Abstract
Mast cells play an important role in host defense against various pathogens, but their role in viral infection has not been clarified in detail. dsRNA, synthesized by various types of viruses and mimicked by polyinosinic-polycytidylic acid (poly(I:C)) is recognized by Toll-like receptor 3 (TLR3). In this study, we demonstrate that poly(I:C) injection in vivo potently stimulates peritoneal mast cells to up-regulate a number of different costimulatory molecules. Therefore, we examined the expression and the functional significance of TLR3 activation in mast cells. Mast cells express TLR3 on the cell surface and intracellularly. After stimulation of mast cells with poly(I:C) and Newcastle disease virus (NDV), TLR3 is phosphorylated and the expression of key antiviral response cytokines (interferon β, ISG15) and chemokines (IP10, RANTES) is upregulated. Interestingly, mast cells activated via TLR3-poly(I:C) potently stimulate CD8+ T-cell recruitment. Indeed, mast-cell–deficient mice (KitW/KitW-v) given an intraperitoneal injection of poly(I:C) show a decreased CD8+ T-cell recruitment, whereas granulocytes normally migrate to the peritoneal cavity. Mast-cell reconstitution of KitW/KitW-v mice normalizes the CD8+ T-cell influx. Thus, mast cells stimulated through engagement of TLR3 are potent regulators of CD8+ T-cell activities in vitro and in vivo.
Introduction
The magnitude and quality of innate immune response is essential for appropriate adaptive response. The activation of naive CD8+ T cells, the main effector cells in the course of viral infections, their clonal expansion, development of effector cells, and maintenance and expansion of memory CD8+ T cells after antigen reappearance are precisely regulated and dependent on an adequate context of costimulation and cytokine/chemokine environment, provided by cells responding to “first defense line” signals of innate immunity.1-4 Mast cells (MCs), pivotal effector cells in immunoglobulin (Ig) E–associated disorders, have recently been recognized as important elements in innate immune defenses.5 Strategically located at host/environment interfaces like the skin, airways, and gastrointestinal and urogenital tracts,6,7 MCs are equipped with a large variety of receptors to detect signs of infections including Toll-like receptors (TLRs), CD48, and complement receptors.8 MCs activated via these receptors secrete a large number of proinflammatory products including granule-associated preformed mediators (histamine, serotonin, proteases, proteoglycans), lipid mediators (leukotrienes B4 and C4 and prostaglandins), as well as cytokines, chemokines, and growth factors.9
In contrast to the well-established crucial role of MCs in induction of host defense responses to bacteria and parasites, the function of MCs in antiviral immune responses remains to be characterized in detail. Recently, a number of reports have shown the functional interplay between MCs and T cells in allergic, infectious, and autoimmune processes, for example, MCs and T cells colocalizing in inflamed tissues, as in the lungs of patients with asthma and in animal models of allergic asthma.10,11 It has also been reported that tumor necrosis factor α (TNF-α) released from MCs contributes substantially to T-cell recruitment to the draining lymph nodes (LNs) in an experimental infection model with Escherichia coli.12 Furthermore, activated MCs induce the chemotaxis of CD8+ T cells through the production of leukotrienes in vitro.13 Further, the recruitment of early effector T cells into the airways in an asthma model is mediated by leukotriene B4 (LTB4)14 released by MCs activated by IgE cross-linking.15 However, the mechanisms and relevance of MC–T-cell interactions, as well as the signals required for MC activation and effector functions in settings of viral infections remain largely unknown.
TLRs play an important role in innate immunity recognizing specific, nonself, conserved components shared by different pathogens. Different TLRs are involved in the specific recognition of various microbes.16,17 Human and mouse MCs have been shown to be activated by bacterial peptidoglycan and lipopolysaccharide (LPS) via TLR2 and TLR4, respectively, producing a number of cytokines such as interleukin 5 (IL-5), IL-6, IL-10, IL-13, and TNF-α.18-21
On binding of viral dsRNA or its synthetic counterpart polyinosinic-polycytidylic acid (poly(I:C)), TLR3 activates a signaling pathway, leading to the activation of interferon regulatory factor 3 (IRF3).22,23 This results in transactivation of primary genes, such as interferon β (IFN-β) and interferon stimulated gene 15 [ISG15], thereby initiating autocrine/paracrine activation of secondary genes involved in the antiviral response.24 In this study, we examined the expression and functional activity of MC-TLR3 and characterized the significance of its engagement and activation by poly(I:C) and Newcastle disease virus (NDV). Our data indicate that MCs actively respond to TLR3 activation producing cytokines and chemokines and specifically recruiting CD8+ T cells to sites of poly(I:C) challenge.
Materials and methods
Materials
Recombinant IL-3 was purchased from PeproTech (London, United Kingdom). Poly(I:C), phorbol myristate acetate (PMA), and ionomycin were from Sigma (St Louis, MO). LPS, kindly provided by H. Brade (Research Center Borstel, Borstel, Germany) was derived from Salmonella enterica serovar Friedenau and prepared by the hot phenol-water procedure, repeated ultracentrifugation, and electrodialysis.25 Poly(I:C) was tested for endotoxin activity by the Coamatic Chromo-LAL kit (Hemochrom Diagnostica, Essen, Germany) with a resolution limit of 0.005 EU (endotoxin units)/mL. This was equal to 0.732 EU/mL corresponding to 50 pg/mL per 10 μg/mL poly(I:C). Stimulation of bone marrow–derived cultured mast cells (BMMCs) with 50 pg/mL LPS does not lead to chemokine production at all. Furthermore, treatment of poly(I:C) with RNAse A (Boehringer Mannheim, Mannheim, Germany) abolished poly(I:C)–induced chemokine production. Antibodies against IRF3 (FL-425), TLR3 (Q-18), and Myc (9E10) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse antiphosphotyrosine (anti–p-Tyr) antibody (RC20) was from BD Transduction Laboratories (Heidelberg, Germany). Goat anti–mouse, goat anti–rabbit, and rabbit anti–goat horseradish peroxidase conjugates (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) were used as secondary antibodies.
Animals and reconstitution experiments
C57BL/6 mice were purchased from Harlan Winkelman (Borchen, Germany). Genetically MC-deficient WBB6F1-KitW/KitW-v (KitW/KitW-v and congenic wild-type WBB6F1-Kit+/+ (Kit+/+) mice were bred at the Department of Dermatology, Mainz, Germany. OTI and OTII TCR transgenic mice were bred in the Animal Care Facility of the Research Center Borstel. All mice were housed in specific pathogen-free facilities. Sex- and age-matched (6-8 weeks of age) mice were used for experiments, performed according to institutional guidelines. Poly(I:C) (10 μgin200 μL phosphate-buffered saline [PBS]) was injected intraperitoneally and controls were injected with PBS only. Reconstitution experiments were performed as described.26 Briefly, 8-week-old KitW/KitW-v mice were reconstituted with 3 × 106 BMMCs injected intraperitoneally, challenged with poly(I:C), and analyzed 6 weeks after reconstitution.
Cell isolation
Peritoneal cells from C57BL/6 mice were isolated by peritoneal lavage with 10 mL cold 0.9% NaCl solution. For some experiments, peritoneal mast cells (PMCs) were enriched by centrifugation using a Percoll gradient (MC purity, 60%-80%) as described.27 Peripheral and mesenterial LNs were excised and single-cell suspensions were prepared by homogenization in PBS.
