Abstract
The primary infection of Epstein-Barr virus (EBV) may result in fatal infectious mononucleosis or hemophagocytic syndrome (HPS) in 2 diseases; that is, X-linked lymphoproliferative disorder (XLP) and hemophagocytic lymphohistiocytosis (HLH). XLP is linked to mutations of the SAP/SH2D1A gene with dysregulated T-cell activation in response to EBV infection. Patients with sporadic HLH, however, usually have no mutation of the SAP/SH2D1A gene, and EBV latent membrane protein-1 (LMP1) can up-regulate Th1 cytokines in EBV-infected T cells. Since both diseases share common manifestations of HPS, it is important to clarify whether a cross-talk exists between signaling lymphocyte activation molecule (SLAM)–associated protein (SAP) and LMP1-mediated pathways to explain the common pathogenesis of HPS. In this study, no mutation of the SAP/SH2D1A gene at exon 2/3 was detected in 7 HLH cases. Interestingly, EBV LMP1 could transcriptionally inhibit the expression of SAP/SH2D1A and activate downstream molecules ERK and interferon-γ (IFN-γ). LMP1-mediated SAP/ERK/IFN-γ signals appear to act via the TNF receptor–associated factor (TRAF)2,5/nuclear factor κB (NF-κB) pathway, since dominantnegative TRAF2/5 and NF-κB inhibitor could rescue SAP expression and downregulate IFN-γ. Although HLH is genetically distinct from XLP, our data suggest that both diseases share a common signal pathway, through either the mutation or LMP1-mediated suppression of the SAP gene, leading to overt T-cell activation and enhanced Th1 cytokine secretion in response to EBV infection.
Introduction
The Epstein-Barr virus (EBV) can infect B cells, epithelial cells, and T or natural killer (NK) cells, resulting in a spectrum of benign and malignant human diseases.1-4 Although primary EBV infections usually run an uneventful course, a fatal form of infectious mononucleosis (IM) or hemophagocytic syndrome (HPS) characterized by hepatosplenomegaly, coagulopathy, pancytopenia, hypercytokinemia, and a systemic proliferation of macrophages with hemophagocytosis may develop in young children.5-9 Among EBV-associated HPS, 2 distinct disease entities, the X-linked lymphoproliferative disorder (XLP)10 and a sporadic-type hemophagocytic lymphohistiocytosis (HLH),2 are of particular interest since they share common clinical phenotype and pathologic features of HPS.
XLP is a genetic disorder that affects young boys and is characterized by an inappropriate immune response to primary EBV infection.11 A few children may survive the initial episode of IM, but later develop dysgammaglobulinemia or progress to Burkitt lymphoma.10,12 Although the exact pathobiology of XLP remains to be established, it has been proposed to result from an excessive or dysregulated T-cell immune response to EBV-infected B lymphocytes.13-16 Recently, the majority of patients with XLP were demonstrated to have mutations of an SH2D1A gene that encodes a signaling lymphocyte activation molecule (SLAM)–associated protein (SAP).17-19 SLAM is a member of the immunoglobulin superfamily that is expressed on T and B cells and is capable of homotypic interaction. The antibody-mediated ligation of SLAM may induce T-cell activation and enhance Th1 cytokine production.20,21 SAP may physically associate with SLAM via the Src homology 2 (SH2) domain and may displace the SH2 domain–bearing proteins such as SHP-2 from SLAM.22-24 By this sense, SAP acts as a natural suppressor or modulator of the SH2 domain–mediated interactions in SLAM-mediated T-cell activation signals.25 The mutation or deletion of the SAP gene will result in absence or nonfunctioning of SAP protein and lead to abnormal T-cell activation and enhanced Th1 cytokine production as demonstrated in patients with XLP.20,25,26
The pathomechanism of sporadic-type HPS or HLH, however, is basically distinct from XLP in the lack of SAP/SH2D1A gene mutation and gender predilection.27,28 Furthermore, EBV directly infects T or NK cells in HLH, rather than the B cells in XLP.29-32 The EBV latent membrane protein-1 (LMP1) has been reported to activate T cells, up-regulate tumor necrosis factoralpha (TNF-α) and interferon-gamma (IFN-γ), and in turn activate macrophages and lead to an enhanced cytokine storm and hemophagocytosis.16,33,34
Albeit there exist distinct differences for these 2 disease entities as described above, they share common manifestations of dysregulated T-cell activation, enhanced Th1 cytokine secretion, and the subsequent development of fatal HPS. Therefore, the potential involvement of the SAP/SH2D1A molecule in the LMP1-mediated signal pathway in HLH deserves to be elucidated. This study was therefore designed to clarify the potential role of SAP/SH2D1A in the pathomechanism of HLH in EBV-infected T cells. Interestingly, we demonstrated that EBV LMP1 can inhibit the expression of SAP/SH2D1A at the transcriptional level. Tissue samples obtained from 7 Taiwanese patients with HLH were evaluated for the mutation studies of the SAP/SH2D1A gene. Since EBV LMP1 has been demonstrated to exert its function through the aggregation of cytoplasmic c-terminal domains, carboxyl terminal activating region 1 (CTAR1) and CTAR2, to recruit TNF receptor–associated factor (TRAF) molecules in B and epithelial cells, resulting in the activation of transcriptional factors,35,36 the potential involvement of TRAF/NF-κB in LMP1-mediated SAP signaling in T cells was studied.
