linking reagent. CD19 F or APC, NK1.1-PE or APC, DX-5-PE and CD3ϵ-PercP (Becton Dickinson/PharMingen), and 4E5-FITC were used for flow cytometry analysis.
Flow cytometry analysis
Cells were stained as previously described16 and analyzed on either a FACSort or an LSR flow cytometer (Becton Dickinson, San Jose, CA). Cells were directly stained using FITC-, phycoerythrin (PE-), Per-CP-, and APC-labeled primary antibodies. The BDCBA Phospho Flex Set (Becton Dickinson) is a bead-based immunoassay measuring mouse signal-regulated kinases (p38, ERK1/2) was analyzed in denatured cell lysate samples.
Cytokine measurement
Cytokines were measured using IFN-γ and chemokine ELISA kits (R&D Systems, Minneapolis, MN). Cell stimulations were performed at cell concentrations of 1-5 × 106 cells/mL. Antibodies were added at a concentration of 1 μg/106 cells for 30 minutes at 4°C. Cells were then washed and plated on 24-well Costar (Corning, NY) plates that were precoated with 2 μg/well rabbit F(ab')2 anti-rat IgG and blocked with media containing 10% fetal calf serum. Unless otherwise stated, samples were collected after 5 to 6 hours of incubation (37°C, 5% CO2) and were measured in duplicate against the standard curve of the assay and reported as picograms per milliliter. In all assays, the standard deviation was less than 5 pg/mL.
Ribonuclease protection assay
The multiprobe RNAse Protection Assay (RPA) was performed using the mck-1 or mck-5 template set (PharMingen, San Diego, CA). Total cellular RNA was extracted using Trizol (Life Technologies, Gaithersburg, MD), and 1-5 μg total mRNA was hybridized with a 33-P UTP-labeled RNA probe (1 × 106 cpm/sample) prepared according to the manufacturer's directions (PharMingen, La Jolla, CA) using the PharMingen RiboQuant In Vitro Transcription kit. After hybridization, the samples were treated with RNAse A and T1 according to the procedure provided by PharMingen. The RNAse was inactivated and precipitated with the use of a master cocktail containing 200 μL Ambion (Austin, TX) RNAse inactivation reagent, 50 μL ethanol, 5 μg yeast tRNA, and 1 μL Ambion GycoBlue coprecipitate per RNA sample. The samples were mixed well, incubated at -70°C for 15 minutes, and centrifuged at 16 100g for 15 minutes at room temperature. Pellets were suspended in 3 μL PharMingen sample buffer and subjected to polyacrylamide gel electrophoresis, as recommended by the manufacturer (PharMingen).
Quantification of variant forms of Nkg2d mRNA
Specific forward and common reverse primers for quantification of short and long forms of Nkg2d mRNA were designed using the Primer Pick 3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) from the published mRNA sequences for the long (GenBank accession number AF054819) and short (GenBank accession number AF030313) forms of Nkg2d mRNA, as follows: Nkg2d reverse 5′ ctc gaa caa cga aca ttg ga 3′ (R); Nkg2d long-form specific 5′gcattgattcgtgatcgaaa 3′ (L); Nkg2d short-form specific 5′ acaagaaaca ggatctccct (S) (for the location of the primers, see Supplemental Figure S1, available at the Blood website; click on the Supplemental Figures link at the top of the online article).
Generation of positive controls for real-time PCR assays
For isolating the cDNA segment specific for the long form of the Nkg2d mRNA, the previously published18 forward primer (P2 primer: 5′caggaagcagaggcagattatctc) was used. Plasmid DNA containing cDNA segments specific for the long and short forms of the Nkg2d mRNA was isolated as follows. Amplicons were isolated from RNA obtained from either spleen or IL-2-treated spleen. cDNA was synthesized using the Superscript III kit from Invitrogen (Carslbad, CA) according to the manufacturer's protocol using either the oligodT or the Nkg2d R primer described here. Amplicons were amplified by PCR using P2/R and S/R primers using Advantage2 polymerase from BD Biosciences (Franklin Lakes, NJ). PCR conditions used were 94°C for 2 minute, 30 cycles of 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 60 seconds, and a final extension at 72°C for 7 minutes. cDNA fragments were cloned into the Topo TA cloning vector (Invitrogen), and the cDNA inserts in the plasmid DNA were verified by DNA sequencing (data not shown).
Real-time PCR assays
For quantification of the variant form of Nkg2d mRNA, plasmid DNA isolated as described was used to generate a standard curve. The real-time PCR assays for Nkg2d plasmid DNA or cDNA were carried out in 10-μL reactions using S/R and L/R primers, SYBR Green master mix (Qiagen, Valencia, CA), and run on an ABI 7900 (Applied Biosystems, Foster City, CA). PCR conditions used were 95°C for 15 minutes, 40 cycles of 95°C for 15 seconds, 60°C for 1 minute. This was followed by a dissociation curve at 95°C for 15 seconds, 60°C for 15 seconds, and a 2% ramp rate to 95°C for 15 seconds. Standard mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) plasmid DNA was purchased from Serologicals (Gaithersburg, MD). Real-time PCR assays for GAPDH plasmid DNA were carried out in 10-μL reactions using the mouse GAPDH control kit (Applied Biosystems) and run on an ABI 7900. PCR conditions used were 95°C for 15 minutes, 40 cycles of 95°C for 15 seconds, 60°C for 1 minute, and then a dissociation curve at 95°C for 15 seconds, 60°C for 15 seconds, and a 2% ramp rate to 95°C for 15 seconds. Sensitivity and linear dynamic range were checked on the serial dilutions (10-106 copies per reaction) of Nkg2d short and long and GAPDH plasmid DNA and were found to be greater than 99% efficient to have a slope of -3.6. The Nkg2d short form of mRNA expression was normalized to the GAPDH expression in multiplex and was quantified with its own standard curve, greater than 97% efficiency with a slope of -3A8. Similarly, the Nkg2d long form of mRNA had greater than 97% efficiency and a slope of -3.5.