Generation of BMMCs
The BMMCs were generated by cultivation of bone marrow cells from C57BL/6 mice in the presence of recombinant murine IL-3. Cells were maintained in complete medium consisting of 10% heat-inactivated fetal calf serum (Biochrom, Berlin, Germany), 50 μM β-mercaptoethanol (Gibco, Rockville, MD), nonessential amino acids, 2 mM l-glutamine, penicillin, streptomycin, and 1 mM sodium pyruvate (all from Gibco) in Iscove modified Dulbecco medium (IMDM; PAA, Linz, Austria) supplemented with 5 ng/mL IL-3.28 After 4 weeks of culture, BMMCs represented more than 98% of the total cells according to toluidine blue staining and fluorescence-activated cell sorting (FACS) analysis of cell-surface expression of CD117 (c-Kit) and FcϵRI29 and were negative for CD11c, B220, F4/80, Gr-1, and CD3 expression.
Infection with NDV
NDV was kindly provided by R. Zawatzky (DKFZ, Heidelberg, Germany). BMMCs and PMCs were infected with 0.5 hemagglutinating units (HA)/mL and 2.4 HA/mL NDV in complete medium. It was demonstrated that in these concentrations NDV strongly activates human blood monocytes30 and murine dendritic cells.31
Evaluation of cytokine concentrations
To analyze cytokine production, BMMCs (2 × 106/mL) were stimulated with poly(I:C) or infected with NDV for 48 hours at 37°C and 5% CO2. Supernatants were collected and cytokines and chemokines were measured by specific enzyme-linked immunosorbent assays (ELISAs) using specific antibodies and standard proteins from R&D Systems (Wiesbaden-Nordenstadt, Germany).
Flow cytometry
Cells were washed twice in FACS buffer (2% newborn calf serum, 0.1% NaN3, 2 mM EDTA [ethylenediaminetetraacetic acid] in PBS) and stained with phycoerythrin (PE)–, allophycocyanin (APC)–, or FITC-conjugated antibodies. To detect FcϵRI, cells were preincubated first with purified mouse IgE (C38-2), then washed twice, and incubated with biotinylated α-IgE (R35-118; both from BD PharMingen, San Diego, CA). This was followed by incubation with streptavidin labeled with PE (Dianova, Hamburg, Germany) or APC (BD PharMingen). Monoclonal antibodies against CD4 (L3T4, RM4-5), CD8α (Ly-2, 53-6.7), Gr-1 (Ly-6C and G, 53-6.7), CD11b (integrin αM chain, M1/70), CD117 (c-Kit, 2B8), CD16/32 (FcγIII/II, 2.4G2), CD28 (37.51) CD80 (B7.1, 16-10A1), CD21/35 (CR1/CR2, 7G6), I-A/I-E (M5/114152), all from BD PharMingen; CD86 (B7.2, GL1; Southern Biotechnology, Birmingham, AL); and T1/ST2 (DJ8; Morwell Diagnostics, Zurich, Switzerland) were used. Antibodies to mouse TLR3 (40C1285) from Qbiogene-Alexis (Grünberg, Germany) were used for surface and intracellular staining. They were either labeled with Alexa Fluor 647 monoclonal antibody labeling kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions or detected by biotinylated secondary anti–mouse IgG1 antibodies (Southern Biotechnology) and APC-labeled streptavidin. To prevent unspecific binding, all samples were preincubated with Fc-Block or unlabeled isotype-matched unspecific antibodies (BD PharMingen). Samples were analyzed using a FACScalibur flow cytometer (BD Biosciences) according to standard protocols. For surface staining, gates on viable cells were set according to exclusion of propidium iodide staining. For intracellular staining, cells were stained first for surface expression of c-Kit and CD16/32, then fixed and permeabilized by Cyto Fix/Perm Kit (BD PharMingen), and stained for intracellular expression of TLR3. Surface-stained c-Kit+, CD16/32+ double-positive cells were gated and TLR3 expression was analyzed.
Degranulation assay
Release of histamine-containing granules in vitro was quantified by measuring β-hexosaminidase in supernatants of BMMCs as described.32 Briefly, BMMCs were loaded with 5 μg/mL dinitrophenyl (DNP)–specific IgE (SPE-1, Sigma) for 30 minutes, washed, and resuspended in Tyrode buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] buffer at pH 7.4, 130 mM NaCl, 5 mM KCl, 1.4 mM CaCl2, 1 mM MgCl2, 5.6 mM glucose and 0.1% bovine serum albumin [BSA]). Degranulation was induced by stimulation with DNP–human serum albumin (HSA), PMA, and ionomycin. After 20 minutes samples were centrifuged, supernatants were collected, and cell pellets were solubilized with 0.5% Triton X-100 in Tyrode buffer. Enzymatic activities of β-hexosaminidase were assessed after incubation for 40 minutes at 37°C with p-nitrophenyl-N-acetyl-β-d-glucosaminide (Sigma) in 0.1 M sodium citrate (pH 4.5). Production of p-nitrophenol was stopped by addition of 0.2 M glycine (pH 10.7) and quantified by absorbance measurement at 405 nm. The extent of degranulation was calculated by dividing p-nitrophenol absorbance in the supernatants by the sum of absorbance in the supernatant and cell lysate.
Chemotaxis assays
Chemotaxis of granulocytes or CD4+ and CD8+ cells was measured by assessing migration through a polycarbonate filter of 5-μm pore size in 24-well transwell chambers (Corning Life Sciences, Schipol-Rijk, Netherlands) with the use of complete IMDM. BMMCs (3.3 × 106 cells/mL) were loaded into the lower chambers and incubated with or without poly(I:C) (10 μg/mL) and 1 ng/mL IL-3 for different periods of time. Complete IMDM with or without poly(I:C) was used to determine the spontaneous migration. Then, 3 × 106 bone marrow cells freshly isolated from C57BL/6 mice or 4 × 106 LN cells from OTI or OTII TCR transgenic mice were added into the upper chamber in a volume of 200 μL and incubated at 37°C and 5% CO2 for indicated time intervals. After incubation, the total cell number in the lower chamber was determined microscopically by staining with trypan blue (Biochrom), and the percentages of granulocytes (Gr1+ and CD11b+ double-positive cells), CD4+ cells, and CD8+ cells were determined by staining with specific antibodies and subsequent FACS analysis. Chemotactic indices were determined by dividing absolute migrated cell numbers in probes by absolute migrated cell numbers in controls with medium only. Poly(I:C) alone did not induce chemotaxis in any of the cell populations tested.