Patients, materials, and methods
Patients and tissue samples
A total of 35 cases of childhood HPS were encountered in National Taiwan University Hospital and National Cheng Kung University Hospital during the period of 1991 to 2002.37,38 Tissues or freshly frozen cells were available for DNA and RNA extraction in 7 cases, including lymph nodes in 2 cases, spleen in 1 case, skin in 1 case, and bone marrow in 3 cases. As controls, 10 specimens of reactive lymphoid hyperplasia (5 cases), IM (2 cases), and nasal type NK/T-cell lymphoma (3 cases) were included. Informed consent was received from the patients according to the Declaration of Helsinki. Approval for these studies was obtained from the National Health Research Institutes (NHRI), Taiwan.
PCR amplification and reverse-transcriptase–polymerase chain reaction analysis of the SAP/SH2D1A gene
In order to detect the mutations on the SAP/SH2D1A gene, we analyzed sequences of both genomic DNA and SAP gene transcripts. For detecting SAP gene mutation on the exon 2/3 as demonstrated previously on cases of XLP,17 the genomic DNA of 7 HLH cases was extracted for polymerase chain reaction (PCR) and automatic sequencing with the same primer sets described previously.17
For detecting the gene expression of SAP in tissues and cells, the gene transcripts of SAP were detected by reverse transcriptase (RT)–PCR. For reverse transcription, total RNA samples were extracted by TRIzol (Invitrogen, Carlsbad, CA) and were reversely transcripted by Superscript II (Invitrogen). The resulted cDNA products were next amplified by PCR with primer sets listed as follows: SAP forward 5′-CGGGATCCAGGCCATGGACGCAGTG-3′; SAP reverse, 5′-CGGGATCCTCATGGGGCTTTCAGGCAGAC-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GAAGGTGAAGGTCGGAGTCAA-3′; GAPDH reverse, 5′-GCAGAGGGGGCAGAGATGAT-3′. The resulting products were resolved on a 2% agarose gel and quantitated by a densitometer.
Cell lines and transfection
In this study, 2 EBV-negative T-cell lines (H9 and Jurkat) and one B-cell line (BJAB) were used. All these cell lines were maintained in RPMI-1640 medium (JRH Biosciences, Lenexa, NY) supplemented with 10% (vol/vol) fetal bovine serum (FBS; JRH Biosciences) and penicillin/streptomycin. For the establishment of stable cell lines expressing LMP1 and TRAF2/5 dominant mutants, cells were first transfected with pSG5-LMP1 or TRAF2/5 mutant constructs by electrophoration (Bio-Rad, Hercules, CA). For each electrophoration, a total of 2 × 106 cells and 7.5 μg DNA were used. The constructs containing dominant-negative mutants of TRAF2/5 genes were kindly provided by Professor Toshiki Watanabe of the University of Tokyo, Japan.39 The stable clones were obtained under the selection of neomycin (800 μg/mL) starting from 48 hours after transfection. The concentrations of neomycin were reduced to 400 μg/mL for the maintenance of stable clones.
To restore the expression of SAP, a SAP plasmid was constructed and expressed in a stable virus-producing mouse fibroblast cell line, PT67. The PT67 cells were transfected with retroviral plasmids by LipofectAMINE (Invitrogen). The SAP expression construct was generated by subcloning the previously amplified Jurkat SAP cDNA products into BamHI cutting site of the retroviral vector pRCMV-Hyg. Retrovirus-mediated SAP gene transduction was performed on the established LMP1-H9 cells by coincubating the LMP1-H9 cells with SAP gene–containing PT67 cells. The SAP-transduced LMP1-H9 cells were then collected after 48 hours of incubation and maintained under the selection of 40 μg/mL Hygromycin (Sigma, St Louis, MO).
Extraction of cytoplasmic and nuclear proteins
To study the protein expression in the cytoplasmic and nuclear fractions, 106 cells were added to 50 μL buffer A containing 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)–KOH (potassium hydroxide), 1.5 mM MgCl2, and 10 mM KCl. After incubating on ice for 10 minutes, the mixtures were spun down at 9 500 g for 10 seconds. The supernatant fraction after the centrifugation represents the cytoplasmic protein lysate. The pellets were next resuspended with 100 μL buffer B containing 20 mM HEPES-KOH, 1.5 mM MgCl2, 0.2 mM EDTA (ethylenediaminetetraacetic acid), and 25% glycerol. The mixtures were incubated on ice for 20 minutes and spun down at 9 500 g for 2 minutes, and the supernatants represent the nuclear protein fractions. α-actin and histone H1 were used as the controls for correct preparations of the cytoplasmic and nuclear fraction proteins, respectively.
Antibodies and reagents
For Western blotting, the anti-LMP1 mouse monoclonal antibody (CS1-4; Dako, Glostrup, Denmark), and anti-SAP, anti-TRAF1, anti-TRAF2, anti-TRAF5, anti–NF-κB p65, anti–α-actin, anti-ERK, anti-pERK, goat anti-SLAM (Santa Cruz Biotechnology, Santa Cruz, CA), and anti–Histone H1 (Upstate, Waltham, MA) were used. For immunoprecipitation of SLAM, the mouse monoclonal anti-SLAM (R&D Systems, Minneapolis, MN) and protein G PLUS-agarose (Oncogene, San Diego, CA) were used. For inhibition of NF-κB activity on LMP1-H9 cells, BAY 11-7082 was used (Sigma).
