Abstract
To realize the therapeutic potential of human embryonic stem cells (hESCs), it is necessary to regulate their differentiation in a uniform and reproducible manner. We have developed a method in which known numbers of hESCs in serum-free medium were aggregated by centrifugation to foster the formation of embryoid bodies (EBs) of uniform size (spin EBs). These spin EBs differentiated efficiently and synchronously, as evidenced by the sequential expression of molecular markers representing stem cells, primitive streak, and mesoderm. In the presence of hematopoietic growth factors, reproducible differentiation was achieved with blood cells formed in more than 90% of EBs. Using chimeric EBs generated from mixtures of green fluorescence protein–positive (GFP+) and GFP– hESCs in a clonogenic assay, hematopoietic precursor frequency was estimated to be approximately 1:500 input cells. This method of EB formation provides a generally applicable means for modulating and objectively monitoring the directed differentiation of hESCs.
Introduction
Embryoid body (EB) differentiation of murine embryonic stem cells (mESCs) recapitulates many aspects of early mouse embryogenesis.1 Synchronous differentiation is typically achieved by forming mEBs from mESCs seeded at low cell densities.2-4 Because human embryonic stem cells (hESCs) survive poorly as single cells,5 human EB formation has generally been initiated from enzymatically digested colony pieces of different sizes or from high-density suspension cultures.6-11 Cocultivation of hESCs with stromal layers, such as OP9 and S17, has successfully induced hematopoietic differentiation,7,8,12 but these systems are compromised by cell-associated and secreted components derived from the feeder layer and by potential interactions between added factors and the stromal layers.7 The weaknesses inherent in the existing protocols highlight the need for a robust stromal- and serum-free culture system for the differentiation of hESCs.
Study design
Human embryonic stem cells 2, 3, and 4 (hES2, hES3, and hES4)13 and the green fluorescence protein (GFP)–expressing hES3 derivative ENVY14 were cultured essentially as described.5 A detailed protocol for hESC differentiation is provided in the supplemental Document S1 (see the Supplemental Materials link at the top of the online article, at the Blood website). In brief, hESCs were trypsinized into a single-cell suspension, washed in phosphate-buffered saline, and resuspended in serum-free medium (SFM).15 EB formation was induced by seeding the desired number (300-10 000) of hESCs in 100 μL SFM supplemented with growth factors in each well of 96-well, round-bottomed, low-attachment plates (Nunc, Roskilde, Denmark) and centrifuging the plates at 1500 rpm (478g) for 4 minutes at 4°C to aggregate the cells. After 10 to 12 days, the EBs were transferred to 96-well, flat-bottomed tissue culture plates precoated with gelatin in SFM supplemented with growth factors and were allowed to differentiate further.
Results and discussion
In our initial attempts to direct hematopoietic differentiation of hESCs, hEBs were formed from pieces of undifferentiated colonies in the same SFM that we had used to induce mesoderm from mESCs.4 However, only the subset of hEBs that developed from larger colony fragments (approximately 500-1000 cells) regularly formed blood cells, suggesting that a minimum number of hESCs was required to generate hEBs that differentiated into mesodermal lineages.
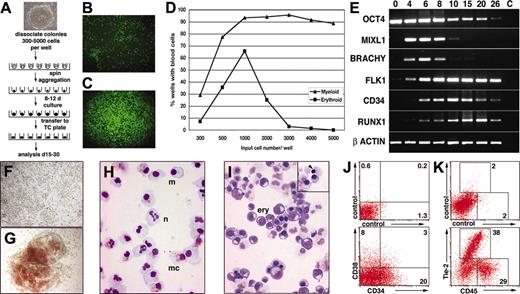
Because it was not possible to reproducibly disaggregate hESC colonies into pieces of exactly the desired size, we attempted to improve the efficiency of hematopoietic differentiation by depositing known numbers of hESCs in round-bottomed, low-adherence, 96-well plates and then aggregating the cells into EBs by centrifugation (Figure 1A). After 8 to 12 days, these so-called spin EBs were transferred to tissue culture–treated plates and differentiated further. The wells were scored for the presence of developing myeloid and hemoglobinized erythroid cells by light microscopy. Visual cell lineage assignments were confirmed by correlating the morphologic appearance of cells in the wells with May-Grünwald-Giemsa–stained cytocentrifuge preparations, flow cytometry, immunocytochemistry, and gene expression analysis in selected cases (Figure 1; Figures S1-S3).
