Abstract
Natural killer (NK) cells activate quickly in response to pathogens, tumors, and allogeneic hematopoietic cell transplants. Modulating the NK cell response are clonally distributed NK cell receptors that survey cells for change in the expression of major histocompatibility complex (MHC) class I and structurally related ligands. Here the enzyme-linked immunospot (ELISPOT) assay, intracellular cytokine staining (ICS), and short-term culture were used to quantify the response of bulk NK cell populations from human donors to HLA class I–deficient 221 cells and to 221 cells transfected with single HLA class I allotypes. NK cells in cultures containing interleukin-2 (IL-2) or IL-12 exhibited specificities of HLA class I–mediated inhibition that correlated well with those previously defined using NK cell clones in long-term culture and with the frequencies of cells expressing particular inhibitory HLA class I receptors. Culture with IL-12, but not IL-2, gave an increased frequency of cells expressing CD94: NKG2A but no change in killer immunoglobulin-like receptor (KIR) expression. For some heterozygote combinations of KIR3DL1 alleles, ICS can be used to compare the functional properties of the 2 allotypes. Thus, both the low-expressing KIR3DL1*005 and the high-expressing KIR3DL1*002 gave similar inhibitory response on challenge with an HLA-B*5801 ligand. The single-cell assays developed here should facilitate future population study and clinical analysis of human NK cell regulation by MHC class I.
Introduction
Natural killer (NK) cells are innate immune lymphocytes active in host defense against pathogens and tumors. They mediate these activities through direct killing of transformed or infected cells and production of cytokines such as interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF).1 NK cell functions are regulated by cytokine feedback loops, contact with dendritic cells, immune complexes, and direct interactions with the infected or transformed cells that serve as targets for their cytotoxicity.2-4 NK cell specificity for diseased cells is determined by a balance of signals generated by diverse stimulatory and inhibitory receptors.5 Several inhibitory NK cell receptors have specificity for major histocompatibility complex (MHC) class I allotypes. These receptors survey for MHC class I molecules on potential targets, the NK cells being inhibited by cells with normal expression but responding to cells with compromised levels of class I on their surface.6,7 In the context of allogeneic hematopoietic cell transplantation, the specificity of inhibitory NK cell receptors for class I can have significant impact on graft rejection, graft versus host disease (GVHD), and graft versus leukemia (GVL) effects.8-10
A key feature of NK cell class I receptor families is their extensive diversity, which plays out within individuals and across populations of individuals and species.11-13 The distribution of multiple receptor families and multiple receptors within a family on NK cell subsets accounts for the diversity within an individual's NK cell receptor repertoire.13-15 In humans, families of lectinlike and immunoglobulin (Ig)–like membrane glycoproteins share the responsibility of detecting host cell class I expression, thereby maintaining NK cell tolerance.13 The inhibitory receptors include the lectinlike receptor CD94:NKG2A that recognizes the nonclassical HLA-E class I molecule and is encoded in the NK complex (NKC) on chromosome 12.16 HLA-E is brought to the surface by association with peptides cleaved from the leader sequences of most classical HLA-A, -B, and -C allotypes.17,18 In this way HLA-E expression is used by CD94:NKG2A as a sensor for the overall HLA class I level of a cell.
The Ig-like receptors are encoded by genes in the leukocyte receptor complex (LRC) of chromosome 19 and are divided into 2 families: killer Ig-like receptors (KIRs) and the leukocyte Ig-like receptors (LILRs).19 The LILRs (previously called immunoglobulin-like transcript [ILT] or leukocyte immunoglobulin-like receptor [LIR]) are broadly reactive toward HLA-A, -B and -C allotypes and, similarly to CD94:NKG2A, allow NK cells to survey for overall class I expression.20-22 By contrast, individual KIR family members exhibit finer specificity for HLA class I allotypes and can distinguish between groups of HLA-A, -B, and -C allotypes.13,23-29 The more focused specificity of inhibitory KIR governs human NK cell alloreactions and likely renders NK cells responsive to selective alterations to class I expression on virally infected and/or transformed cells.30
Stimulatory members of the lectinlike (CD94:NKG2C/D) and Ig-like (KIR2/3DS) receptors have also been described, some of which have overlapping class I specificity and antibody epitopes with their inhibitory counterparts.14 The exact role of these stimulatory receptors remains a subject for debate; however, their capacity to activate NK cells is usually dominated by the negative signals generated through inhibitory KIR and/or CD94:NKG2A class I recognition.13 Demonstration that NKG2D recognizes MHC class I related chain A/B (MICA/B) and Rae1, and that a stimulatory class I receptor of mice recognizes a murine cytomegalovirus (MCMV) class I–like decoy protein, suggests that the stimulatory class I receptors may have evolved to detect class I–like structures encoded by genes located within and away from the MHC.31-34
The specificities of both the inhibitory and stimulatory members of KIR, ILT-2, and CD94:NKG2 receptor families have mainly been assessed with assays of NK cell function, although direct binding studies have also been used.35-39 The functional determination of receptor specificity has usually employed cytolytic assays with NK cell lines or clones as effectors and class I–deficient cells transfected with individual class I alleles as targets. This approach has allowed the identification of the several KIR, LILR, and CD94:NKG2 specificities for HLA class I. Limiting the approach is its need for activated NK cell clones and lines derived from long-term culture in high doses of interleukin-2 (IL-2).13,24-29 Given that a defining aspect of NK cell biology is the ability to work early in the immune response, we have investigated the HLA class I specificity of inhibitory NK cell receptors in short-term (12 to 24 hours) cytokine-cultured NK cells purified from the peripheral blood of human donors. By adapting the enzyme-linked immunospot (ELISPOT) technique for IFN-γ and granzyme B secretion and intracellular cytokine staining (ICS) for IFN-γ, we studied the response of NK cells to class I–deficient cells and their inhibition by HLA class I allotypes under conditions more like those of an early-stage immune response.
Materials and methods
Cells and cytokines
Epstein-Barr virus (EBV)–transformed B-cell lines, MHC class I–deficient 721.221, and 221-B*5801, -Cw*0304, -Cw*0401, and -Cw*0702 class I transfectants were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 μg/mL l-glutamine, and an antibiotic mixture of penicillin and streptomycin. The expression of class I molecules in 721.221 transfectants was checked weekly by using W6/32 (anticlass I) antibody. Recombinant human IL-2 was kindly provided by Dr R. E. Aurigemma, National Cancer Institute, Frederick, MD. Recombinant human IL-12 was purchased from Sigma Aldrich (St Louis, MO).
KIR genotyping and KIR3DL1 allele typing
Peripheral blood mononuclear cells (PBMCs) were purified from buffy coats derived from the blood of healthy donors and obtained from Stanford Blood Center (Stanford, CA) by separation on a Ficoll-Hypaque gradient. Genomic DNA was prepared from PBMCs using a QIAamp Blood kit (Qiagen, Valencia, CA). KIR typing of genomic DNA was performed as described previously.40,41 KIR3DL1 allele typing was performed as described previously42 with the addition of one primer pair (forward: 5′-CCATCGGTCCCATGATGCT-3′, reverse: 5′-ACGTTCATGGGCTCCCCG-3′) that amplifies 3DL1*002.
NK cell purification
Purified, resting NK cells were obtained from frozen PBMCs by magnetic depletion of T and B lymphocytes and monocytes using a cocktail of biotin-conjugated monoclonal antibodies against CD3, CD4, CD14, CD15, CD19, CD36, and CD123, antibiotin microbeads, LS separation columns, and midiMACS magnet (Miltenyi Biotec, Auburn, CA). After depletion, NK cell preparations (CD56+/CD3–) were from 85% to 95% pure, and contamination with CD14+, CD19+, and CD3+ cells was less than 1% and was, for lineage-negative cells, 5% to 15%.