Nuclear and cytoplasmic fractionation and RNA isolation
Approximately 5 × 107 NK cells were pelleted by centrifugation at 300g in a Sorvall H1000B rotor (Sorvall, Asheville, NC), washed once in PBS, resuspended in 3 mL homogenization buffer (15 mM HEPES, pH 7.4, 0.3 M sucrose, 60 mM KCl, 15 mM NaCl, 2 mM EDTA, 0.5 mM EGTA, 0.15 mM spermine, 0.5 mM spermidine, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.6% NP40) and were lysed by incubation on ice for 5 minutes. The homogenate was centrifuged at 800g for 5 minutes at 4°C. The resultant pellet was washed twice in 2 mL nuclei wash buffer (homogenization buffer without detergent) and centrifuged at 800g for 5 minutes at 4°C and used immediately for nuclear RNA isolation by the addition of Trizol to the pellet. Cytoplasmic RNA extraction was performed after cell lysis and centrifugation by the addition of Trizol LS to the supernatant fraction.
Results
ITAM-containing receptor-cytokine synergy
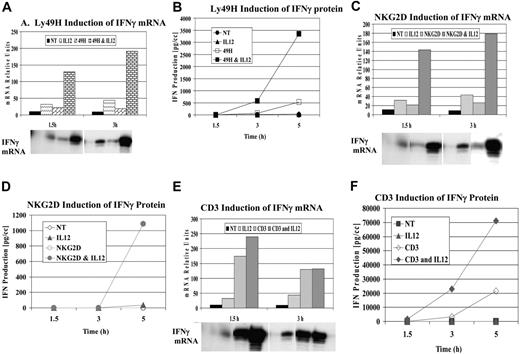
Based on our previous studies, we examined additional ITAM-associated receptors present on NK cells for their ability to synergize with cytokines after ligation. Through the use of untreated mouse liver NK cells or purified NK cells (after magnetic depletion of lineage-positive cells), specific receptor ligation was performed with antibodies specific for Ly49H and NKG2D. Figure 1A-B demonstrates a representative experiment examining the mRNA expression and secretion of IFN-γ after cross-linking Ly49H with or without IL-12. Similar to what has been observed with Ly49D, the DAP-associated Ly49H receptor exhibited strong synergy for IFN-γ mRNA and cytokine production compared with cytokine or receptor cross-linking alone. Next, we examined the effects of cross-linking the DAP-associated NKG2D after cross-linking with NKG2D-specific mAb 3D10 (Figure 1C-D). Unlike Ly49D and Ly49H, receptor cross-linking NKG2D alone did not result in significant IFN-γ secretion unless IL-12 was added. Based on these results, we examined the effects of cross-linking the TCR-associated receptor using the NKT-rich liver leukocytes because the conical ITAM-bearing receptor is CD3. We observed strong synergy for IFN-γ cytokine production when the receptor was cross-linked in the presence of IL-12 compared with either cytokine or receptor cross-linking alone (Figure 1E-F). Thus, with all classes of ITAM-bearing receptors examined, a potent costimulatory signal could be observed with IL-12 (Figure 1) or IL-18 (not shown). Furthermore, cross-linking of NK1.1 (linked to the signaling chain, FcγR) also demonstrated similar synergy with IL-12 and IL-18 (not shown).
Synergy of IFN-γ induction by ITAM-bearing receptors. Liver lymphocyte receptor cross-linking was performed with anti-Ly49H (3D10) (A-B), anti-NKG2D (3A10) (C-D), and anti-CD3 (500A2) (E-F). IFN-γ mRNA (A,C,E) and protein (B,D,F) expression were evaluated at 1.5, 3, and 5 hours. RPA evaluation of IFN-γ mRNA was normalized to control GAPDH. Values are representative of 3 experiments.
Synergy of IFN-γ induction by ITAM-bearing receptors. Liver lymphocyte receptor cross-linking was performed with anti-Ly49H (3D10) (A-B), anti-NKG2D (3A10) (C-D), and anti-CD3 (500A2) (E-F). IFN-γ mRNA (A,C,E) and protein (B,D,F) expression were evaluated at 1.5, 3, and 5 hours. RPA evaluation of IFN-γ mRNA was normalized to control GAPDH. Values are representative of 3 experiments.
In vitro analysis of Nkg2d gene expression in spleen and liver NK cells
Given that the association of the short form of NKG2D with DAP12 is critical for the triggering of cytokine gene expression, we believe it is important to confirm the presence of sufficient short forms in the NK cells to support our data. Our results indicate that the short form of the Nkg2d mRNA level increased by 5- and 19-fold in IL-2-treated spleen and liver NK cells, respectively, whereas the long form of the Nkg2d mRNA increased by 3- and 10-fold, respectively. Because DAP12 associates with the shorter form of Nkg2d, our data support the in vitro data that resting fresh liver NK cells contain sufficient DAP12-associated NKG2D to exhibit synergy with IL-12, as measured by IFN-γ production (see Supplemental Figures).