Western blotting and immunoprecipitation assays
Cells (1 × 106) were lysed for 15 minutes on ice in 1% ODGP cell-extraction buffer: 20 mM Tris (tris(hydroxymethyl)aminomethane)–HCl buffer, pH 8.0, 15 mM NaCl, 2 mM EDTA, 10 mM sodium fluoride, 1 μg/mL pepstatin A, 1 μg/mL leupeptin, 10 mM phenylmethylsulfonyl fluoride (PMSF), and 100 μM sodium vanadate (all reagents from Sigma). The detergent-insoluble materials were removed by centrifugation for 15 minutes at 18 890 g at 4°C. Nuclear extracts from 5 × 106 cells were prepared according to the method of Schreiber et al.33 Protein concentrations were determined using a BSA protein assay kit (Bio-Rad, Munich, Germany), and 50 μg protein was analyzed by electrophoresis in 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). For immunoprecipitation, 500 μg protein was incubated overnight at 4°C with 5 μg/mL antibodies. Immunocomplexes were captured on protein A/G-agarose (Bio-Rad) with gentle mixing for 1 hour at 4°C. Samples were resuspended in SDS-PAGE sample buffer (62.5 mM Tris-HCL, pH 8.0, 1% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.01% bromophenol blue), boiled for 5 minutes, and analyzed in 10% SDS-PAGE. The resolved proteins were transferred onto nitrocellulose (Bio-Rad). Blots were blocked for 1 hour in PBS containing 0.05% Tween-20 (PBS-T) and 3% BSA (Sigma). After incubations with primary and secondary antibodies and washing with PBS-T, visualization of specific proteins was carried out by an enhanced chemiluminescence (ECL) method using ECL Western blotting detection reagents (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Reverse transcription–PCR
RNA was extracted from cells using Trizol reagent (Gibco-Invitrogen, Karlsruhe, Germany). cDNA was synthesized from 5 μg total RNA using random oligonucleotides as primers and SuperScriptII kit (Gibco-Invitrogen). cDNA was amplified by polymerase chain reaction (PCR) in a reaction mixture (20 μL) containing 2 μL 10-fold PCR buffer with 1.5 mM MgCl2, 250 μM of each dNTPs, 200 nM 5′ and 3′ primers, and 1 U Taq DNA polymerase (“Amplitaq,” Applied Biosciences, Warrington, United Kingdom). The following primers were used: mTLR3 (392 bp) sense 5′-CCGCCCTCTTCGTAACTTGACC-3′, antisense 5′-GCGGCCCGAAAACATCCTT-3′; murine IFN-inducible protein 10 (mIP10; 188 bp) sense 5′-GATGACGGGCCAGTGAGAATG-3′, antisense 5′-GGAGCCCTTTTAGACCTTTTT-3′; mRANTES (murine regulated on activation, normal T-cell expressed and secreted; 202 bp) sense 5′-CTGCCCTCACCATCATCCTCACTG-3′, antisense 5′-CACACTTGGCGGTTCCTTC-3′; mIFNβ (462 bp) sense 5′-CTCCAGCTCCAAGAAAGGACG-3′, antisense 5′-GAACTTTCTGGTAAGTCTTCG-3′; murine interferon stimulated gene 15 (mISG15; 430 bp) sense 5′-TGGCCTGGGACCTAAAGGTGAAGA-3′, antisense 5′-TGCACTGGGGCTTTAGGCCATACT-3′; and β-actin sense 5′-GTGGGGCGCCCCAGGCACCA-3′, antisense 5′-CTCCTTAATGTCACGCACGATTTC-3′ (Metabion, Planegg-Martinsried, Germany). Amplification of β-actin message was used to normalize the amount of cDNA. A mock PCR (without cDNA) was included to exclude contamination in all experiments. Samples were amplified in a DNA Thermocycler (Eppendorf, Hamburg, Germany) for 35 cycles. Each cycle consisted of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and elongation at 72°C for 1 minute, preceded by initial denaturation at 94°C for 5 minutes and followed by a final extension step at 72°C for 5 minutes. Aliquots of PCR products were electrophoresed on 1.5% agarose gel and visualized by ethidium bromide staining.
Statistical analysis
All experiments were performed in at least 3 independent assays, which yielded highly comparable results. Data are summarized as mean plus or minus SEM. Statistical analysis of the results was performed by Student t test for unpaired samples. P values below .05 were considered statistically significant.
Results
Poly(I:C) activates PMCs in vivo
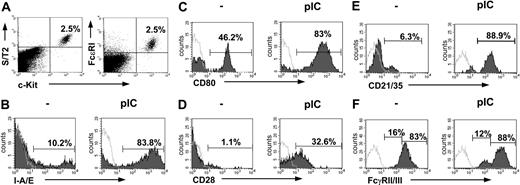
We were interested whether MCs can sense and react to viral products in vivo. Therefore, we injected C57BL/6 mice intraperitoneally with 10 μg poly(I:C) dsRNA, a synthetic mimetic of viral RNA recognized by the TLR3. Peritoneal-cell suspensions were obtained 24 hours thereafter, and MCs, defined as c-Kit, FcϵRI, and S/T2 receptor positive34 (Figure 1A), were analyzed by flow cytometry. As expected, MCs were negative for CD11c, B220, F4/80, Gr-1, and CD3 expression. As shown in Figure 1 and quantified in Table 1, poly(I:C) treatment markedly up-regulated the expression of major histocompatibility complex (MHC) class II molecules (Figure 1B), costimulatory molecules CD80 (Figure 1C) and CD28 (Figure 1D), and complement receptors (Figure 1E). Enrichment of PMCs by centrifugation over Percoll gradient, leading to a considerable increase in the amount of PMCs, also increases the numbers of CD80 and CD28 stained cells (data not shown). The stem-cell factor receptor c-Kit instead shows a down-regulation in peritoneal MCs (mean fluorescence intensity [MFI] 651 ± 91 versus 389 ± 35; n = 5-6; P < .05). Poly(I:C) stimulation induced the generation of 2 different FcγRII/III-positive MC populations. One indicated as high expressing, strongly up-regulates the FcγRII/III; the other one instead, named intermediate, down-regulates the receptor (Figure 1F; Table 1). Taken together these data indicate that MCs can sense and respond to dsRNA.
Murine MCs express TLR3
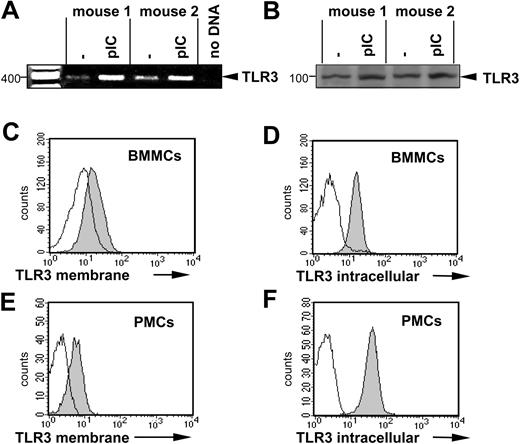
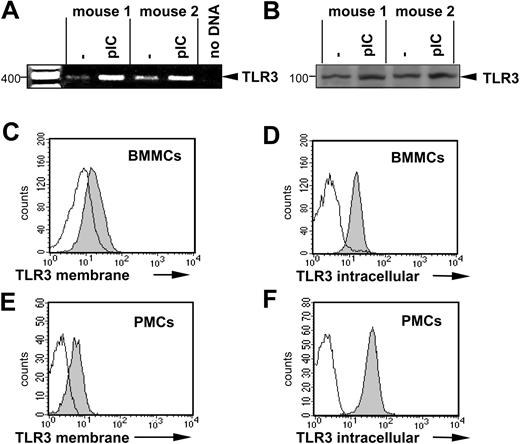
To investigate whether murine MCs express innate immune receptors, well known to detect dsRNA, we examined BMMCs and PMCs for TLR3 expression. BMMCs express and up-regulate the TLR3 message after stimulation with 10 μg/mL poly(I:C) for 1 hour as assessed by reverse transcription (RT)–PCR (Figure 2A). Western blotting confirmed the constitutive expression of TLR3 by BMMCs (Figure 2B) and FACS analyses detected TLR3 on the surface and in the cytoplasm of both BMMCs and PMCs (Figure 2C-F), confirming that BMMCs and PMCs express TLR3.
PMCs dramatically change their phenotype on poly(I:C) stimulation. PMCs were characterized as c-Kit+, FcϵRI+, and S/T2+ cells (A). Expression levels of I-A/E (B), CD80 (C), CD28 (D), CD21/35 (E), and FcγRII/III (F), as analyzed on gated c-Kit+, S/T2+ double-positive cells, were analyzed before and 24 hours after injection of poly(I:C) by flow cytometry. Representative data from one mouse per group (n = 4-10) are shown. Horizontal bars in panels C-F indicate the percentage of MCs expressing the indicated molecules. Shaded and open curves show staining with specific antibodies and isotype-matched control antibodies, respectively.