ELISPOT assay
ELISPOT plates and reagents for IFN-γ and granzyme B were purchased from BD Biosciences (Mountain View, CA). For each set of conditions the number of NK cells per well was established in preliminary experiments. In the presence of IL-2 or IL-12, NK cells were 2 × 103 to 3 × 103 NK cells per well and 2 × 104 to 3 × 104 cells per well for IFN-γ and granzyme B ELISPOT, respectively. In the absence of cytokines, the NK cell number was 2 × 104 to 3 × 104 NK cells per well and 2 × 105 NK cells per well for IFN-γ and granzyme B ELISPOT, respectively. NK cells were mixed with 721.221 target cells (parental or singly transfected with class I alleles) at a ratio of 1:10 and plated in duplicate or triplicate in 200 μL assay medium per well at 37°C and 7.5% CO2 for 20 to 24 hours of stimulation. Control wells were either targets or effectors alone in media in the presence or absence of cytokines. In blocking experiments, target cells (221 or 221-transfectants) were first incubated for 1 hour with monoclonal antibody (mAb) DX17 (anti-HLA class I, kindly provided by Dr L. Lanier, University of California, San Francisco) before the ELISPOT assay was performed. When GL-183 (anti-KIR2DL2/3; Beckman-Coulter, Brea, CA) was used, NK cells were first incubated for 1 hour with mAb and then targets were added. ELISPOT assays were performed according to the manufacturer's instructions. The spots were counted off the images of each well that were obtained from scanning and analysis of the plates with an ImmunoSpot Series 2 Analyzer (Cellular Technology, Cleveland, OH). The ImmunoSpot Series 2 Analyzer consists of an analog camera (Sony DXC-390). The images were scanned with the ImmunoSpot Image Acquisition software version 3 and were analyzed with ImmunoSpot Analysis software version 2.08 with a resolution of 490 × 480 pixels per well.
Antibodies and flow cytometry
For the analysis of NK cell receptor expression, mAbs EB6-PE (anti-KIR2DL1/KIRDS1), GL-183–phycoerythrin (GL-183–PE) (anti-KIR2DL2/KIR2DL3/KIR2DS2) (Beckman-Coulter-Immunotech), DX9-PE (anti-KIR3DL1; BD Biosciences), and DX31-PE (anti-KIR3DL2; generously provided by Dr J. H. Phillips, DNAX Research Institute, Palo Alto, CA) were used in combination with Z199 (anti-NKG2A; Beckman-Coulter-Immunotech) conjugated with Alexa Fluor 488 by using a monoclonal antibody labeling kit (Molecular Probes, Eugene, OR) and with CD85j-CyChrome (anti-LILRB1; BD Biosciences) in 3-color flow cytometry (FACScan; BD Biosciences) of purifed NK cells from each donor. Analysis was performed using CellQuest software (BD Biosciences). Subsets of KIR+ CD94:NKG2A+, KIR+ CD94:NKG2A–, and KIR– CD94:NKG2A+ NK cells were detected in FL1 and FL2 channels. Subsets of LILRB1+ KIR– CD94:NKG2A– NK cells were detected by gating on the KIR– LILRB1+ (FL2 and FL3) and subsequent analysis of that population for expression of CD94:NKG2A and LILRB1 (FL1 and FL3). Depletion of CD94:NKG2A+ NK cells was obtained by sorting of purified NK cells using the Z199 antibody. Experiments with 5(and-6)–carboxyfluorescein diacetate succinimidyl ester (CFSE) were performed by labeling NK cells with 10 μM CFSE (Molecular Probes) followed by incubation at 37°C at a cell density of 2 × 107/mL in Hanks balanced salt solution (HBSS) without FBS. After 10 minutes, cells were washed in cold HBSS supplemented with 20% serum and then cultured in RPMI, 10% FBS at 2 × 105/200 μL per well. Positive controls for the assay were CFSE-labeled PBMCs from the same donor stimulated with 5 μg/mL phytohemagglutinin (PHA). Cells were harvested after 1, 3, and 6 days and analyzed by flow cytometry.
Intracellular staining of IFN-γ
Purified NK cells were mixed with target cells (721.221 or 221 transfected with class I) or without target cells (as control) in medium alone or with IL-2 (1000 to 3000 U/mL) at an effector-target (E/T) ratio of 1:1 in sterile round-bottom 96-well plates, centrifuged at 100 g for 1 minute, and incubated at 37°C and 7.5% CO2 for approximately 12 to 14 hours. Higher doses of IL-2 were required for ICS experiments than those used for ELISPOT studies. Golgi Plug (1:1000) containing brefeldin A (BD Biosciences) was added 1 hour after the mixing of effectors and targets to inhibit cytokine secretion. Cells were prepared for flow cytometry using the protocol developed by BD Biosciences with slight modification. Briefly, the dead cell discrimination reagent (Miltenyi Biotec) that distinguishes dead cells in fixed samples was added to the cells. After incubation for 10 minutes under a light source for photolabeling of dead cells, antibodies for NK cell receptors were added for 30 minutes in the dark at 4°C. Cells were fixed and permeabilized using Cytofix/Cytoperm solution (BD Biosciences), and IFN-γ–fluorescein isothiocyanate (IFN-γ–FITC) (clone B27; BD Biosciences) was added for 30 minutes at 4°C in the dark. Three-color flow cytometry (FACScan; BD Biosciences) was performed. The results were analyzed by using FlowJo software. A gate was drawn on NK cells with exclusion of dead cells (in FL3-positive channel) and target cells (based on side and forward scatter).
Calculating the proportion of NK cells inhibited by 221 cells bearing single class I allotypes
As a modification of the methods used to calculate inhibition of NK clone cytotoxicity by class I–bearing targets, we used 2 simple calculations for our ELISPOT and ICS results. (1) ELISPOT: 1 – (no. of spots following incubation with 221 class I transfected/no. of spots following incubation with 221 parental cells) × 100 = percent inhibition; and (2) ICS: 1 – (% of IFN-γ–positive NK cells following incubation with 221 transfectants/% of IFN-γ–positive NK cells following incubation with 221 parental) × 100 = percent inhibition.
Results
An ELISPOT technique for the study of class I–mediated regulation of NK cells
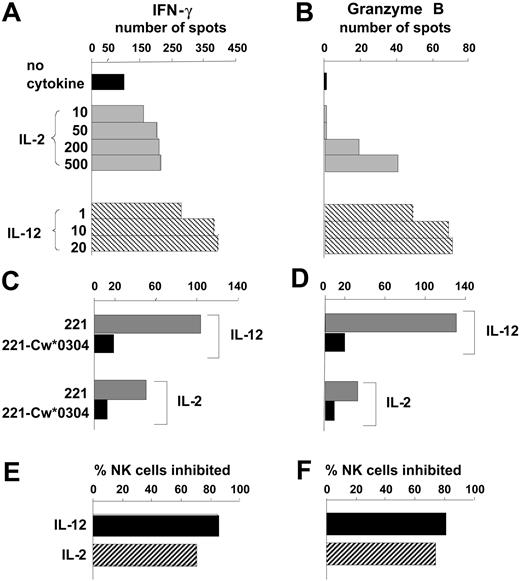
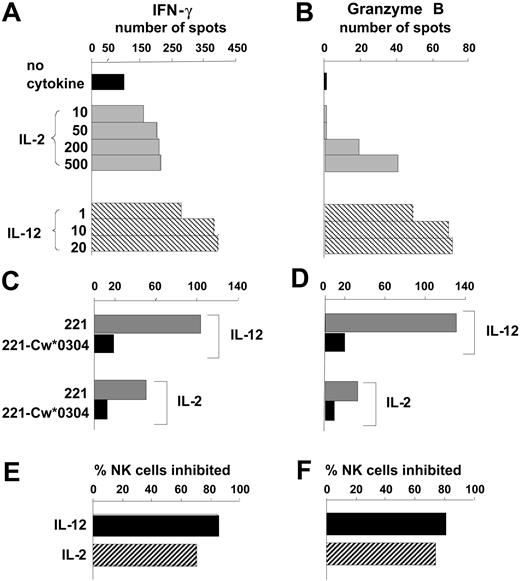
As a new approach for analyzing the regulation of NK cells by MHC class I, we adapted the ELISPOT technique to measure the IFN-γ and granzyme B secreted by individual cells in a bulk NK cell response to the class I–deficient B-cell line 221. Granzyme B and IFN-γ were chosen as markers for the 2 major NK cell functions: cytotoxicity and cytokine secretion. The number of NK cells secreting IFN-γ or granzyme B was increased by inclusion of either IL-2 or IL-12 in the assay culture, with IL-12 consistently being more potent than IL-2. (Figure 1A-B).