The differentiation process was affected by several parameters. If the hESCs were seeded into flat-bottomed wells, they failed to form large aggregates even after centrifugation of the cells (Figure 1B), and both survival and differentiation were unpredictable. Conversely, deposition of hESCs into low-attachment round-bottomed wells followed by brief centrifugation encouraged cellular association into a close pellet (Figure 1C) from which uniform EBs reproducibly formed and differentiated (see examples in Figure 2). Hematopoietic differentiation was also influenced by the number of cells seeded into each well. In the representative experiment shown in Figure 1D, efficient blood formation required in excess of 500 hESCs per well, and optimum erythropoiesis was observed when 1000 cells were seeded per well. In 5 experiments in which 3000 hESCs were deposited per well, EBs developed in 1112 of 1164 (95.5%) wells, with an average of 95.8% ± 1.6% of wells containing viable EBs and 91.8% ± 7.8% of wells containing blood cells per experiment.
Gene expression analysis of hEBs revealed sequential expression of differentiation stage–related genes, similar to the pattern of gene expression observed in cultures of mouse EBs (Figure 1E).4,16-18 During hESC differentiation, gradual attenuation of the expression of the stem cell gene OCT4 was observed. The primitive streak genes MIXL1 and BRACHYURY were strongly expressed from day 4 to day 8, indicating a wave of mesendodermal induction. Vascular endothelial growth factor (VEGF) receptor 2 (FLK1/KDR) expression increased and remained at high levels throughout the differentiation period, consistent with the ventral patterning of mesoderm in these cultures, followed at day 10 and day 20 by peak expression of the hematopoietic and endothelial gene surface marker CD34 and the hematopoietic transcription factor RUNX1, respectively.17,19
Most EBs generated by this protocol generated large numbers of hemoglobinized or nonhemoglobinized blood cells (Figure 1F-G). May-Grünwald-Giemsa–stained cytospins showed neutrophils, macrophages, and mast cells in wells containing myeloid cells (Figure 1H) and maturing erythroid cells in wells containing overtly hemoglobinized cells (Figure 1I). Although most erythroid cells were nucleated, consistent with a primitive, yolk sac lineage, occasional cells undergoing enucleation were observed, suggesting the presence of definitive erythropoiesis (Figure 1I, inset). Flow cytometry of EBs disaggregated at day 11 showed high proportions of CD34+ and CD38+ cells (Figure 1J), confirming the efficiency of differentiation. Analysis at day 26 showed that the cultures supported the expansion of hematopoietic mesoderm because most cells expressed either CD45 or Tie-2, marking hematopoietic or endothelial lineages, respectively (Figure 1K).
Differentiation of defined numbers of hESCs as spin EBs. (A) Schematic diagram of the spin EB method for hESC differentiation. (B-C) Immunofluorescence images of 3000 ENVY hESCs deposited in (B) flat- and (C) round-bottomed wells immediately after centrifugation. Aggregation of ES cells was only induced in the round-bottomed wells. Original magnification, 50×. Objective used was 5 ×/0.12 NA. (D) Percentages of wells containing myeloid or erythroid cells graphed as a function of the input number of cells in each well. Seventy-two wells were assayed at each point. (E) Sequential expression of differentiation stage–related genes in EBs harvested after the indicated number of days of differentiation. C indicates no template control. (F-K) Hematopoietic cells derived from spin EBs. (F-G) Bright-field microscopy of EBs differentiated for 11 days in suspension culture and then plated down for a further 14 days, revealing large numbers of (F) nonhemoglobinized and (G) hemoglobinized blood cells. Original magnification, 100 ×. Objective used was 10 ×/0.30 NA. (H-I) May-Grünwald-Giemsa–stained cytocentrifuge preparations after 28 days of differentiation showing neutrophils (n), macrophages (m), and mast cells (mc) in wells containing (H) myeloid cells and large numbers of maturing erythroid cells (ery) in wells containing (I) overtly hemoglobinized cells. Some cells with condensed chromatin apparently undergoing enucleation were observed (inset, arrow). Original magnification, 400 ×. Objective used was 40 ×/1.30 NA. (J,K) Flow cytometry of EBs dissociated (J) at day 11 and stained for expression of CD34 and CD38 and (K) at day 26 and stained for expression of Tie-2 and CD45. Percentages of cells within the indicated quadrants or regions are shown. All images were captured with a Zeiss Axiocam mounted on an Axiovert 200 inverted microscope using Axiovision software (Carl Zeiss, Göttingen, Germany). Image montages were created in Adobe Photoshop (Adobe Systems, San Jose, CA).