Regulatory role of NKRs in cytokine gene expression
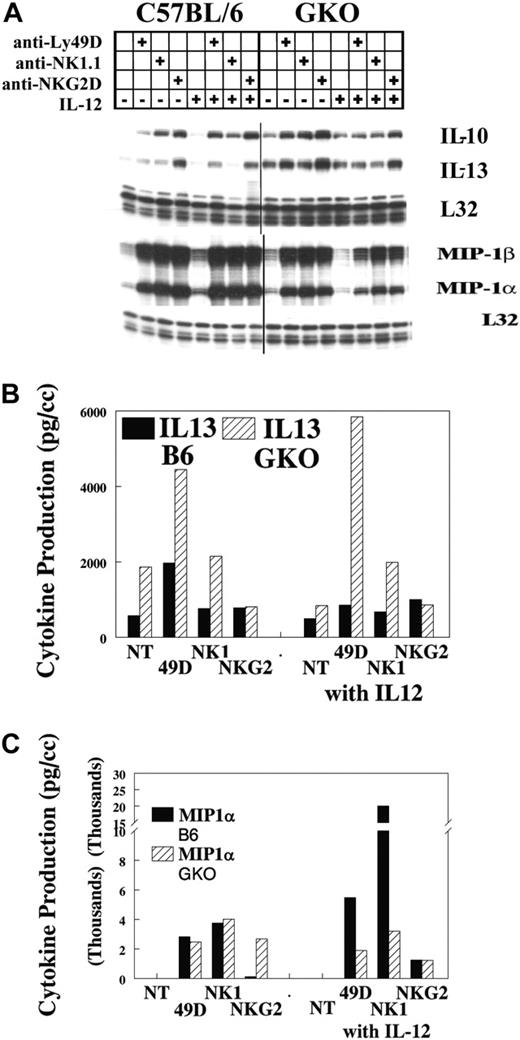
Our previous studies17,19 demonstrated that IFN-γ was predominately induced by IL-12 when Ly49D was coengaged. To examine whether IFN-γ might alter the production or costimulation of other cytokines on NKR ligation, we performed IL-12 costimulation in IFN-γ-deficient mice (GKO). Representative results of several experiments are shown in Figure 2. In B6 mice, MIP1α and MIP1β were strongly increased by Ly49, NK1.1, and NKG2D cross-linking. IL-10 mRNA was increased to a lesser degree, whereas IL-13 mRNA was only weakly increased. When GKO mice were evaluated, a strong TH2 bias was observed. NKRs strongly increased MIP1α and MIP1β, whereas IL-10 and IL-13 had high basal levels of mRNA that were only modestly increased by NKR ligation (Figure 2A). Cotreatment with IL-12 did not significantly alter the expression of any of these genes. When cytokine protein levels were analyzed, results indicated that IFN-γ did not alter the response of NK cells to receptor cross-linking with respect to other types of chemokine/cytokine production (Figure 2B-C) in the presence or absence of IL-12. Of possible interest is the strong synergy also observed in MIP1α production when IL-12 is combined with cross-linking NKRs. This was not seen in the GKO mouse and may indicate a role for IFN-γ in this synergy. This observation awaits further analysis.
IL-12 synergy in GKO mice. Lymphocytes were obtained from the livers of untreated B6 mice and were stimulated with the specific anti-NKRs. Cells were evaluated for IFN-γ mRNA (A) and cytokine production of IL-13 and MIP1α in 3-hour supernatants (B-C). Values are representative of 3 experiments.
IL-12 synergy in GKO mice. Lymphocytes were obtained from the livers of untreated B6 mice and were stimulated with the specific anti-NKRs. Cells were evaluated for IFN-γ mRNA (A) and cytokine production of IL-13 and MIP1α in 3-hour supernatants (B-C). Values are representative of 3 experiments.
Regulatory role of IL-12 in NKT cells
Because the liver is a rich source of NKT cells and our data with anti-CD3 cross-linking indicated that TCR-ITAM ligation synergized with IL-12, we examined the response of NKT cells to costimulation. Ligation of the TCR resulted in the production of IFN-γ, TNFα, and IL-2 (Figure 3A). As expected, cytokine stimulation was rapid and direct, with mRNA increases observed by 1 hour and maximally by 3 hours, followed by rapid protein expression in culture supernatants. As in NK cells, IL-12 synergy was only seen with respect to IFN-γ expression (Figure 3B) because TNFα (Figure 3B) and IL-2 (Figure 3C) expression were not affected by the addition of IL-12 (Figure 3B, D). Identical results were obtained when primary NKT cells were stimulated with αgalactosyl ceramide (αGalCer) (Figure 3D) in the presence or absence of IL-12. Thus, IL-12 appears to be a key cytokine for maximizing the response of NKT cells to cell surface receptor cross-linking.
Effects of IL-12 and IL-18 on the synergistic response
To further evaluate the synergistic response, we sought to determine whether multiple interactions with different costimulatory cytokines would result in an even greater response to NKR activation. Therefore, a dosing checkerboard of IL-12 and IL-18 was added to NK cells pretreated with control IgG or anti-Ly49D (4E5) antibodies. As expected, IL-12 and IL-18 demonstrated potent costimulation without NKR cross-linking for IFN-γ production in NK cells; however, when doses of IL-18 reached 0.1 ng/mL, limited activation by IL-12 was observed at 1 and 0.1 ng/mL (Table 1; see IFN-γ production in control IgG). When these same suboptimal costimulating doses of IL-12 and IL-18 were analyzed with NK cells stimulated through the Ly49D NKR, strong, near maximal IFN-γ production was observed (29 000 and 25 200 pg/mL; values in bold) compared with 10-ng treatment results. These physiologic levels of IL-12 and IL-18 could sensitize NK cells to produce a greatly enhanced response to ligation of NKRs.
Receptor cross-linking
With the recent emergence of other IL-12 family members, we sought to evaluate whether IL-23 and IL-27 would result in costimulation similar to that of IL-12. Figure 4A depicts a typical result with these cytokines and cross-linking of Ly49D. Both IL-23 and IL-27 costimulated IFN-γ production upon Ly49D ligation, but they were quantitatively less potent than IL-12. In contrast to IL-12, cytokines alone did not result in significant levels of IFN-γ (not shown). In addition, we sought to determine whether other cytokines that interacted with NK cells at the time of NKR ligation and IL-12 interaction might alter subsequent synergy. Therefore, NK cells were pretreated with other inflammatory cytokines, such as IFN-α, IFN-β, IFN-γ, IL-1, IL-10, IL-13, and TNFα. None of these cytokines demonstrated strong and consistent alteration in the ability of IL-12 to synergize with NKR ligation for the production of IFN-γ (not shown), nor did they synergize with receptor cross-linking when cytokine expression was analyzed.