PMCs dramatically change their phenotype on poly(I:C) stimulation. PMCs were characterized as c-Kit+, FcϵRI+, and S/T2+ cells (A). Expression levels of I-A/E (B), CD80 (C), CD28 (D), CD21/35 (E), and FcγRII/III (F), as analyzed on gated c-Kit+, S/T2+ double-positive cells, were analyzed before and 24 hours after injection of poly(I:C) by flow cytometry. Representative data from one mouse per group (n = 4-10) are shown. Horizontal bars in panels C-F indicate the percentage of MCs expressing the indicated molecules. Shaded and open curves show staining with specific antibodies and isotype-matched control antibodies, respectively.
Poly(I:C) stimulation induces signal transduction and transactivation of a set of primary genes in BMMCs
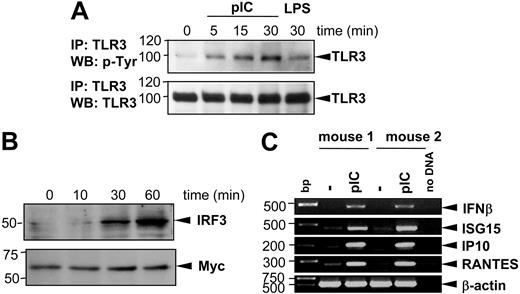
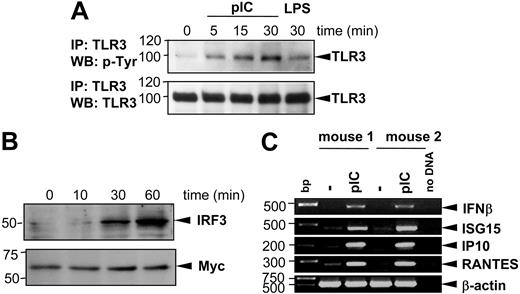
On dsRNA stimulation, TLR3 undergoes tyrosine phosphorylation.35,36 Furthermore, the transcriptional up-regulation of several primary genes, primarily IFN-β, depends on IRF3.24 To better characterize the downstream events in the poly(I:C)–mediated MC activation, we first assessed the extent and kinetics of TLR3 phosphorylation in BMMCs. To this end, BMMCs were stimulated for 5, 15, or 30 minutes with 10 μg/mL poly(I:C) and TLR3 was immunoprecipitated from cells lysates. Tyrosine phosphorylation of TLR3 was analyzed on 10% SDS-PAGE. Stimulation with 100 ng/mL LPS for 30 minutes was used as negative control. As demonstrated in Figure 3A, poly(I:C) induced phosphorylation of TLR3 after 5 to 30 minutes of stimulation, with LPS showing no significant phosphorylation induction. Secondly, we analyzed whether poly(I:C) treatment results in activation of the transcriptional factor IRF3 by investigating its translocation into the nucleus. Figure 3B shows that after 30 to 60 minutes of poly(I:C) stimulation IRF3 accumulates in nuclei extracts of activated BMMCs. Nuclear translocation of IRF3 after poly(I:C) stimulation leads to the transcription of primary response genes—IFNβ, IP10, ISG15, RANTES, and IFIT1.24 Therefore, to evaluate the influence of poly(I:C) stimulation on cytokine and chemokine expression, BMMCs were stimulated with 10 μg/mL poly(I:C) and harvested for RNA extraction. RT-PCR analyses were performed to assess the transcription level of several primary response genes, including IFNβ, ISG15, IP10, and RANTES. As shown in Figure 3C, poly(I:C) is a potent stimulus, which up-regulates the transcription of IFN-β, ISG15, IP10, and RANTES after 1 hour of stimulation, demonstrating that MCs react to dsRNA via activation of the TLR3 signaling cascade.
Expression of TLR3 in murine MCs. (A) BMMCs from 2 mice were left unstimulated or were stimulated for 1 hour with 10 μg/mL poly(I:C). RT-PCR was performed as described in “Materials and methods.” The amplified products were electrophoresed on 1.5% agarose gel. A mock PCR (without cDNA) was included to exclude contamination. The amount of cDNA analyzed was similar in different samples, as shown by PCR amplification of β-actin cDNA. (B) BMMCs were treated with 10 μg/mL poly(I:C) for 24 hours (pIC) or left untreated. Expression of TLR3 protein was analyzed by Western blotting in cell lysates using specific anti-TLR3 antibodies. Position of TLR3 is indicated on the right, and molecular mass is indicated on the left. Expression of TLR3 was shown by surface staining of BMMCs (C) or PMCs (E) as described in “Materials and methods.” Intracellular staining of BMMCs (D) or PMCs (F) for TLR3 was performed after fixation and permeabilization of cells. For surface staining of BMMCs, anti-TLR3 antibodies and biotinylated secondary anti–mouse IgG1 antibodies and APC-labeled streptavidin were used. For surface staining of PMCs and intracellular staining, A647-labeled anti-TLR3 antibodies were used. Gray and white areas show staining with anti-TLR3 and isotype-matched control antibodies, respectively. Results are representative of 4 independent experiments.
Expression of TLR3 in murine MCs. (A) BMMCs from 2 mice were left unstimulated or were stimulated for 1 hour with 10 μg/mL poly(I:C). RT-PCR was performed as described in “Materials and methods.” The amplified products were electrophoresed on 1.5% agarose gel. A mock PCR (without cDNA) was included to exclude contamination. The amount of cDNA analyzed was similar in different samples, as shown by PCR amplification of β-actin cDNA. (B) BMMCs were treated with 10 μg/mL poly(I:C) for 24 hours (pIC) or left untreated. Expression of TLR3 protein was analyzed by Western blotting in cell lysates using specific anti-TLR3 antibodies. Position of TLR3 is indicated on the right, and molecular mass is indicated on the left. Expression of TLR3 was shown by surface staining of BMMCs (C) or PMCs (E) as described in “Materials and methods.” Intracellular staining of BMMCs (D) or PMCs (F) for TLR3 was performed after fixation and permeabilization of cells. For surface staining of BMMCs, anti-TLR3 antibodies and biotinylated secondary anti–mouse IgG1 antibodies and APC-labeled streptavidin were used. For surface staining of PMCs and intracellular staining, A647-labeled anti-TLR3 antibodies were used. Gray and white areas show staining with anti-TLR3 and isotype-matched control antibodies, respectively. Results are representative of 4 independent experiments.
Poly(I:C) stimulation induces TLR3 phosphorylation, translocation of IRF3 into the nucleus, and increased expression of primary response genes (IFNβ, ISG15, IP10, RANTES) in BMMCs. BMMCs were stimulated with 10 μg/mL poly(I:C) for different time intervals as indicated. (A) TLR3 protein was precipitated from protein lysates of activated BMMCs using anti-TLR3 antibodies and subjected to 10% SDS-PAGE. Blots were analyzed by anti–p-Tyr antibodies (top blot). To prove that equal amounts of TLR3 protein were loaded in each sample, blots were stripped and reprobed with anti-TLR3 antibodies (bottom blot). Stimulation of BMMCs with 100 ng/mL LPS was used as negative control. Position of TLR3 is indicated on the right, and molecular mass is indicated on the left. IP indicates immunoprecipitation; WB, Western blotting. (B) Nuclear extracts from BMMCs were analyzed in 10% SDS-PAGE with anti-IRF3 antibodies (top blot). For loading control, expression of Myc protein was detected on the same membrane after stripping (bottom blot). (C) BMMCs generated from 2 mice were stimulated with 10 μg/mL poly(I:C) for 1 hour. Total RNA was extracted from cells, reverse transcribed, and subjected to PCR amplification using specific primers for IFN-β, ISG15, IP10, and RANTES as indicated. The amount of cDNA was equalized by PCR amplification of β-actin. A mock PCR (no DNA) was included as a negative control. The data represent 3 separate experiments with comparable results.