Quantifying the effects of IL-12 and IL-2 on human NK cell responses to HLA class I–deficient cells. Purified NK cells were tested by ELISPOT assay for secretion of IFN-γ (A) and granzyme B (B) in a 20- to 24-hour response to the HLA class I–deficient B-cell line 221. Comparison was made between assays performed in the absence of cytokines (▪) with those made in the presence of different concentrations of IL-2 (▦) and IL-12 (▧). Cytokine concentrations are in units per milliliter for IL-2 and nanograms per milliliter for IL-12. Data are presented as the mean number of spots counted in duplicate wells and are representative of at least 5 independent experiments. Using single concentrations of cytokine (IL-12 at 10 ng/mL, IL-2 at 250 U/mL), similar assays compared NK cell secretion of IFN-γ (C) and granzyme B (D) in response to 221 transfectants expressing HLA-Cw*0304 (▪) with that in response to untransfected 221 cells (▦). From the results in panels C and D the proportion of NK cells that responded to 221 but were inhibited by 221-Cw*0304 was calculated (E and F, respectively). ▪ shows the inhibition of NK cells incubated with IL-12; ▨ shows the inhibition of NK cells incubated with IL-2.
Quantifying the effects of IL-12 and IL-2 on human NK cell responses to HLA class I–deficient cells. Purified NK cells were tested by ELISPOT assay for secretion of IFN-γ (A) and granzyme B (B) in a 20- to 24-hour response to the HLA class I–deficient B-cell line 221. Comparison was made between assays performed in the absence of cytokines (▪) with those made in the presence of different concentrations of IL-2 (▦) and IL-12 (▧). Cytokine concentrations are in units per milliliter for IL-2 and nanograms per milliliter for IL-12. Data are presented as the mean number of spots counted in duplicate wells and are representative of at least 5 independent experiments. Using single concentrations of cytokine (IL-12 at 10 ng/mL, IL-2 at 250 U/mL), similar assays compared NK cell secretion of IFN-γ (C) and granzyme B (D) in response to 221 transfectants expressing HLA-Cw*0304 (▪) with that in response to untransfected 221 cells (▦). From the results in panels C and D the proportion of NK cells that responded to 221 but were inhibited by 221-Cw*0304 was calculated (E and F, respectively). ▪ shows the inhibition of NK cells incubated with IL-12; ▨ shows the inhibition of NK cells incubated with IL-2.
Human NK cell clones and lines are frequently susceptible to inhibition by HLA-C, because members of all 3 inhibitory NK cell receptor families recognize HLA-C allotypes.43 KIRs distinguish 2 groups of HLA-C allotypes defined by presence of either asparagine or lysine at position 80. KIR2DL1 recognizes allotypes having lysine at position 80, which are represented in this study by Cw*0401. KIR2DL2 and KIR2DL3 recognize allotypes having asparagine at position 80, which are represented here by Cw*0304 and Cw*0702.24 Both LILRB1 (previously called LIR-1 or ILT2) and CD94:NKG2A recognize a broad range of HLA-C allotypes, the latter through HLA-C leader peptides bound to HLA-E.17,20,21,44 Because of the prevalence of HLA-C–mediated inhibition, it was first examined using the ELISPOT system.
By comparison with untransfected cells, 221 cells transfected with HLA-Cw*0304 (221-Cw*0304) stimulated fewer NK cells to secrete IFN-γ (Figure 1C) or granzyme B (Figure 1D). For both the IFN-γ and granzyme B response, the HLA-C–mediated inhibition was slightly less for NK cells cultured with IL-2 than for those cultured with IL-12 (Figure 1D-E). IL-12 was also more effective than IL-2 in raising the frequency of NK cells that responded to 221 cells (Figure 1A-B). Analogous experiments, in which 221 transfected with HLA-Cw*0401 (221-Cw*0401) was used to inhibit the NK cells from several donors, gave similar results (data not shown). These observations show that cytokine production (eg, IFN-γ) and cytotoxicity (eg, granzyme B) are inhibited to similar effect when inhibitory NK cell receptors (KIR, CD94:NKG2A, LILRB1) engage HLA-C ligands on target cells. Because it required fewer NK cells than the granzyme B ELISPOT, the IFN-γ ELISPOT was used as the main assay of effector function for most subsequent experiments in this study.
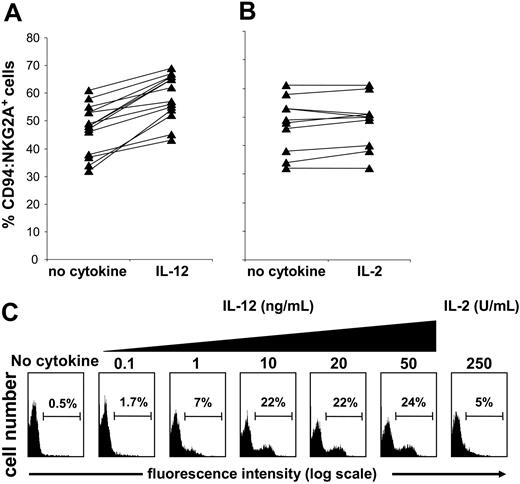
IL-12 increases the frequency of CD94:NKG2A+ NK cells
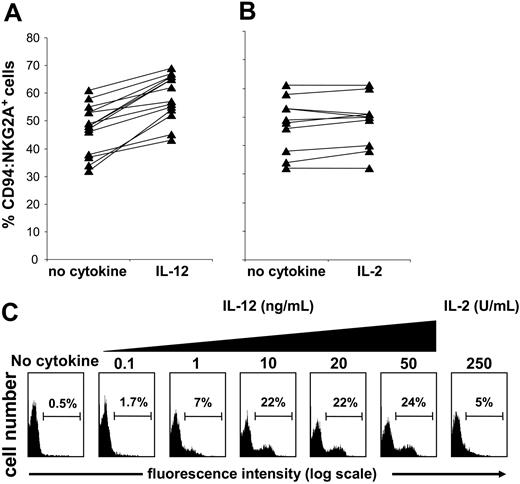
Finding increased sensitivity of IL-12–treated NK cell populations to class I inhibition led us to seek for changes in inhibitory NK cell receptor expression that occur on culture with IL-12. Comparison of KIR, CD94:NKG2A, and LILRB1 expression by purified NK cells following incubation with IL-12 revealed an increased frequency of CD94:NKG2A expression (Figure 2A) but not for KIR or LILRB1 (data not shown). When NK cells were similarly incubated with IL-2 there was no significant change in CD94: NKG2A expression (Figure 2B) or in KIR or LILRB1 (data not shown).
Short-term culture with IL-12 increases the frequency of CD94:NKG2A+NK cells. Purified NK cells from different donors were cultured in medium alone or medium supplemented with IL-12 (10 ng/mL) (A) or IL-2 (250 U/mL) (B) for 24 hours. The NK cells were then stained with anti-CD94:NKG2A (mAb Z199) and analyzed by flow cytometry. Culture with IL-12 gave a median increase in CD94:NKG2A-expressing NK cells of about 12% (n = 14; P < .000 03), whereas culture with IL-2 had no significant effect (n = 11; P > .05). Purified NK cells were depleted of the CD94:NKG2A+ population. The depleted cell population was cultured for 24 hours in the absence of cytokine (control), with 250 U/mL IL-2, or with increasing doses of IL-12 (0.1 and 50 ng/mL) and then assayed for CD94:NKG2A expression (C).