Inhibitory receptors and IL-12
Our previous studies17 demonstrate that IL-12 can override the ability of Ly49G2 to inhibit Ly49D activation. Because Ly49G2+ NK cells also coexpress NKG2D and NK1.1 and consist 50% to 60% of Ly49D+, we examined whether IL-12 could also coregulate their inhibition. As shown in Figure 4B, cotreatment with IL-12 strongly stimulated IFN-γ production regardless of whether Ly49G2 was coengaged. The coengagement of Ly49G2 on cells not treated with IL-12 demonstrated a 50% to 70% inhibition of IFN-γ production, regardless of which activating receptor was cross-linked. Thus, costimulation with IL-12 with activating receptors results in the diminution of the ability of multiple inhibitory receptors to attenuate the NK cell cytokine response.
IL-12 synergy with TCR on NKT cells. Highly purified NKT cells were sorted from untreated liver lymphocytes (CD3+, NK1.1+), then expanded for 4 to 5 days in IL-2. Cells were evaluated for synergy, as described, using anti-CD3. IFN-γ mRNA (A) or cytokines TNFα and IFN-γ (B) or IL-2 (C) were measured in 1- and 3-hour supernatants, respectively (B-D). Values are representative of 3 experiments. (D) Liver NKT cells were obtained from untreated B6 mice and stimulated with the specific ligand αGalCer after loading into a CD1d-positive cell line A20. Cells were evaluated for synergy of cytokine production with IL-12 and αGalCer at 3 hours. TNFα, IFN-γ, or IL-2 was measured in 3-hour supernatants. Values are representative of 3 experiments.
IL-12 synergy with TCR on NKT cells. Highly purified NKT cells were sorted from untreated liver lymphocytes (CD3+, NK1.1+), then expanded for 4 to 5 days in IL-2. Cells were evaluated for synergy, as described, using anti-CD3. IFN-γ mRNA (A) or cytokines TNFα and IFN-γ (B) or IL-2 (C) were measured in 1- and 3-hour supernatants, respectively (B-D). Values are representative of 3 experiments. (D) Liver NKT cells were obtained from untreated B6 mice and stimulated with the specific ligand αGalCer after loading into a CD1d-positive cell line A20. Cells were evaluated for synergy of cytokine production with IL-12 and αGalCer at 3 hours. TNFα, IFN-γ, or IL-2 was measured in 3-hour supernatants. Values are representative of 3 experiments.
In vivo synergy
To determine whether the synergy of IL-12 with the NKG2D NKRs examined in this study occurred in vivo, B6 mice were injected intrasplenically (a route that results in rapid migration of tumor cells to the liver, a rich environment for NK and NKT cells) with cells that did or did not express Rae1γ, an NKG2D ligand, and then were injected with IL-12 protein. As shown in Figure 4C, when either the parent tumor line (Baf3), Rae1γ-expressing Baf3, or IL-12 was injected alone, less than 10 pg/mL IFN-γ was detected in the serum. However, when Rae1γ-expressing Baf3 cells were injected with IL-12, significant levels of IFN-γ were detected at 24 hours. These results indicate that the in vivo response to the NKG2D receptor-ligand interaction can be significantly enhanced on exposure to IL-12.
Role of IL-12 in the synergistic response
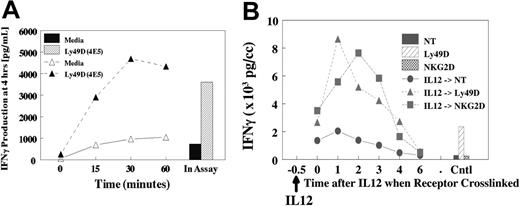
The potent synergy that is observed in vitro and in vivo17 for NKR activation and production of cytokines suggests an important regulatory role for IL-12 in vivo. Therefore, we evaluated whether pretreatment of NK cells with IL-12 (Figure 5A) or IL-18 (not shown) would result in subsequent synergy if NKRs were subsequently ligated. Cells were pretreated with IL-12 for 15 to 60 minutes at 37°C, stimulated on ice with anti-Ly49D (4E5), then evaluated for costimulation of IFN-γ. As shown in Figure 5A, pretreatment for as little at 15 minutes resulted in near-maximal production of IFN-γ at 4 hours compared with IL-12 addition during the assay. Furthermore, the activation of phospho-STAT4 by IL-12 was similar in the presence or absence of cross-linking (not shown). These results suggested that a brief encounter with costimulatory cytokines in vivo is sufficient to result in potent subsequent synergy with NKRs. We next determined how long the cells retained their sensitivity after IL-12 treatment because cells were treated with IL-12 for 30 minutes, followed by delayed NKR cross-linking (Figure 5B). Results demonstrated that synergy with IL-12 was still observed, even if NKR cross-linking was delayed for 2 to 3 hours. However, synergy was lost if cross-linking was delayed for 4 to 6 hours.
We next investigated the role of STAT4 in the synergistic response by comparing the response in BALB/c wild-type (WT) and STAT4 knockout (KO) mice using the NKRs NKG2D and Ly49L (data not shown). All synergy—whether between IL-2 and IL-12 or NKR and IL-12—was ablated in cells from the STAT4 KO mice, demonstrating the obligatory role for STAT4 in priming the cells for a maximal response to NKR cross-linking.