Immunofluorescence staining
H9 cells were first washed twice in Tris-buffer saline (TBS) with 2% bovine serum albumin (BSA), transferred on glass slides by cytospinning, and then fixed with 100% methanol for 10 minutes. After fixation, cells were washed 3 times in TBS. Samples were first incubated with a mixture of anti-LMP1 and anti-TRAF2 or anti-TRAF5 antibodies at the concentration of 5 μg/mL for each. After incubation with fluorescence-labeled secondary antibodies for 30 minutes at room temperature, samples were washed 3 times with TBS and covered with a FluoroGuard antifade reagent (Bio-Rad). Fluorescence signals were detected under confocal microscopy (Radiance 2000; Bio-Rad).
NF-κB promoter reporter assays
To evaluate NF-κB activity, a reporter assay using an NF-κB site-dependent luciferase vector p6x κB-Luc was performed as previously described.40 Luciferase expression vector driven by the herpes simplex virus thymidine kinase promoter (pRL-TK; Promega, Madison, WI) was cotransfected to standardize each experiment. For each transfection, 5 μg p6x κB-Luc, 2 μg pRL-TK, and 5 μg of the indicated expression vector were used. Luciferase activity was measured by Dual Luciferase assay kit (Promega).
Enzyme-linked immunosorbent assay
In order to examine the effects of SAP and dominant-negative TRAF2/5 on IFN-γ secretion, secreted IFN-γ cytokine levels in culture supernatants of H9 cells were measured by IFN-γ enzyme-linked immunosorbent assay (ELISA) kit (Biosource International, Camarillo, CA) as described previously.33 Culture supernatants were harvested by filtration through a 0.45 μm pore size filter (Costar, Cambridge, MA) to exclude cell debris.
Results
No detectable mutations on exon 2/3 of the SAP/SH2D1A gene in 7 patients with sporadic HLH
The pertinent clinical and laboratory data of 7 HLH cases (4 male and 3 female patients; mean age, 16.7 years) are summarized in Table 1. Three patients responded to the therapy using a protocol of intravenous immunoglobulin (IVIG)/VP-16,38 2 had chronic active diseases, and 2 had rapidly progressive diseases.
Sequence analyses of the SAP/SH2D1A gene and its transcripts on HLH cases revealed no mutation on exons 2/3, nor on those of the control specimens.
LMP1 suppressed the expression of the SAP gene at the transcriptional level in T-cell lines
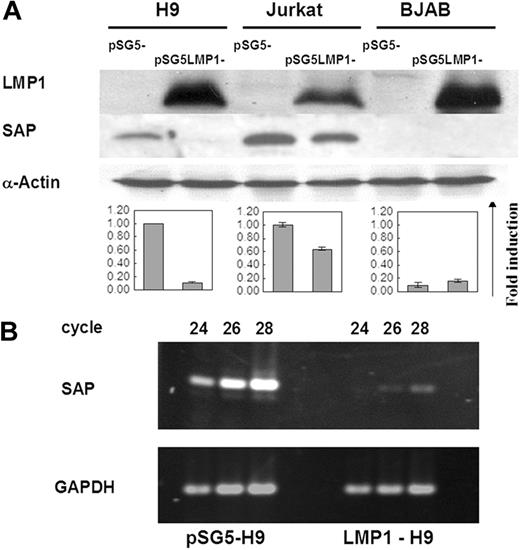
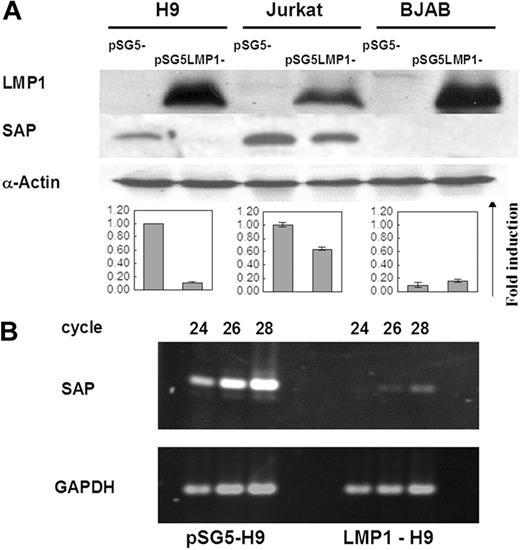
To determine whether LMP1 can regulate the expression of the SAP gene in T- and B-cell lines, we examined the SAP gene expression in LMP1 transfectants by Western blotting. As previously reported,24 SAP protein was not expressed in the B-cell line BJAB, and was not affected by LMP1 expression on the LMP1-BJAB line. The 2 T-cell lines H9 and Jurkat, however, showed endogenous SAP protein expression. Induction of LMP1 expression on these 2 T-cell lines revealed inhibitory effects on the SAP expression, reduced by 90% on LMP1-H9 and 40% on LMP1-Jurkat cells (Figure 1A). In order to clarify whether the regulation of the SAP gene by LMP1 was at the transcriptional or translational level, the transcripts of SAP in H9 and LMP1-H9 cells were further evaluated by RT-PCR analysis. The RNA levels of the SAP gene on the LMP1-H9 cells were reduced 16-fold (Figure 1B). These results indicate that LMP1 could suppress SAP gene expression on T cells at the transcriptional level.