Short-term culture with IL-12 increases the frequency of CD94:NKG2A+NK cells. Purified NK cells from different donors were cultured in medium alone or medium supplemented with IL-12 (10 ng/mL) (A) or IL-2 (250 U/mL) (B) for 24 hours. The NK cells were then stained with anti-CD94:NKG2A (mAb Z199) and analyzed by flow cytometry. Culture with IL-12 gave a median increase in CD94:NKG2A-expressing NK cells of about 12% (n = 14; P < .000 03), whereas culture with IL-2 had no significant effect (n = 11; P > .05). Purified NK cells were depleted of the CD94:NKG2A+ population. The depleted cell population was cultured for 24 hours in the absence of cytokine (control), with 250 U/mL IL-2, or with increasing doses of IL-12 (0.1 and 50 ng/mL) and then assayed for CD94:NKG2A expression (C).
To assess the phenomenon further, purified NK cells were first depleted of CD94:NKG2A+ cells and then incubated with increasing concentrations of IL-12 for 1 day. After this treatment, more than 20% of the cells expressed CD94:NKG2A (Figure 2C). Extending the IL-12 treatment for several days led to further increase in the frequency of CD94:NKG2A+ NK cells (data not shown). Uncertain was whether the effect of IL-12 was to induce expression of CD94:NKG2A in NK cells that lack this receptor or to drive the proliferation of residual CD94: NKG2A+ NK cells that escaped depletion. When CFSE labeling was used to assess for proliferation in cultures depleted of CD94:NKG2A+ NK cells, we obtained no evidence that IL-12 had induced cell division in the population as a whole or any particular subset (data not shown). In contrast, IL-2 did induce cell division in such cultures but without a major increase in the frequency of CD94:NKG2A+ NK cells. These results, and the short exposure (20 to 24 hours) necessary to see the effect, favor the interpretation that IL-12 induces CD94:NKG2A expression on a subset of NK cells lacking detectable levels of this receptor.
Correlations between class I receptor expression and functional inhibition of resting and short-term–activated NK cells
To assess the contribution of different receptors to HLA-C–mediated inhibition of NK cell function, we sought donors with KIR genotyes that lack the stimulatory KIRs (KIR2DS1, 2DS2, and 2DS3). These stimulatory KIRs could have complicated the analysis in 2 ways: first, by interacting with HLA-C to activate NK cells and, second, by cross-reacting with the monoclonal antibodies used to detect inhibitory HLA-C–specific KIRs.45
NK cells from the selected donors (Figure 3) were purified and challenged with either 221, 221-Cw*0304 (Figure 4A), or 221-Cw*0401 (Figure 4C) in the presence of IL-2, IL-12, or no cytokine. NK cell secretion of IFN-γ in these cultures was assayed by ELISPOT (Figure 4A,C). The purified NK cells (CD3–CD56+) from each donor were also subjected to 3-color flow cytometric analysis to determine the frequency of NK cells expressing HLA-C–reactive inhibitory receptors: KIR, CD94:NKG2A, and LILRB1 (data not shown). The proportion of cells having one or more inhibitory receptors that can bind to the inhibiting HLA-C allotype gave a prediction for the frequency of NK cells that would be inhibited when challenged by that allotype. Such predictions were made from analysis performed on freshly isolated NK cells as well as cells cultured for 20 to 24 hours with cytokines. The proportion of NK cells expressing at least one inhibitory receptor for class I did not change following incubation in the absence of cytokines or with IL-2, whereas culture with IL-12 gave higher values for some HLA class I allotypes (eg, for donor 11; see the legend to Figure 4) because of the increased frequency of expression of CD94:NKG2A induced by IL-12 (Figure 2).
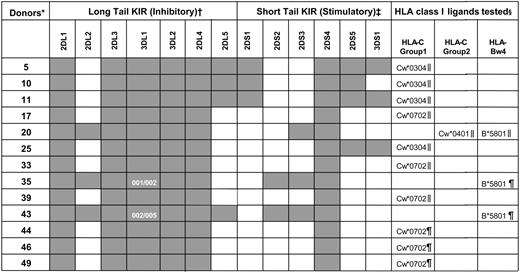
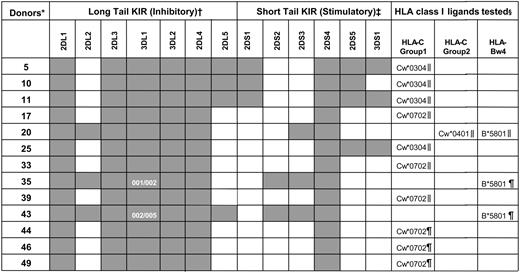
KIR genotypes for the human NK cells used in functional studies
KIR genotyping was performed on the donor-derived blood cells used in the functional studies (see “Materials and methods”). Dark boxes indicate the presence of a KIR gene and white boxes their absence. KIR3DL1 allele typing for donors 35 and 43 is also shown.
*Donors were coded numerically.
†Inhibitory KIR genes (2DL1-5 and 3DL1-2) encode for class I receptors with long cytoplasmic tails containing immunoreceptor tyrosine-based inhibitory motifs (ITIMs).16 Despite possessing a long cytoplasmic tail with ITIMs, KIR2DL4 has potential for both inhibitory and stimulatory function.46-49
‡Stimulatory KIR genes (2DS1-5 and 3DS1) encode receptors with short cytoplasmic tails that lack ITIMs.
§ELISPOT (∥) or ICS (¶) analyses were used to determine the proportion of NK cells inhibited by the indicated class I allotypes.
KIR genotypes for the human NK cells used in functional studies
KIR genotyping was performed on the donor-derived blood cells used in the functional studies (see “Materials and methods”). Dark boxes indicate the presence of a KIR gene and white boxes their absence. KIR3DL1 allele typing for donors 35 and 43 is also shown.
*Donors were coded numerically.
†Inhibitory KIR genes (2DL1-5 and 3DL1-2) encode for class I receptors with long cytoplasmic tails containing immunoreceptor tyrosine-based inhibitory motifs (ITIMs).16 Despite possessing a long cytoplasmic tail with ITIMs, KIR2DL4 has potential for both inhibitory and stimulatory function.46-49
‡Stimulatory KIR genes (2DS1-5 and 3DS1) encode receptors with short cytoplasmic tails that lack ITIMs.
§ELISPOT (∥) or ICS (¶) analyses were used to determine the proportion of NK cells inhibited by the indicated class I allotypes.