IL-12, IL-23, and IL-27 regulation of ITAM receptors. (A) IL-23 and IL-27 synergy of IFN-γ induction by NK receptors. Enriched NK cells were evaluated for synergy with media, IL-12, IL-23, and IL-27 by addition into the assay for 4 hours after precoating with anti-Ly49D (4E5). Values are representative of 3 experiments. (B) Reversal of the dominant inhibitory signal by IL-12. Highly enriched Ly49G2+ NK cells were expanded for 4 days with IL-2 after selection by antibody-coated magnetic beads. Cells were depleted of CD3, CD19, and CD24, then selected into Ly49G2+ subsets. Cells were greater than 95% Ly49G2+, 52% Ly49D+, 88% NKG2D+, and greater than 95% NK1.1+. NK cells were precoated with NKR antibodies, then cross-linked for 4 hours with or without IL-12. Values are representative of 2 experiments. (C) In vivo evaluation of NKG2D synergy with IL-12. B6 mice were injected intrasplenically with either Baf3- or Rae1γ-expressing Baf3 cells (5 × 105 cells). After 15 minutes, spleens were surgically removed. After 1 hour, mice were injected intraperitoneally with 10 ng IL-12 protein, and serum was collected for indicated times to 48 hours. Serum was evaluated for cytokine production using the CBA TH1/TH2 kit (Becton Dickinson). Values represent mean ± SE, with 5 mice per group.
IL-12, IL-23, and IL-27 regulation of ITAM receptors. (A) IL-23 and IL-27 synergy of IFN-γ induction by NK receptors. Enriched NK cells were evaluated for synergy with media, IL-12, IL-23, and IL-27 by addition into the assay for 4 hours after precoating with anti-Ly49D (4E5). Values are representative of 3 experiments. (B) Reversal of the dominant inhibitory signal by IL-12. Highly enriched Ly49G2+ NK cells were expanded for 4 days with IL-2 after selection by antibody-coated magnetic beads. Cells were depleted of CD3, CD19, and CD24, then selected into Ly49G2+ subsets. Cells were greater than 95% Ly49G2+, 52% Ly49D+, 88% NKG2D+, and greater than 95% NK1.1+. NK cells were precoated with NKR antibodies, then cross-linked for 4 hours with or without IL-12. Values are representative of 2 experiments. (C) In vivo evaluation of NKG2D synergy with IL-12. B6 mice were injected intrasplenically with either Baf3- or Rae1γ-expressing Baf3 cells (5 × 105 cells). After 15 minutes, spleens were surgically removed. After 1 hour, mice were injected intraperitoneally with 10 ng IL-12 protein, and serum was collected for indicated times to 48 hours. Serum was evaluated for cytokine production using the CBA TH1/TH2 kit (Becton Dickinson). Values represent mean ± SE, with 5 mice per group.
Effects on IFN-γ mRNA
To investigate whether a possible mechanism for the observed synergy was stabilization of the IFN-γ mRNA, we analyzed mRNA half-life after treatment alone or in combination (Figure 6A). With all 3 treatments (NKR, IL-12, and combination), the half-life of IFN-γ mRNA was between 1 and 2 hours, indicating that the increased production of IFN-γ was not caused by effects on mRNA stability. Previous studies from our laboratory have demonstrated that IL-12 causes an accumulation of IFN-γ-unprocessed mRNA in the nucleus that is processed and driven out of the nucleus by costimulation with IL-2. Using this model and probes that uniquely recognized unprocessed mRNA (detecting intron/exon boundaries), we examined the effects of costimulation on IFN-γ mRNA processing and cellular compartmentalization. The results (Figures 6B, S2) indicated that stimulation with IL-12 plus Ly49D resulted in a synergistic increase in cytoplasmic IFN-γ mRNA, with a 10- to 25-fold increase over that observed in untreated cells. The examination of nuclear RNA indicated that similar and parallel increases in exon 3/intron 3-, exon 1/intron 1-, exon 3-, and exon 1-containing RNAs occurred, demonstrating that IL-12 plus Ly49D treatment increased nuclear accumulation of unprocessed and processed forms of IFN-γ mRNA. Additionally, nucleocytoplasmic transport of the IFN-γ mRNA was not altered because similar increases in nuclear and cytoplasmic accumulation of IFN-γ mRNA was observed after treatment with IL-12 plus Ly49D.
Pretreatment with IL-12 and response upon NKR activation. (A) Liver lymphocytes were obtained from untreated liver cells from B6 mice. Cells were pretreated with IL-12 at 37°C for specified times. Cells were washed, chilled, and coated with anti-NKRs for 20 minutes. Cells were then washed and cultured for 4 hours. IL-12 was added to the control at the time of assay initiation, and IL-12 remained in assay for 4 hours. (B) Delayed NKR activation. Liver-enriched NK cells (greater than 90% NK1.1+) were pretreated at 37°C with IL-12, then held for specified times, and cells were coated with anti-Ly49D or anti-NKG2D at 4°C for 15 minutes, then washed and cross-linked on plastic dishes coated with antirat or antihamster secondary antibodies. Supernatants were collected at 4 to 5 hours.
Pretreatment with IL-12 and response upon NKR activation. (A) Liver lymphocytes were obtained from untreated liver cells from B6 mice. Cells were pretreated with IL-12 at 37°C for specified times. Cells were washed, chilled, and coated with anti-NKRs for 20 minutes. Cells were then washed and cultured for 4 hours. IL-12 was added to the control at the time of assay initiation, and IL-12 remained in assay for 4 hours. (B) Delayed NKR activation. Liver-enriched NK cells (greater than 90% NK1.1+) were pretreated at 37°C with IL-12, then held for specified times, and cells were coated with anti-Ly49D or anti-NKG2D at 4°C for 15 minutes, then washed and cross-linked on plastic dishes coated with antirat or antihamster secondary antibodies. Supernatants were collected at 4 to 5 hours.