TRAF2/5 signals were involved in LMP1-mediated suppression of the SAP gene on T-cell lines
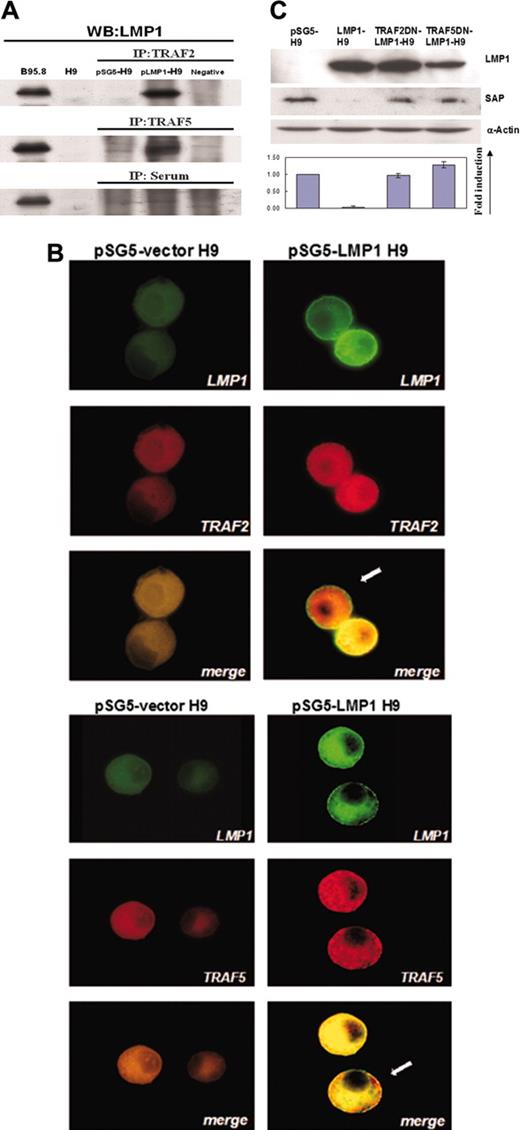
TRAF signals have been demonstrated to be essential for the LMP1-mediated signaling in B cells and epithelial cells.41 To investigate whether TRAF signals were also involved in the LMP1-mediated SAP suppression in T cells, we first tested the potential interaction between LMP1 and TRAF proteins on LMP1-H9 cells. With immunoprecipitation using antibodies against TRAF1, TRAF2, and TRAF5, the interaction was examined by immunoblotting with antibody against LMP1. On LMP1-H9 cells, LMP1 interacted with both TRAF2 and TRAF5 (Figure 2A), but not TRAF1 (data not shown).
Suppression of SAP expression by LMP1 in T cell lines. (A) Protein expression of SAP in 2 T-cell lines (H9 and Jurkat) and 1 B-cell line (BJAB) transfected with pSG5-LMP1 constructs. SAP protein could be detected on both H9 and Jurkat cells, but not on BJAB cells. The expression of SAP was strongly reduced on LMP1-H9 cells and mildly decreased on LMP1-Jurkat cells. Cells transfected with the pSG5 construct were used as vector control groups. The quantitative results of SAP expression adjusted with α-actin are shown below the blots. Error bars representing the standard deviation of 3 independent experiments are shown. (B) The mRNA expression of SAP in H9 cells. SAP gene transcripts on H9 and LMP1-H9 cells were detected by RT-PCR, and products collected at different PCR cycles were resolved on the agarose gels for quantitation.
Suppression of SAP expression by LMP1 in T cell lines. (A) Protein expression of SAP in 2 T-cell lines (H9 and Jurkat) and 1 B-cell line (BJAB) transfected with pSG5-LMP1 constructs. SAP protein could be detected on both H9 and Jurkat cells, but not on BJAB cells. The expression of SAP was strongly reduced on LMP1-H9 cells and mildly decreased on LMP1-Jurkat cells. Cells transfected with the pSG5 construct were used as vector control groups. The quantitative results of SAP expression adjusted with α-actin are shown below the blots. Error bars representing the standard deviation of 3 independent experiments are shown. (B) The mRNA expression of SAP in H9 cells. SAP gene transcripts on H9 and LMP1-H9 cells were detected by RT-PCR, and products collected at different PCR cycles were resolved on the agarose gels for quantitation.
To further confirm the recruitment of TRAF2/5 proteins by LMP1 protein on LMP1-H9 cells, the intracellular localization of TRAF2/5 and LMP1 proteins was examined with immunofluorescence microscopy. As compared with pSG5-H9 cells, the expression levels of TRAF2/5 were increased on LMP1-H9 cells, and TRAF2/5 was observed to colocalize with LMP1 (Figure 2B).