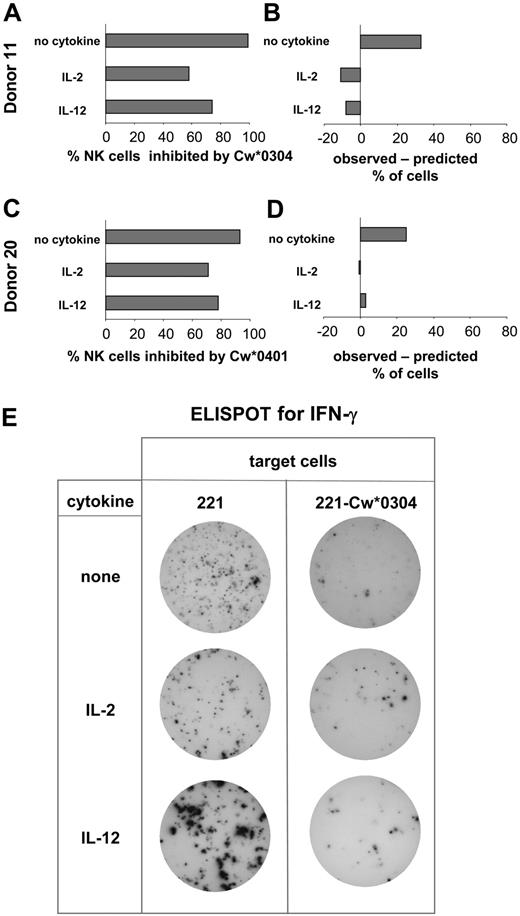
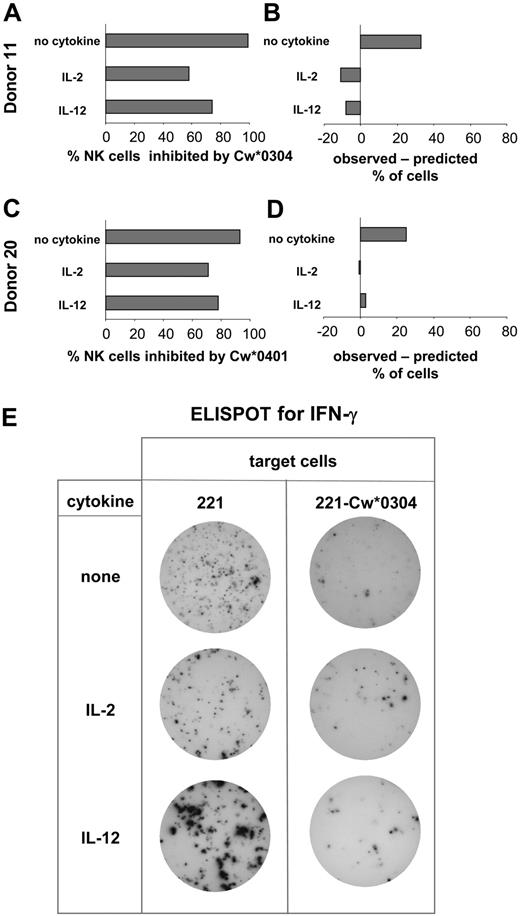
Comparison of the NK cell expression of specific inhibitory receptors and functional inhibition by HLA-C. Based upon KIR genotype, donors were selected for having inhibitory KIR2DL with specificity for HLA-C and no cross-reactive stimulatory KIR2DS. NK cells from these donors were assayed by ELISPOT for secretion of IFN-γ. Assays were performed in the presence and absence of IL-2 or IL-12. Target cells were either 221, 221-Cw*0304 (A), or 221-Cw*0401 (C). Of the cells responding to 221, the percentage inhibited by presence of HLA-C ligand was calculated. For each donor these observed values were compared with predictions based upon the percentage of NK cells expressing either the cognate inhibitory KIRs or other inhibitory HLA-C–reactive receptors (CD94:NKG2A and LILRB) as determined by flow cytometry using specific monoclonal antibodies (B,D). (A,B) The analysis of inhibition of NK cells by 221-Cw*0304. The data shown for donor 11 is representative of that obtained with 4 donors (donors 5, 10, 11, and 25) (Figure 3). For donor 11 the frequency of NK cells expressing inhibitory HLA-Cw*0304–reactive receptors was 66% after culture without cytokine; 69% after culture with IL-2; and 82% in the IL-12–treated cultures (where CD94:NKG2A expression increases; Figure 2). These values were obtained by summing the frequencies of the following NK cell subsets: KIR2DL3+ CD94:NKG2A+, KIR2DL3+ CD94:NKG2A–, KIR2DL3– CD94: NKG2A+, and KIR2DL3– CD94:NKG2A– LILRB1+. (C-D) The inhibition of NK cells from donor 20 (Figure 3) by HLA-Cw*0401; 2 independent experiments gave similar results. For donor 20 the frequency of NK cells expressing inhibitory HLA-Cw*0401–reactive receptors was 64% after culture without cytokine, 71% after culture with IL-2, and 75% after culture with IL-12. These values were obtained by summing the frequencies of the following NK cell subsets: KIR2DL1+ CD94:NKG2A+, KIR2DL1+ CD94:NKG2A–, and KIR2DL1– CD94:NKG2A+. Because LILRB1 recognizes HLA-Cw*0304 but not HLA-Cw*0401,44 the KIR2DL1– CD94:NKG2A– LILRB1+ subset (22% of NK cells of donor 20) was not included in the NK cells predicted to be inhibitable by HLA-Cw*0401. (E) Representative data obtained in the ELISPOT assay. NK cells producing IFN-γ were identified in each well of a 96-well microtiter plate. Analysis was performed on duplicate wells of NK cells challenged with 221 or 221-Cw*0304 cells. To obtain sufficient spots from NK cells cultured without cytokines, 10-fold more NK cells were plated per well than for NK cells cultured with cytokine.
Comparison of the NK cell expression of specific inhibitory receptors and functional inhibition by HLA-C. Based upon KIR genotype, donors were selected for having inhibitory KIR2DL with specificity for HLA-C and no cross-reactive stimulatory KIR2DS. NK cells from these donors were assayed by ELISPOT for secretion of IFN-γ. Assays were performed in the presence and absence of IL-2 or IL-12. Target cells were either 221, 221-Cw*0304 (A), or 221-Cw*0401 (C). Of the cells responding to 221, the percentage inhibited by presence of HLA-C ligand was calculated. For each donor these observed values were compared with predictions based upon the percentage of NK cells expressing either the cognate inhibitory KIRs or other inhibitory HLA-C–reactive receptors (CD94:NKG2A and LILRB) as determined by flow cytometry using specific monoclonal antibodies (B,D). (A,B) The analysis of inhibition of NK cells by 221-Cw*0304. The data shown for donor 11 is representative of that obtained with 4 donors (donors 5, 10, 11, and 25) (Figure 3). For donor 11 the frequency of NK cells expressing inhibitory HLA-Cw*0304–reactive receptors was 66% after culture without cytokine; 69% after culture with IL-2; and 82% in the IL-12–treated cultures (where CD94:NKG2A expression increases; Figure 2). These values were obtained by summing the frequencies of the following NK cell subsets: KIR2DL3+ CD94:NKG2A+, KIR2DL3+ CD94:NKG2A–, KIR2DL3– CD94: NKG2A+, and KIR2DL3– CD94:NKG2A– LILRB1+. (C-D) The inhibition of NK cells from donor 20 (Figure 3) by HLA-Cw*0401; 2 independent experiments gave similar results. For donor 20 the frequency of NK cells expressing inhibitory HLA-Cw*0401–reactive receptors was 64% after culture without cytokine, 71% after culture with IL-2, and 75% after culture with IL-12. These values were obtained by summing the frequencies of the following NK cell subsets: KIR2DL1+ CD94:NKG2A+, KIR2DL1+ CD94:NKG2A–, and KIR2DL1– CD94:NKG2A+. Because LILRB1 recognizes HLA-Cw*0304 but not HLA-Cw*0401,44 the KIR2DL1– CD94:NKG2A– LILRB1+ subset (22% of NK cells of donor 20) was not included in the NK cells predicted to be inhibitable by HLA-Cw*0401. (E) Representative data obtained in the ELISPOT assay. NK cells producing IFN-γ were identified in each well of a 96-well microtiter plate. Analysis was performed on duplicate wells of NK cells challenged with 221 or 221-Cw*0304 cells. To obtain sufficient spots from NK cells cultured without cytokines, 10-fold more NK cells were plated per well than for NK cells cultured with cytokine.
For NK cells cultured with IL-2 or IL-12 there was fair agreement between the frequency of inhibited NK cells observed in the ELISPOT assay and the frequency of inhibitable cells predicted from the flow cytometric analysis (Figure 4B,D). In contrast, the observed inhibition of NK cells cultured without cytokine was greater than predicted from the distribution of inhibitory HLA-C–reactive receptors (Figure 4B,D).
In continuing to study this difference, we further simplified the experimental system so that the action of a single inhibitory receptor could be studied. This was achieved by using HLA-Cw*0702 as the inhibitory ligand; it interacts with inhibitory KIR but does not furnish an HLA-E binding peptide to make a CD94:NKG2A ligand.13,17 In addition, LILRB1 does not interact with HLA-Cw*0702.44 When 221 transfectants expressing this allotype were used as inhibitors for NK cells from donors selected for KIR genotype that lacked cross-reactive stimulatory receptors, inhibition was predicted to be due to a single inhibitory KIR.