Poly(I:C) stimulation induces TLR3 phosphorylation, translocation of IRF3 into the nucleus, and increased expression of primary response genes (IFNβ, ISG15, IP10, RANTES) in BMMCs. BMMCs were stimulated with 10 μg/mL poly(I:C) for different time intervals as indicated. (A) TLR3 protein was precipitated from protein lysates of activated BMMCs using anti-TLR3 antibodies and subjected to 10% SDS-PAGE. Blots were analyzed by anti–p-Tyr antibodies (top blot). To prove that equal amounts of TLR3 protein were loaded in each sample, blots were stripped and reprobed with anti-TLR3 antibodies (bottom blot). Stimulation of BMMCs with 100 ng/mL LPS was used as negative control. Position of TLR3 is indicated on the right, and molecular mass is indicated on the left. IP indicates immunoprecipitation; WB, Western blotting. (B) Nuclear extracts from BMMCs were analyzed in 10% SDS-PAGE with anti-IRF3 antibodies (top blot). For loading control, expression of Myc protein was detected on the same membrane after stripping (bottom blot). (C) BMMCs generated from 2 mice were stimulated with 10 μg/mL poly(I:C) for 1 hour. Total RNA was extracted from cells, reverse transcribed, and subjected to PCR amplification using specific primers for IFN-β, ISG15, IP10, and RANTES as indicated. The amount of cDNA was equalized by PCR amplification of β-actin. A mock PCR (no DNA) was included as a negative control. The data represent 3 separate experiments with comparable results.
Mast cells release MIP-1β, KC, and RANTES, but do not degranulate, after activation via TLR3
Induction of primary response genes by poly(I:C) prompted us to analyze the cytokine and chemokine secretion by BMMCs stimulated with poly(I:C) in vitro. For this purpose, we stimulated BMMCs with 10 μg/mL poly(I:C) and analyzed the cytokine and chemokine production in their supernatants 48 hours later. As shown in Figure 4, poly(I:C) induces a strong production of macrophage inflammatory protein 1β (MIP-1β; Figure 4A), RANTES (Figure 4B), and keratinocyte-derived chemokine (KC) (Figure 4C). In contrast, BMMCs did not show an increased release of the proinflammatory cytokine IL-6 (Figure 4E), the TH2 cytokine IL-13 (Figure 4D), and the chemokine monocyte chemoattractant protein 1 (MCP-1; Figure 4F) after poly(I:C) treatment. Because LPS via TLR4 engagement is known to be a strong inductor of such responses, it was used here as a positive control. Thus, MCs on poly(I:C) treatment release a defined set of chemoattractants.
In response to FcϵRI cross-linking, adenosine triphosphate (ATP), or thrombin, MCs degranulate.37 In contrast it is controversial whether MCs degranulate in response to the TLR engagement.18,20,38-40 Therefore we investigated if TLR3 stimulation leads to degranulation. For this purpose we assayed the BMMC degranulation activity on poly(I:C) stimulation. Notably, poly(I:C) did not alter the levels of released β-hexosaminidase, a preformed MC granule component, in the supernatant of stimulated BMMCs (Figure 4G) or isolated peritoneal MCs (data not shown). Stimulation of BMMCs with PMA and ionomycin, IgE, and specific antigen, led to the expected degranulation. Taken together, activation of MCs via TLR3 induces the production of specific cytokines/chemokines not affecting MCs degranulation.
MCs are activated following NDV infection
Because MCs are activated by the recombinant dsRNA, poly(I:C), we further investigated whether similar effects could be induced by natural dsRNA viruses. For this purpose we infected BMMCs with NDV and investigated the TLR3 engagement, transcriptional activation of primary response genes, surface receptor expression, and chemokine production.
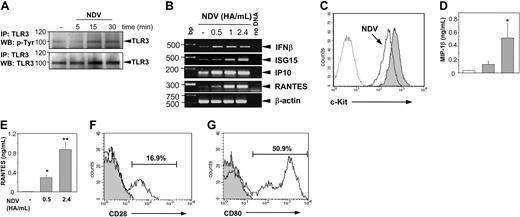
As shown in Figure 5A, NDV infection induces tyrosine phosphorylation of the TLR3 similarly to the poly(I:C) stimulation. Transcription of primary response genes is induced in a dose-dependent manner (Figure 5B). Interestingly, virus infection leads to a down-regulation of c-Kit expression (Figure 5C) as well. Furthermore, NDV-infected BMMCs are potent releasers of MIP-1β (Figure 5D) and RANTES (Figure 5E), a phenomenon depending on the virus concentrations used. Also, infection of PMCs with NDV in vitro leads to up-regulation of costimulatory molecules CD28 (Figure 5F) and CD80 (Figure 5G). Incubation of PMCs for 16 hours in the absence of NDV did not change the initial expression pattern of CD80 and CD28 shown in Figure 1C-D (data not shown).
Induction of chemokine production, but not degranulation, by stimulation of BMMCs with poly(I:C) in vitro. BMMCs were stimulated with 10 μg/mL poly(I:C) for 48 hours, supernatants were collected, and concentrations of indicated cytokines were measured by ELISA. MIP-1β (A), RANTES (B), KC (C), IL-13 (D), IL-6 (E), and MCP-1 (F) were measured in supernatants from poly(I:C)–treated BMMCs (▩) or left untreated (□). BMMCs were generated from 4 mice (*P < .05; **P < .001). Stimulation with 100 ng/mL LPS (D-F; ▦) was used as positive control. (G) Degranulation of BMMCs was induced by PMA and ionomycin (IO), and by IgE plus antigen (AG), but not by poly(I:C), as assessed by measuring released β-hexosaminidase in supernatants. BMMCs generated from 3 mice were analyzed in 3 independent experiments (data derived from one representative experiment; *P < .05).
Induction of chemokine production, but not degranulation, by stimulation of BMMCs with poly(I:C) in vitro. BMMCs were stimulated with 10 μg/mL poly(I:C) for 48 hours, supernatants were collected, and concentrations of indicated cytokines were measured by ELISA. MIP-1β (A), RANTES (B), KC (C), IL-13 (D), IL-6 (E), and MCP-1 (F) were measured in supernatants from poly(I:C)–treated BMMCs (▩) or left untreated (□). BMMCs were generated from 4 mice (*P < .05; **P < .001). Stimulation with 100 ng/mL LPS (D-F; ▦) was used as positive control. (G) Degranulation of BMMCs was induced by PMA and ionomycin (IO), and by IgE plus antigen (AG), but not by poly(I:C), as assessed by measuring released β-hexosaminidase in supernatants. BMMCs generated from 3 mice were analyzed in 3 independent experiments (data derived from one representative experiment; *P < .05).