To further confirm the role of TRAF2/5 in the LMP1-mediated suppression of SAP, 2 dominant-negative mutants of TRAF2/5 were used. These dominant-negative mutants contained TRAF2/5 genes either without the C-terminal binding region (pTRAF2-DN) or without the cytoplasmic region (pTRAF5-DN), both regions of which were essential for LMP1-mediated signal transduction. When H9 cells were transfected with TRAF2/5 dominant-negative mutants, the suppression of SAP by LMP1 was reversed on LMP1-H9 cells (Figure 2C), indicating the involvement of TRAF2/5 in LMP1-mediated suppression of SAP.
Involvement of NF-κB in LMP1-mediated suppression of SAP in H9 T cells
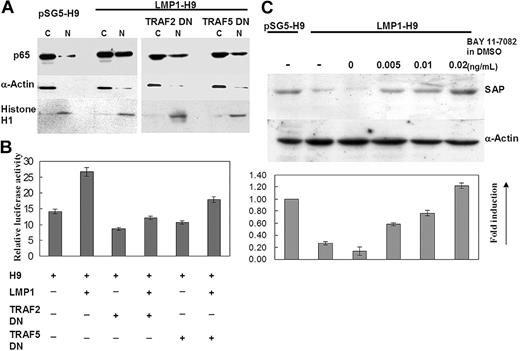
Many reports have shown that the signaling of LMP1/TRAF2/5 involves NF-κB activation in B cells and epithelial cells.35,42 Therefore, we examined NF-κB activation in the LMP1-mediated suppression of SAP in H9 T cells. NF-κB activation was first evaluated by detecting the nuclear translocation of p65Rel protein on LMP1-H9 cells and then by examining the NF-κB–driven luciferase activity. As compared with parental H9 cells, translocated nuclear NF-κB p65 was significantly enhanced by LMP1 expression (Figure 3A). The dominant-negative TRAF2/5 mutants showed mild to moderate inhibitory effects on NF-κB nuclear translocation. The same results were also observed on the reporter assays (Figure 3B).
Involvement of TRAF2/5 in LMP1-mediated suppression of SAP on H9 cells. (A) The association between TRAF2/5 and LMP1 on H9 cells. H9 cells were transfected with either pSG5 or pSG5-LMP1 constructs. Cell lysates were immunoprecipitated with antibodies against TRAF2 or TRAF5. The immunoprecipitates were examined by immunoblot with LMP1 antibody. (B) Double immunofluorescence staining revealed colocalization of TRAF2/5 and LMP1 on H9 T cells. Cells were immunostained by antibodies of TRAF2/5 and LMP1, and were observed under confocal microscopy. In LMP1-H9 cells, aggregation and colocalization of TRAF5 or TRAF2 with LMP1 proteins near the cell membrane are shown as indicated by arrows. Original magnification: × 1000 (objective lens: 200 ×). Cells were imaged under Bio-Rad MRC-100 confocal laser scanning (Bio-Rad Microscience Division, Cambridge, MA). The system was connected to a Nikon Diaphot 200 inverted microscope (Nikon, Melville, NY). Images were collected using COMOS software (Bio-Rad) and assembled for publication using Adobe Photoshop 6.0 software (Adobe Systems, San Jose, CA). (C) Recovery of SAP expression on LMP1-H9 cells by TRAF2/5 dominant (DN) mutants. H9 cells were transfected with pSG5 and pSG5-LMP1 along with TRAF2/5 dominant-negative mutant constructs. The expressions of SAP protein were reduced by LMP1 and this inhibitory effect could be rescued by cotransfection with TRAF2/5 dominant-negative mutants. Error bars representing the standard deviation of 3 independent experiments are shown.
Involvement of TRAF2/5 in LMP1-mediated suppression of SAP on H9 cells. (A) The association between TRAF2/5 and LMP1 on H9 cells. H9 cells were transfected with either pSG5 or pSG5-LMP1 constructs. Cell lysates were immunoprecipitated with antibodies against TRAF2 or TRAF5. The immunoprecipitates were examined by immunoblot with LMP1 antibody. (B) Double immunofluorescence staining revealed colocalization of TRAF2/5 and LMP1 on H9 T cells. Cells were immunostained by antibodies of TRAF2/5 and LMP1, and were observed under confocal microscopy. In LMP1-H9 cells, aggregation and colocalization of TRAF5 or TRAF2 with LMP1 proteins near the cell membrane are shown as indicated by arrows. Original magnification: × 1000 (objective lens: 200 ×). Cells were imaged under Bio-Rad MRC-100 confocal laser scanning (Bio-Rad Microscience Division, Cambridge, MA). The system was connected to a Nikon Diaphot 200 inverted microscope (Nikon, Melville, NY). Images were collected using COMOS software (Bio-Rad) and assembled for publication using Adobe Photoshop 6.0 software (Adobe Systems, San Jose, CA). (C) Recovery of SAP expression on LMP1-H9 cells by TRAF2/5 dominant (DN) mutants. H9 cells were transfected with pSG5 and pSG5-LMP1 along with TRAF2/5 dominant-negative mutant constructs. The expressions of SAP protein were reduced by LMP1 and this inhibitory effect could be rescued by cotransfection with TRAF2/5 dominant-negative mutants. Error bars representing the standard deviation of 3 independent experiments are shown.