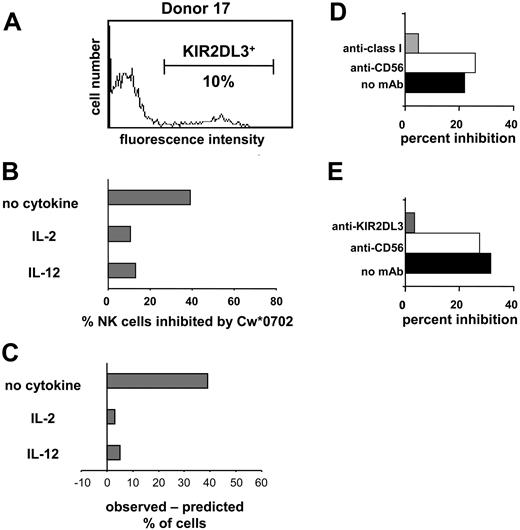
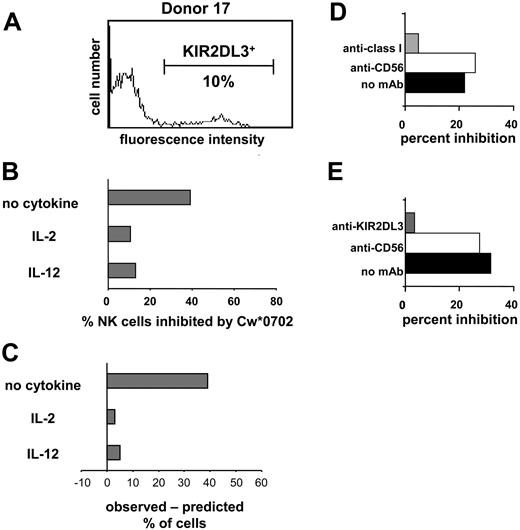
HLA-Cw*0702 is a ligand for KIR2DL2 and 2DL3.13,25,50 Because donor 17 lacked genes for KIR2DL2 and the stimulatory receptors KIR2DS2/2DS3, the GL-183 monoclonal antibody could be used to enumerate NK cells expressing KIR2DL3. An experiment analogous to that shown in Figure 4 was performed with 221-Cw*0702 cells as inhibitors, and similar results were obtained. For NK cells incubated with IL-2 or IL-12, the observed Cw*0702-mediated inhibition of NK cells corresponded well with that predicted from the frequency of KIR2DL3 expression (Figure 5). For NK cells incubated without cytokine the observed inhibition was much greater than that predicted. That inhibition in this assay was dependent upon both HLA class I and KIR2DL3 was demonstrated by adding mAb specific to either class I (Figure 5D) or KIR2DL3 (Figure 5E), both of which reversed the inhibitory effect. Our results with unstimulated (resting) NK cells indicate that, in the absence of proinflammatory cytokines, NK cell populations are more susceptible to inhibition by class I as compared with that observed previously for NK cell lines and clones.
Inhibition of the IFN-γ response of NK cells through cognate interaction between HLA class I and KIR. Donor 17 expresses KIR2DL3 but not the serologically cross-reactive KIR2DL2 and KIR2DS2 (Figure 3). Flow cytometric analysis with the GL-183 antibody (anti-KIR2DL2/3, 2DS2) was used to determine the frequency (10%) of NK cells of donor 17 that express KIR2DL3 (A) and are potentially inhibitable by HLA-Cw*0702. NK cells from donor 17 were cultured in medium alone, IL-2, or IL-12; challenged with 221 or 221-Cw*0702 cells; and assayed for IFN-γ by ELISPOT. (B) The proportion of NK cells that made IFN-γ in response to 221 but were inhibited by 221-Cw*0702. (C) The difference between the observed frequency of NK cells inhibited by HLA-Cw*0702 and that predicted from the frequency of NK cells expressing KIR2DL3. (D-E) Blocking of the 221-Cw*0702–mediated inhibition with anti-HLA class I (DX17; ▦) (D) or anti-KIR2DL3 antibody (GL-183; ▦) (E). Anti-CD56 (Leu19; □) was included as a control antibody, which binds to NK cells but does not affect their function. ▪ indicates no mAb. NK cells were cultured in IL-2 with target cells, and antibodies were added at 10 μg/mL. NK cells from donors 33 and 39 were used for the experiments shown in panels D and E, respectively. (KIR2DL3+ NK cells were 22% and 36% for donors 33 and 39, respectively.)
Inhibition of the IFN-γ response of NK cells through cognate interaction between HLA class I and KIR. Donor 17 expresses KIR2DL3 but not the serologically cross-reactive KIR2DL2 and KIR2DS2 (Figure 3). Flow cytometric analysis with the GL-183 antibody (anti-KIR2DL2/3, 2DS2) was used to determine the frequency (10%) of NK cells of donor 17 that express KIR2DL3 (A) and are potentially inhibitable by HLA-Cw*0702. NK cells from donor 17 were cultured in medium alone, IL-2, or IL-12; challenged with 221 or 221-Cw*0702 cells; and assayed for IFN-γ by ELISPOT. (B) The proportion of NK cells that made IFN-γ in response to 221 but were inhibited by 221-Cw*0702. (C) The difference between the observed frequency of NK cells inhibited by HLA-Cw*0702 and that predicted from the frequency of NK cells expressing KIR2DL3. (D-E) Blocking of the 221-Cw*0702–mediated inhibition with anti-HLA class I (DX17; ▦) (D) or anti-KIR2DL3 antibody (GL-183; ▦) (E). Anti-CD56 (Leu19; □) was included as a control antibody, which binds to NK cells but does not affect their function. ▪ indicates no mAb. NK cells were cultured in IL-2 with target cells, and antibodies were added at 10 μg/mL. NK cells from donors 33 and 39 were used for the experiments shown in panels D and E, respectively. (KIR2DL3+ NK cells were 22% and 36% for donors 33 and 39, respectively.)
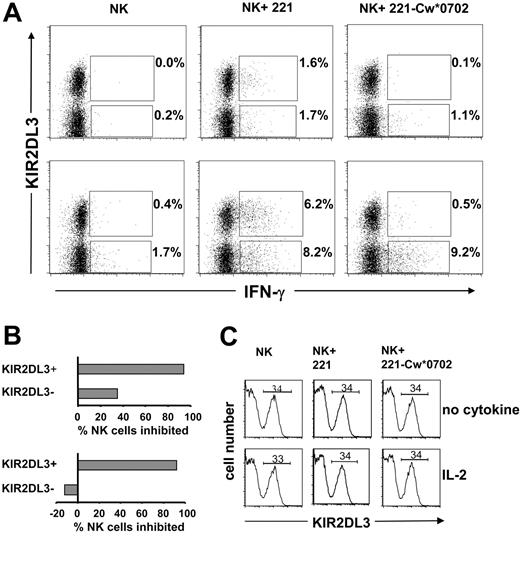
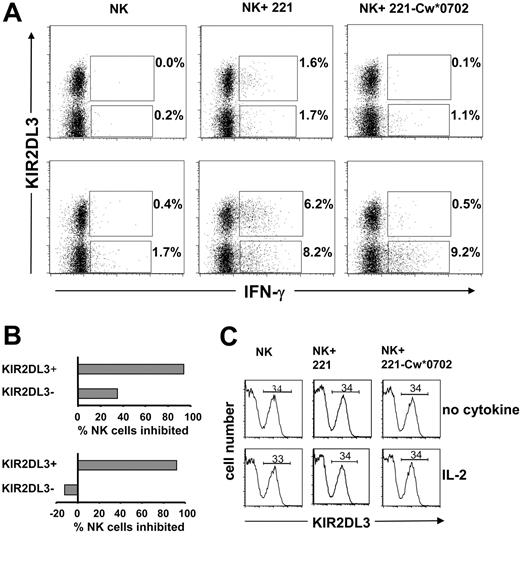
A flow cytometric approach was also used to assess the frequency and phenotype of NK cells secreting IFN-γ following a 12-hour incubation with target cells. As shown in Figure 6, this method was used to examine the responses of KIR2DL3+ and KIR2DL3– NK cells to 221 cells and their inhibition by HLA-Cw*0702. Similar proportions of KIR2DL3+ and KIR2DL3– NK cells made IFN-γ in response to 221 cells (Figure 6A, center panels), whereas few cells responded in the absence of target cells (Figure 6A, left panels). NK cells incubated with (Figure 6A, lower panels) or without IL-2 (Figure 6A, upper panels) showed the effect, but the frequency of cells responding with IL-2 was higher, as expected from previous ELISPOT analysis (Figures 1, 4, and 5). On culture with 221-Cw*0702 cells, the frequency of responding KIR2DL3– NK cells was comparable to that against 221, whereas the KIR2DL3+ NK cells were mostly inhibited (Figure 6A, right panels). The results for assays containing IL-2 demonstrate that Cw*0702-mediated inhibition of NK cells was restricted to the subpopulation expressing the cognate KIR2DL3 receptor (Figure 6B, lower panel). Although a similar bias was seen for NK cells challenged in the absence of cytokine, there was also some Cw*0702-mediated inhibition of KIR2DL3– NK cells (Figure 6B, upper panel). In these assays the expression of KIR2DL3 was not affected by the presence or the nature of the target cell or by the presence or absence of IL-2 (Figure 6C). Neither was HLA class I expression by the target cells affected by the presence or absence of IL-2 (data not shown).