Stimulation of MCs with NDV induces TLR3 phosphorylation, c-Kit down-regulation, chemokine production, and up-regulation of costimulatory molecules. Stimulation of BMMCs with NDV (2.4 HA/mL) for indicated time points leads to TLR3 phosphorylation (A). TLR3 protein was precipitated from protein lysates and subjected to 10% SDS-PAGE. Blots were analyzed by anti–p-Tyr antibodies (top blot). To prove that equal amounts of TLR3 protein were loaded in each sample, blots were stripped and reprobed with anti-TLR3 antibodies (bottom blot). Position of TLR3 is indicated on the right, and molecular mass is indicated on the left. (B) Transcription of primary response genes in BMMCs is induced after stimulation with NDV. BMMCs were stimulated with NDV for 1 hour in concentrations as indicated. Total RNA was extracted from cells, reverse transcribed, and subjected to PCR amplification using specific primers for IFN-β, ISG15, IP10, and RANTES. The amount of cDNA was equalized by PCR amplification of β-actin. A mock PCR (no DNA) was included as a negative control. The data are representative from 2 separate experiments with comparable results. (C) Stimulation of BMMCs with NDV (2.4 HA/mL) for 24 hours leads to down-regulation of c-Kit expression (open histogram) compared to unstimulated control (gray shaded histogram). Dotted histogram shows staining with isotype-matched control antibodies. Results are representative of 4 independent experiments. Productions of MIP-1β (D) and RANTES (E) were analyzed in supernatants of BMMCs in response to virus infection. BMMCs were stimulated with NDV in concentrations as indicated for 48 hours, supernatants were collected, and concentrations of MIP-1β and RANTES were measured by ELISA. NDV infection significantly increased the production of RANTES (▦) as compared with controls (□; *P < .05; **P < .001). BMMCs were generated from 3 individual mice. Production of chemokines was analyzed in 2 independent experiments; data from one experiment are shown. Stimulation of PMCs with NDV (2.4 HA/mL) for 16 hours led to up-regulation of CD28 (F) and CD80 expression (G). Expression levels were analyzed on gated c-Kit+, S/T2+ double-positive cells. Gray shaded histograms show staining with isotype-matched control antibodies. Results are representative of 2 independent experiments. PMCs isolated from 3 to 4 individual mice were analyzed in each experiment.
Stimulation of MCs with NDV induces TLR3 phosphorylation, c-Kit down-regulation, chemokine production, and up-regulation of costimulatory molecules. Stimulation of BMMCs with NDV (2.4 HA/mL) for indicated time points leads to TLR3 phosphorylation (A). TLR3 protein was precipitated from protein lysates and subjected to 10% SDS-PAGE. Blots were analyzed by anti–p-Tyr antibodies (top blot). To prove that equal amounts of TLR3 protein were loaded in each sample, blots were stripped and reprobed with anti-TLR3 antibodies (bottom blot). Position of TLR3 is indicated on the right, and molecular mass is indicated on the left. (B) Transcription of primary response genes in BMMCs is induced after stimulation with NDV. BMMCs were stimulated with NDV for 1 hour in concentrations as indicated. Total RNA was extracted from cells, reverse transcribed, and subjected to PCR amplification using specific primers for IFN-β, ISG15, IP10, and RANTES. The amount of cDNA was equalized by PCR amplification of β-actin. A mock PCR (no DNA) was included as a negative control. The data are representative from 2 separate experiments with comparable results. (C) Stimulation of BMMCs with NDV (2.4 HA/mL) for 24 hours leads to down-regulation of c-Kit expression (open histogram) compared to unstimulated control (gray shaded histogram). Dotted histogram shows staining with isotype-matched control antibodies. Results are representative of 4 independent experiments. Productions of MIP-1β (D) and RANTES (E) were analyzed in supernatants of BMMCs in response to virus infection. BMMCs were stimulated with NDV in concentrations as indicated for 48 hours, supernatants were collected, and concentrations of MIP-1β and RANTES were measured by ELISA. NDV infection significantly increased the production of RANTES (▦) as compared with controls (□; *P < .05; **P < .001). BMMCs were generated from 3 individual mice. Production of chemokines was analyzed in 2 independent experiments; data from one experiment are shown. Stimulation of PMCs with NDV (2.4 HA/mL) for 16 hours led to up-regulation of CD28 (F) and CD80 expression (G). Expression levels were analyzed on gated c-Kit+, S/T2+ double-positive cells. Gray shaded histograms show staining with isotype-matched control antibodies. Results are representative of 2 independent experiments. PMCs isolated from 3 to 4 individual mice were analyzed in each experiment.
MCs contribute to CD8+ T-cell recruitment after poly(I:C) injection in vivo
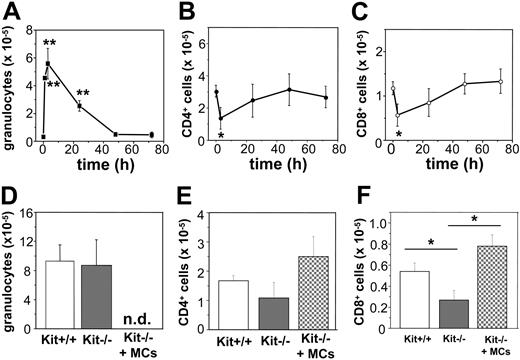
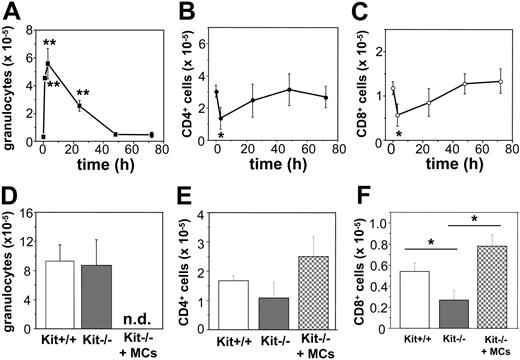
The chemokines we found to be produced in response to poly(I:C) or virus-infected MCs are known to be importantly involved in the recruitment of granulocytes and T cells to sites of antiviral host responses. Thus, we investigated whether the recruitment of such cells is MC dependent after intraperitoneal injections of poly(I:C) in C57BL/6 mice. Notably, injection of 10 μg poly(I:C) was followed by a rapid and pronounced intraperitoneal influx of granulocytes, which peaked 3 hours after injection. Afterward the number of granulocytes started to continuously drop, reaching normal levels at 48 hours after injection (Figure 6A). In contrast, intraperitoneal CD4+ and CD8+ T-cell populations diminished during the first 3 hours (Figure 6B-C), recovered, and reached normal levels 48 hours after injection.
To test whether MCs contribute to granulocyte recruitment or specifically modulate T-cell numbers in response to poly(I:C), we investigated genetically MC-deficient KitW/KitW-v mice46 and normal Kit+/+ mice for granulocyte and T-cell numbers at time 0 and 3 and 48 hours after poly(I:C) injection. KitW/KitW-v and Kit+/+ mice showed comparable and nonsignificant different numbers of T cells (CD4+: 1.30 ± 0.23 × 105 versus 1.82 ± 0.64 × 105; CD8+: 0.43 ± 0.10 × 105 versus 0.47 ± 0.13 × 105 in Kit+/+ and KitW/KitW-v mice [n = 4-7], respectively) and basically no granulocytes (data not shown) before treatment. After 3 hours, as shown in Figure 6D, KitW/KitW-v and Kit+/+ mice injected with poly(I:C) exhibited similar numbers of granulocytes, indicating that the absence of MCs does not significantly influence the early recruitment of granulocytes. On the other hand, 48 hours after poly(I:C) injection, KitW/KitW-v mice showed a slightly reduced number of CD4+ T cells as well as a significantly reduced numbers of CD8+ T cells compared with controls (Figure 6E-F). In fact, MC-deficient mice treated with poly(I:C) exhibited a 48% reduction in the CD8+ T-cell compartment compared with Kit+/+ mice (P < .05 at 48 hours). Reconstitution of KitW/KitW-v mice with BMMCs restores the intraperitoneal CD8+ T-cell compartment but exerts no significant effect on CD4+ T cells (Figure 6E-F), thus demonstrating that the recruitment of CD8+ T cells 48 hours after poly(I:C) injection, but not the granulocyte influx 3 hours after poly(I:C) injection, is strongly modulated by MCs.