To further determine the role of NF-κB in the LMP1-mediated inhibition of SAP expression, NF-κB inhibitor BAY 11-7082 was added and the SAP expression was observed in LMP1-H9 cells. The suppression of SAP by LMP1 could be reversed in a dose-dependent manner (Figure 3C).
The SLAM downstream molecules ERK and IFN-γ were activated and induced by LMP1-mediated SAP suppression
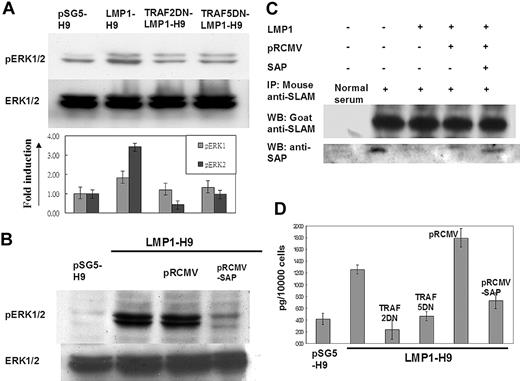
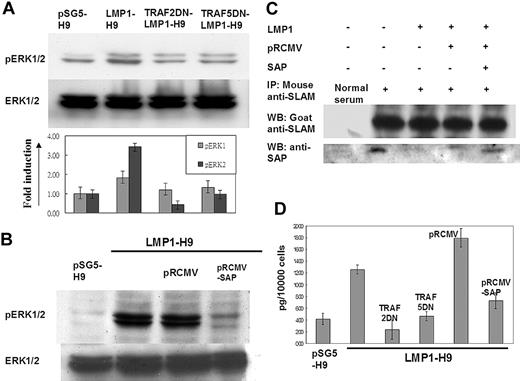
As a physiologic inhibitor or adaptor of SLAM, mutations on the SAP gene as demonstrated in XLP have been shown to lose their ability to inhibit SLAM-mediated ERK/IFN-γ signals.21,43 In order to further confirm the involvement of LMP1 in SLAM/SAP signaling, the downstream molecules ERK and IFN-γ were evaluated. On H9 cells, LMP1 expression enhanced the phosphorylation of ERK1/2. This induction of ERK1/2 phosphorylation could be reduced by cotransfection with TRAF2/5 dominant-negative mutants in LMP1-H9 cells (Figure 4A). In addition, reconstitution of SAP expression by retrovirus gene transduction could reverse the ERK1/2 phosphorylation induced by LMP1 (Figure 4B).
The inhibitory effect of SAP on SLAM-associated signals was mediated by its direct binding on the C-terminus of SLAM.18,44 We next clarified the potential interaction between SLAM and SAP on H9 cells by immunoprecipitation. On H9 cells, SAP was shown to be associated with SLAM protein (Figure 4C). Without affecting the SLAM protein level, the SAP proteins associated with SLAM were markedly reduced on the LMP1-H9 cells. Furthermore, reconstitution of SAP protein expression on LMP1-H9 cells could increase SLAM-bound SAP proteins, suggesting that the reduced binding of SLAM-SAP on LMP1-H9 cells was due to the reduced SAP expression but not the interference of protein-protein interaction (Figure 4C).
In order to understand the effects of SAP signals on the physiologic function of H9 T cells under LMP1 expression, the downstream cytokine IFN-γ was measured by ELISA. The secretion of IFN-γ could be enhanced by LMP1 expression and reversed by the reconstitution of the SAP gene and TRAF2/5 dominantnegative mutants (Figure 4D).
Discussion
For many decades, clinicians and investigators have been puzzled by the differential diagnosis between XLP and sporadic-type HLH when encountered in a young boy presenting with EBV-associated HPS. The recent discovery of the linkage of XLP to the mutations of the SAP/SH2D1A gene provides a major breakthrough in this regard.17,45 However, there still remains a substantial population of HPS cases, either with or without EBV association, which show no detectable mutation on the SAP/SH2D1A gene, as demonstrated in our cases and cases from other reports.28 Given that EBV infects T or NK cells in sporadic HLH, instead of the B cells in XLP, we initially speculated that these 2 diseases may adopt different mechanisms to respond to primary EBV infection. In this study, we demonstrate that the EBV LMP1 protein could effectively suppress the expression of the SAP/SH2D1A gene in T cells, as previously shown in Burkitt lymphoma cells,46 at the transcriptional level, and thereby both XLP and sporadic HLH share a common signal pathway, through either the mutation or the LMP1-mediated suppression of the SAP/SH2D1A gene, finally leading to overt T-cell activation and enhanced Th1 cytokine secretion in response to primary EBV infection.