Differentiation of defined numbers of hESCs as spin EBs. (A) Schematic diagram of the spin EB method for hESC differentiation. (B-C) Immunofluorescence images of 3000 ENVY hESCs deposited in (B) flat- and (C) round-bottomed wells immediately after centrifugation. Aggregation of ES cells was only induced in the round-bottomed wells. Original magnification, 50×. Objective used was 5 ×/0.12 NA. (D) Percentages of wells containing myeloid or erythroid cells graphed as a function of the input number of cells in each well. Seventy-two wells were assayed at each point. (E) Sequential expression of differentiation stage–related genes in EBs harvested after the indicated number of days of differentiation. C indicates no template control. (F-K) Hematopoietic cells derived from spin EBs. (F-G) Bright-field microscopy of EBs differentiated for 11 days in suspension culture and then plated down for a further 14 days, revealing large numbers of (F) nonhemoglobinized and (G) hemoglobinized blood cells. Original magnification, 100 ×. Objective used was 10 ×/0.30 NA. (H-I) May-Grünwald-Giemsa–stained cytocentrifuge preparations after 28 days of differentiation showing neutrophils (n), macrophages (m), and mast cells (mc) in wells containing (H) myeloid cells and large numbers of maturing erythroid cells (ery) in wells containing (I) overtly hemoglobinized cells. Some cells with condensed chromatin apparently undergoing enucleation were observed (inset, arrow). Original magnification, 400 ×. Objective used was 40 ×/1.30 NA. (J,K) Flow cytometry of EBs dissociated (J) at day 11 and stained for expression of CD34 and CD38 and (K) at day 26 and stained for expression of Tie-2 and CD45. Percentages of cells within the indicated quadrants or regions are shown. All images were captured with a Zeiss Axiocam mounted on an Axiovert 200 inverted microscope using Axiovision software (Carl Zeiss, Göttingen, Germany). Image montages were created in Adobe Photoshop (Adobe Systems, San Jose, CA).
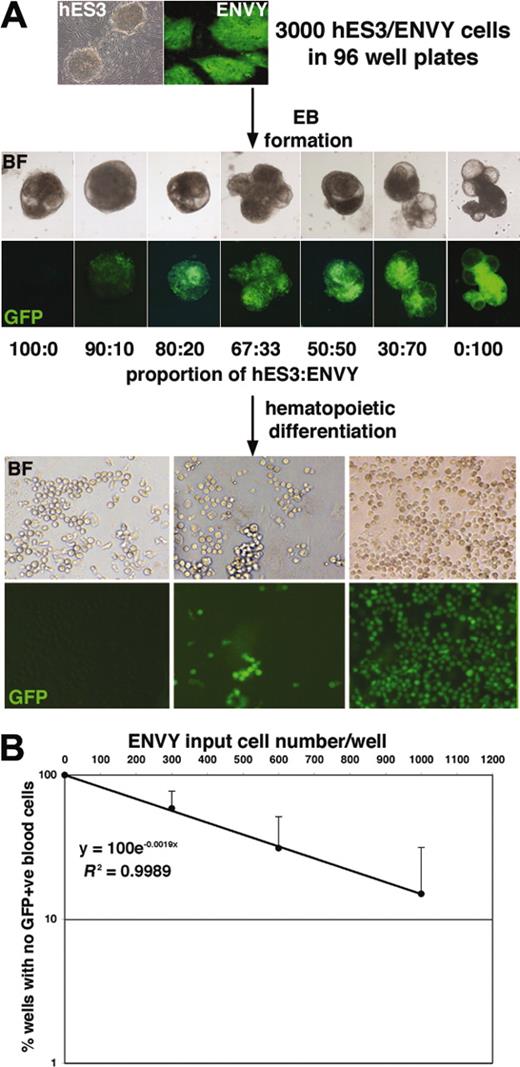
The reproducibility of the spin EB method was exploited to estimate the frequency of hematopoietic precursors arising from a defined number of hESCs. EBs were formed from 3000 hESCs consisting of variable combinations of wild-type hES3 cells and a GFP-expressing derivative, denoted ENVY.14 Aggregation of GFP+ and GFP– hESCs led to the formation of chimeric EBs into which GFP+ cells were integrated in proportion to their input number (Figure 2A). Analysis of these data demonstrated an excellent correlation (R2 > 0.99 for the 3 experiments shown in Figure 2B) between the number of GFP+ input cells and the percentage of EBs forming GFP+ blood cells. The very high correlation coefficient is a testament to the reproducible differentiation observed over many hundreds of wells. Using Poisson statistics, the frequency of hematopoietic precursors was estimated at approximately 1 in 500 input cells.