Analysis of the biochemical pathways involved in the synergistic response
Given that previous studies have shown an important role for p38 in the IL-12-signaling pathway, we sought to examine a potential additional role for ERK (p42/p44) in the synergistic responses. Several recent studies20-23 have shown a direct role for DAP12 signaling through PLCγ and ERK. NK cells were stimulated with control immunoglobulin and anti-Ly49D with or without IL-12, and phospho-ERK and p38 were analyzed by flow cytometry. As expected, IL-12 alone markedly increased p38 (shown at 20 minutes; Figure 7A) and marginally increased phospho-ERK (shown at 30 minutes). However, increased phospho-ERK and p38 was observed on antibody cross-linking, and the levels of these phosphor-proteins were increased and sustained with the addition of IL-12 (Figure 7B). Similar results were seen with NKG2D, except that antibody alone was minimally activating for phospho-ERK (data not shown). This result is consistent with our observation that NKG2D cross-linking by itself was insufficient for IFN-γ production. To directly test whether the synergistic response resulted from the activation of both signal transduction pathways, we analyzed the effects of p38 and ERK inhibitors on the combined treatments. As shown in Figure 7C, the synergistic response between NKR cross-linking and IL-12 was completely abrogated by blocking both MAP kinases, whereas either inhibitor alone had only partial effects. As expected, the response to IL-12 alone was blocked by p38 inhibitors. Collectively, these data indicate that synergy between the MAP kinases p38 and ERK is essential for NKR synergy with IL-12.
Discussion
Our previous studies17 with NK cells demonstrate that costimulation of ITAM-bearing Ly49D and IL-12 resulted in an override of inhibitory signals when cells expressing inhibitory and activating receptors are presented with ligand. This synergy is mediated by and dependent on Ly49D-expressing NK cells and resulted in significant systemic expression of IFN-γ. Thus, triggering of the activating Ly-49 receptors initiates microbial, antiviral, and antitumor immunity, and simultaneous treatment with IL-12 provides a mechanism for the release of activating Ly49 receptors from the inhibitory receptor blockade.17 These results have important implications in the biology of NK cells and their receptors because small amounts of inflammatory cytokines such as IL-12 could result in in vivo activation of activating Ly49D receptors when they are present in an inflammatory environment.
Evaluation of mRNA for IFN-γ. (A) IFN-γ message was analyzed. Liver-enriched NK cells (greater than 90% NK1.1+) were left untreated (▵) or coated with anti-Ly49D at 4°C for 15 minutes, washed, and pretreated at 37°C with IL-12. Cells were washed, actinomycin D was added, and cell lysates were evaluated for IFN-γ mRNA. ▴ indicates anti-Ly49D + actinomycin D; ○, IL-12 + actinomycin D; and •, anti-Ly49D + IL-12 + actinomycin D. Values are expressed relative to mRNA level at 30 minutes before actinomycin D addition (100%), and were derived from digital evaluation of RPA for IFN-γ, relative to L32 control RNA. Values are representative of 2 experiments. (B) Nuclear and cytoplasmic mRNA for IFNγ were analyzed. NK cells were lysed and fractionated, as described in “Materials and methods.” RPA was performed on nuclear and cytoplasmic RNAs using [33P]UTP-labeled exon 1/intron 1 (E1-I1), exon 3/intron 3 (E3-I3), and L32 rRNA riboprobes. The exon/intron probes hybridized and protected unspliced nuclear IFN-γ pre-mRNA, whereas the exon portion of the exon/intron probes recognized only the spliced form of the IFN-γ mRNA in the nucleus and the cytoplasm. L32 was used as a control for RNA input between samples. (Results from a representative RPA are shown in Figure S2). Lanes from left to right represent the following: nontreated (NT), LY49D cross-linked (49D), IL-12-activated (IL-12), and LY49D plus IL-12 (D/12) coactivated NK cells. Graph representation of the quantitation performed by ImageQuaNT analysis on the image shown in panel B. Normalized mRNA values corresponded to the accumulation of exon/intron and exon-only IFN-γ mRNA relative to L32 mRNA in the nuclear (top) and cytoplasmic (bottom) compartments.
Evaluation of mRNA for IFN-γ. (A) IFN-γ message was analyzed. Liver-enriched NK cells (greater than 90% NK1.1+) were left untreated (▵) or coated with anti-Ly49D at 4°C for 15 minutes, washed, and pretreated at 37°C with IL-12. Cells were washed, actinomycin D was added, and cell lysates were evaluated for IFN-γ mRNA. ▴ indicates anti-Ly49D + actinomycin D; ○, IL-12 + actinomycin D; and •, anti-Ly49D + IL-12 + actinomycin D. Values are expressed relative to mRNA level at 30 minutes before actinomycin D addition (100%), and were derived from digital evaluation of RPA for IFN-γ, relative to L32 control RNA. Values are representative of 2 experiments. (B) Nuclear and cytoplasmic mRNA for IFNγ were analyzed. NK cells were lysed and fractionated, as described in “Materials and methods.” RPA was performed on nuclear and cytoplasmic RNAs using [33P]UTP-labeled exon 1/intron 1 (E1-I1), exon 3/intron 3 (E3-I3), and L32 rRNA riboprobes. The exon/intron probes hybridized and protected unspliced nuclear IFN-γ pre-mRNA, whereas the exon portion of the exon/intron probes recognized only the spliced form of the IFN-γ mRNA in the nucleus and the cytoplasm. L32 was used as a control for RNA input between samples. (Results from a representative RPA are shown in Figure S2). Lanes from left to right represent the following: nontreated (NT), LY49D cross-linked (49D), IL-12-activated (IL-12), and LY49D plus IL-12 (D/12) coactivated NK cells. Graph representation of the quantitation performed by ImageQuaNT analysis on the image shown in panel B. Normalized mRNA values corresponded to the accumulation of exon/intron and exon-only IFN-γ mRNA relative to L32 mRNA in the nuclear (top) and cytoplasmic (bottom) compartments.