Recruitment of CD8+ cells after poly(I:C) injection is reduced in MC-deficient WBB6F1-KitW/KitW-v (KitW/KitW-v) mice. Poly(I:C) injection in vivo in C57BL/6 mice leads to recruitment of granulocytes with a maximum after 3 hours (A). Numbers of CD4+ cells (B) and CD8+ cells (C) decreased during the first 3 hours after stimulation and recovered thereafter to reach background levels after 48 hours (*P < 0.05; **P < .001). Five to 10 mice were analyzed for each time point. (D-F) The influx of granulocytes in Kit+/+ (□), KitW/KitW-v mice (▦) 3 hours after poly(I:C) injection was comparable. Seven mice of each genotype were analyzed (D). Recovering of CD8+ T-cell numbers (F), but not CD4+ T-cell numbers (E), 48 hours after poly(I:C) injection was significantly reduced in KitW/KitW-v mice (▦; *P < .05) as compared to Kit+/+ mice (□) or KitW/KitW-v mice reconstituted with BMMCs (▩). Five to 8 mice of each genotype were analyzed; n.d. indicates not determined.
Recruitment of CD8+ cells after poly(I:C) injection is reduced in MC-deficient WBB6F1-KitW/KitW-v (KitW/KitW-v) mice. Poly(I:C) injection in vivo in C57BL/6 mice leads to recruitment of granulocytes with a maximum after 3 hours (A). Numbers of CD4+ cells (B) and CD8+ cells (C) decreased during the first 3 hours after stimulation and recovered thereafter to reach background levels after 48 hours (*P < 0.05; **P < .001). Five to 10 mice were analyzed for each time point. (D-F) The influx of granulocytes in Kit+/+ (□), KitW/KitW-v mice (▦) 3 hours after poly(I:C) injection was comparable. Seven mice of each genotype were analyzed (D). Recovering of CD8+ T-cell numbers (F), but not CD4+ T-cell numbers (E), 48 hours after poly(I:C) injection was significantly reduced in KitW/KitW-v mice (▦; *P < .05) as compared to Kit+/+ mice (□) or KitW/KitW-v mice reconstituted with BMMCs (▩). Five to 8 mice of each genotype were analyzed; n.d. indicates not determined.
MCs activated by poly(I:C) are chemotactic for CD8+ T cells
To understand whether the CD8+ T-cell chemotaxis shown in vivo is directly mediated by MCs, transwell double-chamber migration assays were performed.
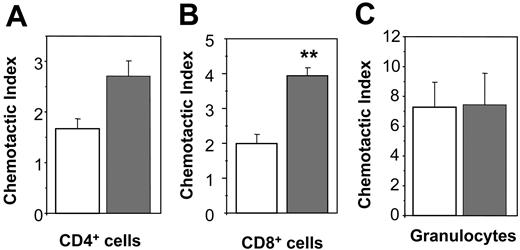
BMMCs stimulated with poly(I:C) induced a moderately increased migration of CD4+ T cells, that is, LN cells derived from MHC class II–restricted T-cell receptor transgenic (OTII) mice, compared to untreated BMMCs (Figure 7A). However, this difference was not statistically significant (CD4-chemotactic index: 2.7 ± 0.3 versus 1.7 ± 0.2 [P = .08] of poly(I:C) and unstimulated BMMCs, respectively). In contrast, chemotaxis induced by poly(I: C)–stimulated BMMCs in CD8+ T cells, that is, LN cells derived from MHC class I–restricted T-cell receptor transgenic mice (OTI mice), was found to be significantly increased compared to chemotactic responses elicited by unstimulated BMMCs (Figure 7B; 3.9 ± 0.2 versus 2.0 ± 0.3 for poly(I:C) stimulated versus unstimulated BMMCs, respectively; P < .01).
The chemotactic responses of granulocytes to untreated or 48-hour poly(I:C)–stimulated BMMCs were similar (Figure 7C). Taken together, these data confirm that CD8+ T cells are selectively recruited by MCs activated via TLR3 receptors in vitro, whereas granulocyte chemotaxis is constitutively induced by MCs and is independent of TLR3 activation.
Discussion
In this paper, we demonstrate that murine peritoneal as well as in vitro–generated BMMCs express TLR3, a member of the evolutionary conserved TLR family responsible for recognition of pathogen-associated molecular patterns.15 Both MC populations investigated express intracellular and membrane TLR3. Responses to the synthetic TLR3 ligand poly(I:C) involve the TLR3 downstream signaling cascade, resulting in the transcription of primary response genes, that is, IFNβ, ISG15, and IP10, the production and release of RANTES, MIP-1β, and KC, and up-regulation of MCH class II molecules, costimulatory molecules, and complement receptors. Furthermore, NDV-stimulated BMMCs phosphorylate TLR3, activate a set of primary TLR3-response genes, and release MIP-1β as well as KC. Most importantly, we found that poly(I:C)/TLR3-activated MCs elicit strong chemotactic responses in CD8+ T cells in vitro and that CD8+ T-cell recruitment to sites of poly(I:C) challenge is significantly impaired in the absence of MCs in vivo.
Induction of chemotaxis of CD8+ T cells, but not CD4+ T cells or granulocytes, by poly(I:C)–stimulated BMMCs in vitro. BMMCs were preincubated in the absence (□) or in the presence of 10 μg/mL poly(I:C) (▦) for 48 hours. To analyze migration of CD4+ T cells (A) or CD8+ T cells (B), LN cells from OTII or OTI mice were used, respectively. Percentages of migrated CD4+ or CD8+ T cells were determined by FACS and expressed as “chemotactic index” (**P < .001). One representative experiment of 3 is shown. Bone marrow cells were used to analyze migration of granulocytes. (C) Percentages of migrated granulocytes (Gr1+, CD11b+ double-positive cells) were determined by FACS. The “chemotactic index” was calculated as described in “Materials and methods.” One representative experiment of 3 is shown. BMMCs generated from 3 mice were analyzed in each experiment.
Induction of chemotaxis of CD8+ T cells, but not CD4+ T cells or granulocytes, by poly(I:C)–stimulated BMMCs in vitro. BMMCs were preincubated in the absence (□) or in the presence of 10 μg/mL poly(I:C) (▦) for 48 hours. To analyze migration of CD4+ T cells (A) or CD8+ T cells (B), LN cells from OTII or OTI mice were used, respectively. Percentages of migrated CD4+ or CD8+ T cells were determined by FACS and expressed as “chemotactic index” (**P < .001). One representative experiment of 3 is shown. Bone marrow cells were used to analyze migration of granulocytes. (C) Percentages of migrated granulocytes (Gr1+, CD11b+ double-positive cells) were determined by FACS. The “chemotactic index” was calculated as described in “Materials and methods.” One representative experiment of 3 is shown. BMMCs generated from 3 mice were analyzed in each experiment.