Formation of hES3/ENVY chimeric EBs enables the frequency of hematopoietic precursors to be estimated. (A) Bright-field (BF) and fluorescence (GFP) images of day 7 chimeric EBs formed from a total of 3000 hESCs consisting of the indicated ratios of GFP– hES3 and GFP+ ENVY cells. Original magnification, 50 ×; objective used was 5 ×/0.12 NA. Further differentiation of the EBs led to the formation of hematopoietic cells in each well, which were all GFP– (left panels) or all GFP+ (right panels) or included a mixture of GFP– and GFP+ cells (middle panels), depending on the proportion of ENVY cells contributing to the EBs. Original magnification, 100 ×; objective used was 10 ×/0.30 NA. Camera and microscope used are as in Figure 1. (B) Plotting the frequency of wells with no GFP+ cells against the ENVY input cell number/well demonstrated an excellent correlation (R2 > 0.99). Results shown are the mean of 3 experiments with error bars representing the SD. The clonogenic frequency of hematopoietic precursors (1:523) was estimated by determining the ENVY input cell number that resulted in 37% of the wells containing only GFP– blood cells.
Formation of hES3/ENVY chimeric EBs enables the frequency of hematopoietic precursors to be estimated. (A) Bright-field (BF) and fluorescence (GFP) images of day 7 chimeric EBs formed from a total of 3000 hESCs consisting of the indicated ratios of GFP– hES3 and GFP+ ENVY cells. Original magnification, 50 ×; objective used was 5 ×/0.12 NA. Further differentiation of the EBs led to the formation of hematopoietic cells in each well, which were all GFP– (left panels) or all GFP+ (right panels) or included a mixture of GFP– and GFP+ cells (middle panels), depending on the proportion of ENVY cells contributing to the EBs. Original magnification, 100 ×; objective used was 10 ×/0.30 NA. Camera and microscope used are as in Figure 1. (B) Plotting the frequency of wells with no GFP+ cells against the ENVY input cell number/well demonstrated an excellent correlation (R2 > 0.99). Results shown are the mean of 3 experiments with error bars representing the SD. The clonogenic frequency of hematopoietic precursors (1:523) was estimated by determining the ENVY input cell number that resulted in 37% of the wells containing only GFP– blood cells.
In preliminary experiments examining the development of methylcellulose colony-forming cells (CFCs), we observed CFCs from day 6 spin EBs giving rise to primitive erythroid and macrophage colonies at a frequency of 92 per 104 cells and day 10 CFCs at a frequency of 23 per 104 cells. Taken together, the high frequencies of day 11 CD34+ cells and hematopoietic precursors observed in our system compared favorably with the results reported using conventional EB or stromal cocultivation methods for hESC differentiation6-8,10,12 and were superior to the precursor frequencies reported by others for human EBs differentiated in serum-free cultures.7 At a molecular level, the spin EBs passed through a transient in vitro gastrulation stage that antedated the expression of mesodermal genes and the emergence of hematopoietic cells, similar to differentiating mESCs.4,16-18 The ability to faithfully reproduce early human embryonic differentiation in vitro provides a powerful tool for the molecular and cellular dissection of this previously inaccessible phase.
We believe that modulating medium composition, extracellular matrix substrate, and hESC numbers will enable spin EBs to be readily adapted to the generation of many cell types. In addition, the ability to form chimeric EBs with marked cells, such as ENVY, will permit an estimation of clonogenic frequency in cell types for which simple precursor assays are unavailable.
Prepublished online as Blood First Edition Paper, May 24, 2005; DOI 10.1182/blood-2005-03-0987.
Supported by the National Health and Medical Research Council (NHMRC) of Australia, the Juvenile Diabetes Research Foundation, and the Australian Stem Cell Centre. A.G.E. is an NHMRC Senior Research Fellow.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.