NF-κB involvement in LMP1-mediated suppression of the SAP gene. (A) Nuclear translocation of NF-κB/p65 was increased on LMP1-H9 cells. H9 cells were transfected with pSG5 and pSG5-LMP1 with or without TRAF2/5 dominant-negative (DN) mutants. Both nuclear (N) and cytoplasmic (C) p65RelA protein in H9 cells were detected by Western blotting. α-actin and histone H1 were used as controls for cytoplasmic and nuclear fractions, respectively. The nuclear-cytoplasm ratio of NF-κB/p65 protein was significantly increased on LMP1-H9 cells. Cotransfection with TRAF2/5 dominantnegative mutants could reduce this effect. (B) NF-κB activation was enhanced by expressing LMP1 in H9 cells. H9 cells and its sublines were transfected with NF-κB–driven luciferase constructs. Constitutive activation of NF-κB was clearly demonstrated in LMP1-expressing H9 T cells, but decreased in those cells coexpressed with dominant-negative TRAF2 or TRAF5 constructs. (C) Inhibition of LMP1-mediated suppression of SAP by NF-κB inhibitor. LMP1-H9 cells were first treated with 0.005, 0.01, or 0.02 μg/μL Bay11-7082 for 2 hours, washed by PBS, and then incubated with normal medium for an additional 6 hours. Dosage-dependent induction of SAP protein by Bay11-7082 in LMP1-H9 cells was shown. Error bars representing the standard deviation of 3 experiments are shown.
NF-κB involvement in LMP1-mediated suppression of the SAP gene. (A) Nuclear translocation of NF-κB/p65 was increased on LMP1-H9 cells. H9 cells were transfected with pSG5 and pSG5-LMP1 with or without TRAF2/5 dominant-negative (DN) mutants. Both nuclear (N) and cytoplasmic (C) p65RelA protein in H9 cells were detected by Western blotting. α-actin and histone H1 were used as controls for cytoplasmic and nuclear fractions, respectively. The nuclear-cytoplasm ratio of NF-κB/p65 protein was significantly increased on LMP1-H9 cells. Cotransfection with TRAF2/5 dominantnegative mutants could reduce this effect. (B) NF-κB activation was enhanced by expressing LMP1 in H9 cells. H9 cells and its sublines were transfected with NF-κB–driven luciferase constructs. Constitutive activation of NF-κB was clearly demonstrated in LMP1-expressing H9 T cells, but decreased in those cells coexpressed with dominant-negative TRAF2 or TRAF5 constructs. (C) Inhibition of LMP1-mediated suppression of SAP by NF-κB inhibitor. LMP1-H9 cells were first treated with 0.005, 0.01, or 0.02 μg/μL Bay11-7082 for 2 hours, washed by PBS, and then incubated with normal medium for an additional 6 hours. Dosage-dependent induction of SAP protein by Bay11-7082 in LMP1-H9 cells was shown. Error bars representing the standard deviation of 3 experiments are shown.
In this study, the 7 HLH cases showed no detectable mutations of the SAP/SH2D1A gene at exons 2/3. The EBV has been previously shown to infect T cells in these cases as demonstrated by double-labeled immunohistochemistry and EBER1 in situ hybridization in 6 of our HLH cases.47 Taken together, these results further support the distinction of our 7 HLH cases from XLP. Recently, a familial disease entity of chronic active EBV infection with HPS has been shown to have mutations of the perforin gene.48 Although no study of perforin gene mutation was performed in this study, the absence of familial aggregation in our patients may reasonably exclude familial HLH cases.
The SLAM-SAP signaling is an important pathway in T-cell activation when engaged with ligands of T-cell receptor or in response to virus infection. It has been demonstrated that SAP can bind to the C-terminus of SLAM and reduce Th1 cytokines secretion by recruiting several adaptor proteins such as SHIP (SH2-containing inositol phosphatase) and Ras-GAP (Ras-GTPase–activating protein).43,44 Therefore, the mutation of the SAP gene in patients with XLP or the knocking-out of the SAP/SH2D1A gene in mouse T cells may result in high levels of Th1 cytokines.20,49 Recently, SAP has been demonstrated to engage in the control of cytolytic activity by polarization of the lytic machinery involving 2B4 and CD84.50 In the present study, we demonstrated that EBV LMP1 could effectively suppress SAP/SH2D1A gene expression and simultaneously activate its downstream molecules ERK1/2 as well as enhance IFN-γ secretion in T cells. These data indicate that the same SLAM/SAP/ERK activation pathway as observed in XLP could also be achieved by LMP1-mediated suppression of the SAP gene in HLH. The tumor suppressor p53 has been demonstrated to bind to the SAP promoter and activate SAP gene expression in B lymphocytes.51 Since LMP1 could regulate p53 function in B lymphocytes,52,53 the potential interaction between LMP1 and p53 in SAP signaling deserves to be investigated in the future.
The SLAM downstream molecules ERK and IFN-γ were activated by LMP1-mediated SAP suppression. (A) ERK1/2 activation on LMP1-H9 cells. The phosphorylation levels of ERK1/2 in LMP1-expressing T cells were examined by Western blotting. Dominantnegative (DN) mutants of TRAF2/5 could decrease the phosphorylation of ERK1/2 induced by LMP1. (B) ERK1/2 phosphorylation was down-regulated by reconstituting SAP. LMP1-H9 cells were infected with retrovirus for restoring SAP expression. The pRCMV-SAP could decrease the level of ERK1/2 phosphorylation. (C) LMP1 decreased the association of SAP with SLAM. The expression of LMP1 has reduced SAP expression on H9 cells that in turn decreased the level of SAP/SLAM interaction as demonstrated by immunoprecipitation. With an equal SLAM level, gene transduction of SAP by retrovirus in H9 cells could enhance the SAP molecules entrapped by SLAM. These data indicate that LMP1-induced SLAM activation was due to the reduction of SAP protein but did not affect the binding capacity of SAP/SLAM. (D) The induction of IFN-γ by LMP1. H9 cells were transfected with plasmids as indicated. LMP1 could enhance IFN-γ secretion on H9 cells, while TRAF2/5 dominant-negative mutants suppressed the LMP1-mediated IFN-γ secretion. In addition, overexpression of SAP on LMP1-H9 cells could also suppress IFN-γ induction by LMP1. Error bars representing the standard deviation of 3 independent experiments are shown in panels A and D.