In the present study, we demonstrated that other innate and adaptive cells that express ITAM-bearing receptors also responded to IL-12 through enhanced expression of IFN-γ. In NK cells, we demonstrated here that Ly49H (a DAP12-associated receptor), NKRp1, (a TCRζ-associated receptor), and NKG2D (a DAP10- and DAP12-associated receptor) demonstrated potent synergy with IL-12 and IL-18 in the induction of IFN-γ, both alone and in the presence of an inhibitory ITIM signal. In addition, we have found that costimulation with IL-12 results in greatly enhanced IFN-γ production on cross-linking of T cells and NKT cells with CD3 (a TCRζ-associated receptor) or αGalCer. The IL-12 signaling is STAT4 dependent and results in higher levels of total IFN-γ mRNA. However, we did not observe an increase in the IFN-γ mRNA half-life or transport of the mRNA from the nucleus to the cytoplasm. Another interesting aspect of this synergy was the restricted association with expression of the TH1 cytokine, IFN-γ. IFN-γ KO mice demonstrated only minimal costimulation response with respect to IL-13 and MIP1α expression when activating NKRs were cross-linked. Similarly, with T cells, the cellular response to IL-12 costimulation was restricted to IFN-γ and was not observed for TNFα or IL-2. Furthermore, our data demonstrated that the synergy observed required activation through the p38 and the ERK pathways and demonstrated how these pathways converged at a single point (ie, enhanced expression of the Ifng gene).
Analysis of MAP kinases. NK cells stimulated through their NKRs in the presence or absence of IL-12. (A) Flow cytometry analysis of phospho-ERK or p38 when cells were unstimulated (shaded histogram) or stimulated through Ly49D alone (dotted histogram) or with IL12 (heavy-line histogram). (B) Kinetic changes in phospho-ERK and phopho-p38 in cells stimulated with IL-12 only (▵), with anti-Ly49D (○) alone, or in combination with IL12 (•). (C) IFN-γ production from fresh liver NK cells 16 hours after stimulation with IL-12, NKR, or their combination. Cells were stimulated after no treatment (NT; ▪) or in the presence of the p38 inhibitor (SB203580,  ), the ERK inhibitor (U0126,
), the ERK inhibitor (U0126,  ), or both (
), or both ( ).
).
Analysis of MAP kinases. NK cells stimulated through their NKRs in the presence or absence of IL-12. (A) Flow cytometry analysis of phospho-ERK or p38 when cells were unstimulated (shaded histogram) or stimulated through Ly49D alone (dotted histogram) or with IL12 (heavy-line histogram). (B) Kinetic changes in phospho-ERK and phopho-p38 in cells stimulated with IL-12 only (▵), with anti-Ly49D (○) alone, or in combination with IL12 (•). (C) IFN-γ production from fresh liver NK cells 16 hours after stimulation with IL-12, NKR, or their combination. Cells were stimulated after no treatment (NT; ▪) or in the presence of the p38 inhibitor (SB203580,  ), the ERK inhibitor (U0126,
), the ERK inhibitor (U0126,  ), or both (
), or both ( ).
).
The synergy observed with NKG2D, previously reported to be a DAP10-associated receptor9,18 in primary NK cells, could be attributed to a low frequency of DAP12-associated NKG2D receptors. This is an important point. As a result of our analysis, we determined that the reported methods to analyze mRNA expression of these 2 forms were confusing.18 As demonstrated in the supplemental data (Figure S1), the in vivo activation of NK cells with IL-2 resulted in a dramatic shift in DAP12 association of NKG2D, consistent with previous reports.18 Thus, the expression of IFN-γ in response to NKG2D cross-linking was dependent on association with DAP12.
We believe that our data reflect conditions relevant to the in vivo host response because only short (15-minutes) pretreatment with IL-12 can result in maximal activation of the synergy with cross-linking of the ITAM-bearing receptors. These data indicate that new protein synthesis is not required after IL-12 treatment and that posttranscriptional modifications of proteins, including transcription factors, may be a result of brief IL-12 treatment. This may result in enhanced recruitment of AP-1 to the IFN-γ promoter, as has been reported in in vitro models.24 Consistent with this model is the fact that Ly49 cross-linking does result in increased AP-1 activity (D.W. McVicar, personal communication, January 2005). In addition, NKR activation kinetics (Figure 5B) indicated that once lymphocytes were activated with IL-12, they remained sensitive to cross-linking for up to 3 hours. This has important in vivo implications because cells that are in an inflammatory environment can receive costimulation in a number of ways, but the outcome would be release from inhibitory signals as long as cytokine and ITAM receptor signals are received in a proximal time frame. In addition, our demonstration that multiple combinations of IL-12, IL-18, and ITAM receptor signal experiments can result in an optimal response when using picogram levels of cytokines, reflects the in vivo condition in which IL-12 and IL-18 might be simultaneously or sequentially released from effector cells such as macrophages. Finally, the in vivo experiments with tumor cells expressing Rae1γ substantiate our in vitro data and indicate that synergy with cytokines such as IL-12 can occur in inflammatory in vivo environments.
These studies have provided novel results regarding the in vivo function of all potential ITAM-bearing receptors. Our data support the contention that the in vivo regulation of inhibitory and activating receptors occurs not only in NK cells but also in T cells and NKT cells and can be coregulated by stimulation with inflammatory cytokines, including IL-12, IL-23, IL-27, and IL-18. These results are consistent with a model whereby the immune system's ability to regulate innate and adaptive responses is to maximize the IFN-γ response when multiple “warning” signals are received in inflammatory sites.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-04-1579.
Supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
J.R.O. and H.A.Y. designed the experiments, analyzed the data, and wrote the manuscript; J.W. provided new vital reagents; and R.W.-P., M.H., E.W.B., A.T.M., N.B., J.C., M.S., and D.L.H. performed the research.
The publisher or recipient acknowledges the right of the US government to retain a nonexclusive, royalty-free license in and to any copyright covering the article.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Tim Back, John Wine, and Erin Lincoln for their support in animal care and experimentation. We thank Jeff Subleski for his assistance in the generation of mouse-specific RPA probes for IFN-γ. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.