TLR3, one of the most important TLRs in the detection of viral infections,24,47 has been shown to be present on the surface or in the cytoplasm of dendritic cells,48,49 macrophages,50 epithelial cells,51,52 fibroblasts,53 and, most recently, natural killer (NK) cells.54,55 MCs have been shown to express TLR1 and TLR2, and TRL4-TLR9.16,19,38,56 In addition, expression of functional TLR3, TLR7, and TLR9 have recently been demonstrated on skin MCs,57 suggesting that MCs may detect viruses by mechanisms other than TLR3. TLR7-TLR9 are also described as receptors involved in viral recognition. For example, IFN-β production induced by herpes simplex virus 1 (HSV-1) and HSV-2, dsRNA viruses, is abrogated in the absence of TLR9.41,58 TLR9 and TLR3 both are required for an effective response to mouse cytomegalovirus (CMV) infection, and mice homozygous for a missense mutation in the Tlr9 allele show impaired lymphocytic choriomeningitis virus (LCMV) infection–induced secretion of type I interferons and NK-cell activation.42 Otherwise, ssRNA viruses could be recognized by TLR7, and an infection with vesicular somatitis virus (VSV) in vivo induces IFN-α production in a MyD88-dependent manner.43 So far, it is unknown, whether MCs could respond to viral RNA by other TLRs in a TLR3-independent manner and how the TLR3-dependent MC response is regulated by downstream adaptor proteins MyD88 (myloid differentiation factor 88) and TRIF (Toll/IL-1R domain-containing adapter inducing IFN-β). On the other hand, activation of MCs via TLR3 could occur and be relevant not only in antiviral immunity but also contribute to the recognition of endogenous danger signals. Heterologous mRNA, released from or associated with necrotic cells or generated by in vivo transcription,59 or siRNA, delivered extracellularly, stimulates TLR3 activation.60 These observations indicate that TLR3-dependent antiviral functions of MCs could be extended to a number of other processes, for example, clearing up inflammation sites or influencing the onset of autoimmune diseases.
Stimulation of MCs with poly(I:C), mimicking the natural TLR3 ligand dsRNA, is followed by rapid phosphorylation of TLR3 and results in 3 distinct responses, all of which could importantly contribute to a protective immunity against viral infections: (1) the production of IFN-β, (2) the up-regulation of costimulatory molecules, and (3) the release of chemokines including RANTES/CCL5 that are critical in regulating T-cell functions.
Our finding that BMMCs induce IFN-β mRNA expression after stimulation with poly(I:C) is in line with the observation that MCs can express IFN-β61 and that viruses and poly(I:C) induce IFN-β production by human MCs.62 MC-derived IFN-β could be important for normal host defense responses to viruses, as the production of type I IFNs is considered to be crucial for the induction of an antiviral protein production, the up-regulation of TLR3 expression, and the activation of macrophages, NK cells, and cytotoxic CD8+ T cells.63 IFN-β is also known to up-regulate the MHC class I expression, thereby promoting the recognition and destruction of virus-infected cells by T cells.
Here we show that expression of costimulatory molecules by MCs is up-regulated by poly(I:C) and NDV, indicating that MCs could shape adaptive antiviral CD4+ and CD8+ T-cell responses. Up-regulation of costimulatory molecules in response to bacteria and bacterial products is a general feature of antigen-presenting cells, including macrophages and dendritic cells, and strongly regulates the adaptive immune response.31,64 MCs can express MHC class I and II molecules, present antigen to T cells,65-67 and express CD28,68 CD80 and CD86,69 CD40L, and intercellular adhesion molecule 1 (ICAM-1).6 Expression of CD28 by BMMCs could be modulated by LPS.68 Complement receptors on MCs are important links between innate and adaptive immunity and can mediate the MC activation as well.70 Interestingly, the expression of c-Kit, the receptor for stem-cell factor, was found to be down-regulated after poly(I:C) injection in vivo and infection with NDV in vitro, indicating potential direct or indirect regulation of c-Kit expression by TLR3-dependent signals. At least 3 mechanisms for regulating the c-Kit expression on the cell surface are described—internalization after ligand binding,71,72 down-regulation by TH2 cytokines such as IL-10 and IL-4,73 and shedding from the membrane, for example, by tumor necrosis factor α–converting enzyme (TACE or ADAM-17).74 Why and how activated MCs down-regulate c-Kit after poly(I:C) injection or NDV infection remains to be clarified.
The cytokines and chemokines produced by MCs are known to profoundly alter the nature and the strength of the innate immune response. Our observations indicate that TLR3-stimulated MCs secrete a unique profile of chemokines independently of classical MC degranulation. These chemokines could determine the kinetics and extent of T-cell recruitment as well. Expression of chemokine receptors and migratory behavior is different for naive, activated, and memory T cells.75 CCR5 receptors for chemokines RANTES, MIP-1α, and MIP-1β are expressed on human memory and effector CD8+ T cells but not on naive CD8+ T cells.76 On the other hand, the CXCR3 chemokines IP10 and monokine induced by interferon-γ (MIG) act largely on activated T cells.77,78 Transgenic overexpression of IP10 in epithelial cells reportedly leads to increased accumulation of CD8+ cells in the lungs in murine models of asthma.79 RANTES, MIP-1β, and IP10, chemokines that have been shown to contribute to the recruitment of T cells in various pathologic settings,79-81 are induced on protein or mRNA level in BMMCs stimulated with poly(I:C) and NDV. Normal antiviral host responses require CD8+ T cells to migrate to sites of virus infection, where they recognize and kill infected cells. The partial reduction in the recruitment of CD8+ T cells observed after poly(I:C) challenge in the absence of MCs is most likely explained by the lack of MC-derived T-cell chemokines that in this particular model could not be compensated by chemokines produced by other cells but could be reconstituted in MC-deficient mice by reconstitution of the PMC compartment. Influx of granulocytes following poly(I:C) injection, on the other hand, is normal in genetically MC-deficient mice, indicating that MCs in antiviral host defense in this particular model, unlike other inflammatory reactions,82-84 are not required for the accumulation of granulocytes.
MCs play an important role in host defense responses against bacterial and parasitic pathogens, but their contribution to antiviral immunity is largely unknown. Increasing evidence indicates that MCs can be infected by viruses and respond to viral signals.85,86 For example, dengue virus and HIV have been shown to infect MCs and to induce MC cytokine and chemokine production in vitro.85,87,88 In addition, encephalomyocarditis virus infection reportedly results in MC chymase and tryptase production in vivo89 and viral infections have been shown to cause accumulation of MCs in the nasal mucosa during the first days of a symptomatic naturally acquired respiratory infection.90 However, the relevance and underlying mechanisms of MC infection and activation in settings of viral infections remain to be characterized in detail. Taken together, our observations suggest that MCs can detect viral infections via TLR3 and that TLR3-activated MCs contribute to antiviral host defense. Understanding the unique properties of MCs, the regulation of their cytokine and chemokine responses by viruses or viral products, and their interaction with specific T-cell subpopulations will be essential for the therapeutic manipulation of local immune responses and antiviral immunity.
Supported in part by grants SFB 367, Project C11 (S.B.-P.) and MA 1909/4-2 (M. Maurer) from the Deutsche Forschungsgemeinschaft.
Prepublished online as Blood First Edition Paper, April 19, 2005; DOI 10.1182/blood-2004-07-2656.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Katrin Streeck, Mandy Schröder, Martina Hein, and Renate Bergmann for excellent technical assistance; Andra Schromm for labeling of TLR3 antibodies; Florian Schiemann for help with chemotaxis assay; and Jodie Urcioli, Annette Wallisch, and Holger Heine for critical reading of the manuscript.