The SLAM downstream molecules ERK and IFN-γ were activated by LMP1-mediated SAP suppression. (A) ERK1/2 activation on LMP1-H9 cells. The phosphorylation levels of ERK1/2 in LMP1-expressing T cells were examined by Western blotting. Dominantnegative (DN) mutants of TRAF2/5 could decrease the phosphorylation of ERK1/2 induced by LMP1. (B) ERK1/2 phosphorylation was down-regulated by reconstituting SAP. LMP1-H9 cells were infected with retrovirus for restoring SAP expression. The pRCMV-SAP could decrease the level of ERK1/2 phosphorylation. (C) LMP1 decreased the association of SAP with SLAM. The expression of LMP1 has reduced SAP expression on H9 cells that in turn decreased the level of SAP/SLAM interaction as demonstrated by immunoprecipitation. With an equal SLAM level, gene transduction of SAP by retrovirus in H9 cells could enhance the SAP molecules entrapped by SLAM. These data indicate that LMP1-induced SLAM activation was due to the reduction of SAP protein but did not affect the binding capacity of SAP/SLAM. (D) The induction of IFN-γ by LMP1. H9 cells were transfected with plasmids as indicated. LMP1 could enhance IFN-γ secretion on H9 cells, while TRAF2/5 dominant-negative mutants suppressed the LMP1-mediated IFN-γ secretion. In addition, overexpression of SAP on LMP1-H9 cells could also suppress IFN-γ induction by LMP1. Error bars representing the standard deviation of 3 independent experiments are shown in panels A and D.
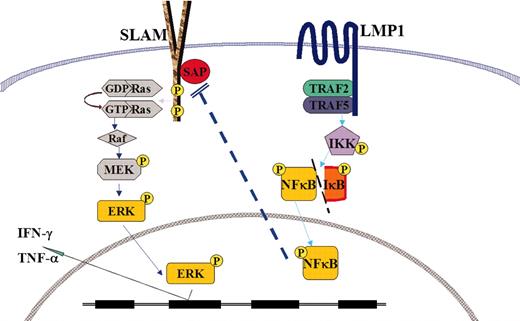
Schematic depiction of the cross-talk between SLAM/SAP/ERK signaling in XLP and LMP1/TRAF/NF-κB. ERK indicates extracellular signal-regulated kinase; MEK, MAPK (mitogen-activated protein kinase)/ERK kinase; GDP, guanosine diphosphate; and GTP, guanosine triphosphate.
Schematic depiction of the cross-talk between SLAM/SAP/ERK signaling in XLP and LMP1/TRAF/NF-κB. ERK indicates extracellular signal-regulated kinase; MEK, MAPK (mitogen-activated protein kinase)/ERK kinase; GDP, guanosine diphosphate; and GTP, guanosine triphosphate.
In this study, we demonstrated that LMP1 could up-regulate IFN-γ via TRAF2/5–NF-κB signals, and EBV LMP1 was shown to aggregate and colocalize with TRAF2/5 proteins in T cells. Similar results of aggregation of TRAF2 and TRAF5 proteins have been observed in B-cell lines and Hodgkin-Reed-Sternberg (H-RS) cell lines.35,42 Cytoplasmic aggregation of TRAF proteins reflects the constitutive activation of CD40/tumor necrosis factor receptor (TNFR) signaling in B cells and CD30 signaling in H-RS cells.39,54,55 EBV LMP1 may therefore use a modified signaling mechanism similar to that adopted by the TNFR/CD40/CD30 superfamily to elicit its pleiotropic activities in T, B, or H-RS cells. The LMP1-mediated suppression of SAP gene and the decrease of IFN-γ secretion by dominant negative mutants of TRAF2/5 suggest that SAP is a downstream signal molecule in LMP1/TRAF2/5–NF-κB signaling pathway. Previously, we demonstrated that EBV infection and LMP1 transfection of T cells could up-regulate TNF-α.33,34 In this study, we further demonstrated that LMP1 expression could enhance IFN-γ secretion. Therefore, both Th1 cytokines TNF-α and IFN-γ could be upregulated by LMP1 expression in T cells.
Although no mutation of the SAP/SH2D1A gene could be detected in EBV-HLH patients, our results showed that the reduced SAP gene expression could be achieved through LMP1-mediated signaling in sporadic HLH. Therefore, our data shown in this study reconcile the difference in the molecular background of XLP and sporadic HLH and both diseases appear to share common pathogenesis of HPS to elicit abnormal T-cell activation and activate macrophages and cytokine storm in response to primary EBV infection, as shown in Figure 5.
Prepublished online as Blood First Edition Paper, July 7, 2005; DOI 10.1182/blood-2005-04-1406.
Supported by grants from the National Health Research Institutes and National Science Council, Taipei, Taiwan (I.J.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.