Analysis of KIR2DL3+and KIR2DL3–NK cell IFN-γ production upon interaction with HLA-Cw*0702. Purifed NK cells from donor 49 were incubated with (bottom row) or without IL-2 (1000 U/mL) (top row) in the presence of 221 or 221-Cw*0702 cells for 12 hours with brefeldin A. To detect the frequency of KIR2DL3+ IFN-γ–positive cells, NK cells were stained with anti-KIR2DL3–PE (GL-183) and anti-IFN-γ–FITC mAbs and analyzed by flow cytometry (A). Cells analyzed were 30 000 for NK cells incubated with target cells and 20 000 for NK cells incubated alone. The gate was drawn to exclude dead NK cells and B-cell targets. The values reported in the upper quadrants are the frequencies of KIR2DL3+ NK cells producing IFN-γ; shown in the lower quadrants are the proportions of KIR2DL3– NK cells producing IFN-γ. The percentage of NK cells inhibited by HLA-Cw*0702 was calculated from the frequencies of KIR2DL3+ or KIR2DL3– NK cells producing IFN-γ observed following culture with 221 cells compared with those observed following culture with 221-Cw*0702 (B). Top graph, no cytokine; bottom graph, incubation with IL-2. For donor 49 the frequency of KIR2DL3+ cells (34%) was determined by flow cytometry using anti-KIR2DL3 (GL-183). The frequency did not change upon a 12-hour incubation with IL-2 or when 221 or 221-Cw*0702 was present (C). Top row, no cytokine; bottom row, incubation with IL-2. Data are representative of results obtained with 3 donors (44, 46, and 49; Figure 3).
Analysis of KIR2DL3+and KIR2DL3–NK cell IFN-γ production upon interaction with HLA-Cw*0702. Purifed NK cells from donor 49 were incubated with (bottom row) or without IL-2 (1000 U/mL) (top row) in the presence of 221 or 221-Cw*0702 cells for 12 hours with brefeldin A. To detect the frequency of KIR2DL3+ IFN-γ–positive cells, NK cells were stained with anti-KIR2DL3–PE (GL-183) and anti-IFN-γ–FITC mAbs and analyzed by flow cytometry (A). Cells analyzed were 30 000 for NK cells incubated with target cells and 20 000 for NK cells incubated alone. The gate was drawn to exclude dead NK cells and B-cell targets. The values reported in the upper quadrants are the frequencies of KIR2DL3+ NK cells producing IFN-γ; shown in the lower quadrants are the proportions of KIR2DL3– NK cells producing IFN-γ. The percentage of NK cells inhibited by HLA-Cw*0702 was calculated from the frequencies of KIR2DL3+ or KIR2DL3– NK cells producing IFN-γ observed following culture with 221 cells compared with those observed following culture with 221-Cw*0702 (B). Top graph, no cytokine; bottom graph, incubation with IL-2. For donor 49 the frequency of KIR2DL3+ cells (34%) was determined by flow cytometry using anti-KIR2DL3 (GL-183). The frequency did not change upon a 12-hour incubation with IL-2 or when 221 or 221-Cw*0702 was present (C). Top row, no cytokine; bottom row, incubation with IL-2. Data are representative of results obtained with 3 donors (44, 46, and 49; Figure 3).
Functional inhibition of NK cells expressing KIR3DL1 allotypes with high and low levels of KIR3DL1
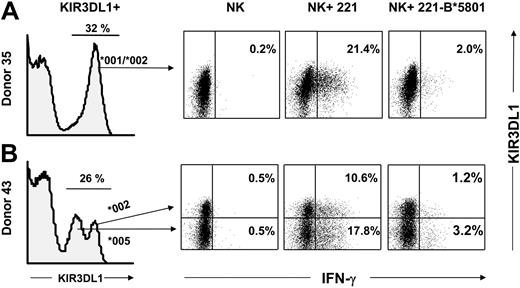
A feature of the polymorphism of KIR3DL1 is that some allotypes are distinguished by different levels of cell surface expression.51,52 Flow cytometric analysis of IFN-γ production by NK cells provided a method for direct comparison of the functional potential of allotypes expressed at different levels. The analysis of 2 donors, both of whom are KIR3DL1 heterozygotes, is shown in Figure 7. Donor 35 is heterozygous for the KIR3DL1*001 and KIR3DL1*002 alleles, both of which are expressed at high level (Figure 6A, left panel). Donor 43 expresses KIR3DL1*002 at high levels and KIR3DL1*005 at low levels. For donor 43, the histogram of NK cell staining with anti-KIR3DL1 mAb (DX9 antibody) comprised 3 subpopulations of cells: negative cells, cells with low mAb binding that express KIR3DL1*005, and cells with high binding that express KIR3DL1*002 (Figure 7B, far left panel). In addition, a small fraction of the high-binding cells are expected to express both alleles.51
Comparable HLA-B*5801–mediated inhibition of NK cell subsets expressing different KIR3DL1 allotypes. Purified NK cells from donors heterozygous for KIR3DL1 allotypes were tested for IFN-γ production in response to 221 or 221-B*5801 in the presence of IL-2 (3000 U/mL). As a control, NK cells were incubated with IL-2 in the absence of any target. NK cells were stained with DX9-PE (anti-KIR3DL1) and CD85j-CyChrome (anti-LILRB1) to gate on the NK subpopulation that expresses KIR3DL1 but not LILRB1. Dead cells and target cells were excluded from the gate. Cells were stained intracellularly with anti-IFN-γ–FITC mAb, and 100 000 cells were analyzed. Results are shown from KIR3DL1+ populations. (A) Donor 35 is heterozygous for KIR3DL1*001/*002 alleles and exhibits a unimodal pattern of KIR3DL1 expression as determined by staining with DX9 mAb. Percentages of IFN-γ–producing cells are reported in the right quadrants. (B) Donor 43 is heterozygous for 3DL1*002/*005 alleles and therefore exhibits a bimodal distribution of NK cells expressing KIR3DL1: the low-expressing cells express 3DL1*005 whereas the high-expressing cells express 3DL1*002 or both alleles.51 IFN-γ–producing cells in each subset are shown in the upper right and lower right quadrants. Donor KIR genotypes are shown in Figure 3.