![Figure 6. Evaluation of mRNA for IFN-γ. (A) IFN-γ message was analyzed. Liver-enriched NK cells (greater than 90% NK1.1+) were left untreated (▵) or coated with anti-Ly49D at 4°C for 15 minutes, washed, and pretreated at 37°C with IL-12. Cells were washed, actinomycin D was added, and cell lysates were evaluated for IFN-γ mRNA. ▴ indicates anti-Ly49D + actinomycin D; ○, IL-12 + actinomycin D; and •, anti-Ly49D + IL-12 + actinomycin D. Values are expressed relative to mRNA level at 30 minutes before actinomycin D addition (100%), and were derived from digital evaluation of RPA for IFN-γ, relative to L32 control RNA. Values are representative of 2 experiments. (B) Nuclear and cytoplasmic mRNA for IFNγ were analyzed. NK cells were lysed and fractionated, as described in “Materials and methods.” RPA was performed on nuclear and cytoplasmic RNAs using [33P]UTP-labeled exon 1/intron 1 (E1-I1), exon 3/intron 3 (E3-I3), and L32 rRNA riboprobes. The exon/intron probes hybridized and protected unspliced nuclear IFN-γ pre-mRNA, whereas the exon portion of the exon/intron probes recognized only the spliced form of the IFN-γ mRNA in the nucleus and the cytoplasm. L32 was used as a control for RNA input between samples. (Results from a representative RPA are shown in Figure S2). Lanes from left to right represent the following: nontreated (NT), LY49D cross-linked (49D), IL-12-activated (IL-12), and LY49D plus IL-12 (D/12) coactivated NK cells. Graph representation of the quantitation performed by ImageQuaNT analysis on the image shown in panel B. Normalized mRNA values corresponded to the accumulation of exon/intron and exon-only IFN-γ mRNA relative to L32 mRNA in the nuclear (top) and cytoplasmic (bottom) compartments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/4/10.1182_blood-2005-04-1579/2/m_zh80040691180006.jpeg?Expires=1763658723&Signature=uGtefFV4Nua5n~7FxzTVhf~pUMEyIo~nG8HzNIu1BzFANqhFcB5qJHGrukoh-xvy0q4zCpN3kcd7ZDJQqhFYZf9eVOY3oi0iSiBLvpo3w78RdrkpJaUyAY73gM3i8KywHMl99TzLzCx1ZpCpQnzfnzXrZetsIAzFodg9Tu85TxTy6ayLnAfxClVfBy~3w4kS0GH518SI9xXh9sGnlJyKwCXx2jrbyIKtRhXM0wt8iBPb0cZ9f9fWfmBydS-m2ZwSZJlpWPPvr3lMIZ8iwcP5VSYctu1CZIkky1h9arYKqnHxy5LJxcR2piFhwMKiMUWtf5uwkK8SGqEwKYqzmBMO2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)







![Figure 6. Evaluation of mRNA for IFN-γ. (A) IFN-γ message was analyzed. Liver-enriched NK cells (greater than 90% NK1.1+) were left untreated (▵) or coated with anti-Ly49D at 4°C for 15 minutes, washed, and pretreated at 37°C with IL-12. Cells were washed, actinomycin D was added, and cell lysates were evaluated for IFN-γ mRNA. ▴ indicates anti-Ly49D + actinomycin D; ○, IL-12 + actinomycin D; and •, anti-Ly49D + IL-12 + actinomycin D. Values are expressed relative to mRNA level at 30 minutes before actinomycin D addition (100%), and were derived from digital evaluation of RPA for IFN-γ, relative to L32 control RNA. Values are representative of 2 experiments. (B) Nuclear and cytoplasmic mRNA for IFNγ were analyzed. NK cells were lysed and fractionated, as described in “Materials and methods.” RPA was performed on nuclear and cytoplasmic RNAs using [33P]UTP-labeled exon 1/intron 1 (E1-I1), exon 3/intron 3 (E3-I3), and L32 rRNA riboprobes. The exon/intron probes hybridized and protected unspliced nuclear IFN-γ pre-mRNA, whereas the exon portion of the exon/intron probes recognized only the spliced form of the IFN-γ mRNA in the nucleus and the cytoplasm. L32 was used as a control for RNA input between samples. (Results from a representative RPA are shown in Figure S2). Lanes from left to right represent the following: nontreated (NT), LY49D cross-linked (49D), IL-12-activated (IL-12), and LY49D plus IL-12 (D/12) coactivated NK cells. Graph representation of the quantitation performed by ImageQuaNT analysis on the image shown in panel B. Normalized mRNA values corresponded to the accumulation of exon/intron and exon-only IFN-γ mRNA relative to L32 mRNA in the nuclear (top) and cytoplasmic (bottom) compartments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/4/10.1182_blood-2005-04-1579/2/m_zh80040691180006.jpeg?Expires=1764334684&Signature=bR-1BDrIIxscBDPmeRmCPQLLfLbWLA6H-ldDr4y3bVEi6sLSasjhhnwxmVcfwg9INM38TSWeeRua7RwOaEaqkjZ88D9TJzI3rMrRwId4BI6~1e~wyu7ZLHQdsIMeHF9ZN9VI-5pEOFk2jZzejwqYF2ZMwFSa4buwQWx7wsity6fUwzON~Nfz1xgaUQIzVM9IBFIq6ar49L81pPPmi8wGiwUlBDAVxHGFtnA9tVQ1lLkLBoqAoyEbEECJIOgxtVROR~0qdhhPTREehWGs0lu6NaSsQ6k8Y1PvgtXVqmGFDfc8sB59DibwCUVbs8U34xLvtNijplxKNwFkcA1Bw5jpFw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

 ), the ERK inhibitor (U0126,
), the ERK inhibitor (U0126,  ), or both (
), or both ( ).
).