Comparable HLA-B*5801–mediated inhibition of NK cell subsets expressing different KIR3DL1 allotypes. Purified NK cells from donors heterozygous for KIR3DL1 allotypes were tested for IFN-γ production in response to 221 or 221-B*5801 in the presence of IL-2 (3000 U/mL). As a control, NK cells were incubated with IL-2 in the absence of any target. NK cells were stained with DX9-PE (anti-KIR3DL1) and CD85j-CyChrome (anti-LILRB1) to gate on the NK subpopulation that expresses KIR3DL1 but not LILRB1. Dead cells and target cells were excluded from the gate. Cells were stained intracellularly with anti-IFN-γ–FITC mAb, and 100 000 cells were analyzed. Results are shown from KIR3DL1+ populations. (A) Donor 35 is heterozygous for KIR3DL1*001/*002 alleles and exhibits a unimodal pattern of KIR3DL1 expression as determined by staining with DX9 mAb. Percentages of IFN-γ–producing cells are reported in the right quadrants. (B) Donor 43 is heterozygous for 3DL1*002/*005 alleles and therefore exhibits a bimodal distribution of NK cells expressing KIR3DL1: the low-expressing cells express 3DL1*005 whereas the high-expressing cells express 3DL1*002 or both alleles.51 IFN-γ–producing cells in each subset are shown in the upper right and lower right quadrants. Donor KIR genotypes are shown in Figure 3.
Incubation of purified NK cells with IL-2 or IL-12 had no effect on the pattern of DX9 binding, indicating that the frequencies and the levels of allelic expression were maintained (data not shown). NK cells were cultured with 221 or 221-B*5801 and assessed by flow cytometry for production of IFN-γ. Costaining was performed with DX9 and anti-LILRB1 (mAb CD85j) so that the analysis could be directed at NK cells that express KIR3DL1 but not LILRB1, to which HLA-B*5801 also binds.53
On incubation of donor 35's NK cells with 221 cells, some 21.4% of the NK cells responded by making IFN-γ, a response not seen in the absence of target cells, and was inhibited by 90% when B*5801-221 was the target. On incubation of donor 43's NK cells with 221 cells, IFN-γ was made by NK cells with high and low levels of KIR3DL1 expression. The frequency of the response was higher for NK cells expressing a low level of KIR3DL1 (17.8%) than for those expressing a high level (10.6%) In contrast, few cells (less than 1%) made IFN-γ in the absence of a target cell. NK cells incubated with 221-B*5801 were largely inhibited in their IFN-γ response, and similar inhibitory effect was observed for cells with high (89% inhibition) and low (82% inhibition) levels of KIR3DL1 (Figure 7, far right panel). This result shows that both the high-expressed allotype (3DL1*002) and the low-expressed allotype (3DL1*005) bind to HLA-B*5801 and deliver inhibitory signals with comparable functional effect.
Discussion
NK cells are nonresponsive to healthy autologous cells, because healthy cells lack the ligands for activating NK cell receptors and their MHC class I molecules engage inhibitory receptors that prevent NK cell activation.7,54,55 Previously, we studied NK cell cytotoxicity and its inhibition by HLA class I using established clones of IL-2–cultured human NK cells clones.13 Here, we have used the ELISPOT and ICS techniques to study NK cell functions and their inhibition by HLA class I in short-term cultures (12 to 24 hours) of NK cells isolated from peripheral blood. Comparison of cytotoxicity and cytokine production, as assessed by granzyme B and IFN-γ secretion, respectively, gave similar results. When NK cells were cultured short-term with IL-2 or IL-12 they were found to have inhibitory specificities for HLA class I that were the same as those previously defined for NK cell clones in long-term cultures.13,23-29 Thus there was concordance between the ELISPOT/ICS approach and the more established methods. That NK cell clones had the same HLA class I specificities as NK cells activated with a tempo closer to that of an in vivo innate immune response increases confidence in the results previously obtained with NK cell clones.
Previous reports have shown that IL-12, IL-15, and transforming growth factor-β (TGF-β) induce the expression of CD94/NKG2A on activated human T lymphocytes.56-58 Analogous to this, we observed that culture with IL-12 increased the frequency of NK cells expressing the CD94:NKG2A receptor, while IL-2 had minimal effect on receptor expression. Although not definitive, our results point to this increase being due to induction of CD94:NKG2A expression rather than selective expansion of cells expressing this receptor. In contrast, no change in either KIR or LILRB expression was detected, as previously reported.59 Both IL-2 and IL-12 activate NK cell functions in similar ways; however, IL-12 acts early on NK cell functions in vivo, whereas IL-2, a product of antigen-specific Th cells, acts later.3,60 IL-12 is produced by diverse populations of antigen-presenting cells, being secreted rapidly and in large quantities by dendritic cell (DC) subsets.61-63 By increasing the abundance of NK cells expressing CD94: NKG2A, an inhibitory HLA class I receptor with broad allotype specificity, the IL-12 made early in the immune response might serve to activate NK cells while simultaneously increasing the inhibitory potential of certain subsets through their increased CD94:NKG2A expression. This dual effect of IL-12, a product of DCs, could impact the outcome of NK-DC encounters by altering the stimulatory/inhibitory balance of the “NK-DC control switch.”64-66 Indeed, CD94:NKG2A-expressing NK cell populations have been shown to interact with autologous DCs differently from that observed for KIR+ subsets.67
For the NK cells cultured with IL-2 or IL-12, the frequency of cells that were inhibited by a particular HLA class I allotype correlated well with the frequency of NK cells that carried an inhibitory KIR specific for the allotype. This was not the case for “control” NK cells cultured in the absence of cytokine. These NK cells secreted IFN-γ in response to class I–deficient 221 cells but less so than cells cultured with IL-2 or IL-12. In addition, these resting populations were also more sensitive to inhibition by HLA class I than their cytokine-treated counterparts. That the frequency of resting NK cells inhibited by a particular HLA class I type was in excess of the frequency of cells expressing an inhibitory KIR specific for the allotype—and this was altered by cytokine treatment—suggests that NK cell sensitivity to class I inhibiton is not fixed. This was seen most clearly in our flow cytometric analyses of NK cell IFN-γ production, where a proportion of resting NK cells lacking the HLA-Cw*0702 inhibitory receptor, KIR2DL3, were still inhibited by 221 target cells expressing HLA-Cw*0702 (Figure 6). Taken as a whole, the ELISPOT and ICS analyses of NK cell populations indicate that the balance of stimulatory and inhibitory signals delivered to NK cells upon target cell encounter is modified rapidly by IL-2 and IL-12. Indeed, after only 12 to 24 hours of culture both cytokines elicit NK cell populations exhibiting class I specificity remarkably similar to that observed for long-term–cultured NK cell lines and clones. From the perspective of NK cell responses in vivo, our results argue that resting NK cell interactions with inhibitory class I allotypes are more likely to maintain tolerance to autologous cells in the absence of pathologic conditions (eg, infection or malignant transformation) when cytokines are not produced. Moreover, resting NK cells might also be more sensitive to inhibition by low levels of class I expression such as those found on nonhematopoietic cells (hepatocytes, neurons).
KIR3DL1 is a highly polymorphic locus for which certain allotypes have different levels of cell surface expression, and the functional implications of these expression levels was not clear. Using ICS to analyze NK cells from KIR3DL1 heterozygotes, we could assess and compare the HLA-B–mediated inhibition for different KIR3DL1 allotypes. Both high-(3DL1*002) and low-expressing allotypes (3DL1*005) gave similar frequencies of NK cell inhibition upon engagement with HLA-B*5801. With the ELISPOT and ICS methods, HLA class I–mediated inhibition and other aspects of NK cell function and specificity can be determined at high resolution using fewer cells in mixed populations and without cloning. In particular, the low cell numbers and the ease and speed of the methods should be amenable to population analysis and clinical studies.
Prepublished online as Blood First Edition Paper, November 4, 2004; DOI 10.1182/blood-2004-08-3174.
Supported by National Institutes of Health (NIH) grants AI22039 and AI57229 (P.P.). M.G. is a Stanford Graduate Fellow and a Pre-doctoral Fellow of the Howard Hughes Medical Institute. Also supported by a grant from the Leukemia Research Foundation (M.Y.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.