Abstract
The transcription repressor BCL-6 is known to play critical roles in B-cell lymphomagenesis, germinal center formation, and balanced Th1/Th2 differentiation. In macrophages, although BCL-6 has also been shown to regulate the expression of several chemokine genes, its function in other aspects of macrophage biology has not been studied. In addition, the precise role of BCL-6 in cell proliferation is poorly understood in general. Here we report that BCL-6-/- macrophages hyperproliferate due to an accelerated G1/S transition accompanied by increased cyclin D2 and c-myc and decreased expression of p27. Crucial to this enhanced proliferation is spontaneous interleukin 6 (IL-6) production and signal transducer and activator of transcription 3 (STAT3) activation in BCL-6-/- macrophages. In colony-forming assays, BCL- 6-/- bone marrow progenitor cells form spontaneous macrophage colonies that can be inhibited by anti-IL-6 antibodies. Gene expression studies demonstrate that BCL-6 binds to several sequence motifs scattered in the IL-6 locus and can repress IL-6 transcription both in 293T cells and in macrophages. In conclusion, our results indicate that BCL-6 negatively regulates proliferation of the monocytic/macrophage lineage by suppressing an autocrine IL-6/STAT3-mediated gene expression program. Our work also suggests that BCL-6 prevents abnormal Th2 differentiation by suppressing basal level IL-6 production in antigen-presenting cells (APCs). (Blood. 2005;105:1777-1784)
Introduction
BCL-6 was originally cloned as the most frequently targeted proto-oncogene in non-Hodgkin lymphomas (NHLs).1,2 In the normal immune system, prominent BCL-6 expression is only found in the germinal center (GC) B cells, although various subsets of T cells and macrophages also express this protein at low levels.3,4 Gene-targeting studies in mice demonstrate that BCL-6 plays important roles in all 3 of these cell types. In BCL-6-/- mice, GC development is completely blocked5-7 ; in addition, differentiation of CD4+ T helper (Th) cells is severely skewed toward Th2 and ultimately results in a fatal hyper-Th2-type inflammatory disease.5,6 Our earlier work indicates that the hyper-Th2 inflammatory disease in BCL-6-/- mice is independent of interleukin 4/signal transducer and activator of transcription 6 (IL-4/STAT6) signaling8 ; furthermore, in in vitro Th polarization assays, the intrinsic Th2 bias of BCL-6-/- T cells can only be revealed when IL-6 but not IL-4 is used as the Th2-inducing cytokine,9 suggesting that BCL-6 regulates the initial priming phase of Th2 differentiation that is dependent on antigen-presenting cell (APC)-T-cell interaction and IL-6 production by activated APCs.10,11 Because our earlier study has shown that the hyper-Th2 inflammatory disease in BCL-6-/- mice is not T-cell autonomous, and has a non-B, non-T-cell component,12 an attractive hypothesis is that BCL-6 may oppose an intrinsic Th2 bias of the CD4+ T cells while at the same time reducing the Th2-promoting ability of APCs.
In vivo, the macrophage is an important type of APC that can significantly modulate the adoptive immune response. We have previously reported that BCL-6 can directly repress transcription of a number of chemokine genes in macrophages including monocyte chemotactic protein 1 (MCP-1).12 Although MCP-1 was shown to be required for Th2 differentiation in vivo,13 it is unlikely to be a major player in the in vivo phenotype because inactivation of MCP-1 does not block the hyper-Th2 disease in BCL-6-/- mice (A. L. Shaffer, personal communication, February 2004). Thus, the hypothesis predicts that the presumed Th2-promoting effect of BCL-6-/- macrophages must be mediated by other immune modulators. In the present work, we characterized gene expression and phenotypic changes in bone marrow (BM)-derived BCL-6-/- macrophages. We show that BCL-6 negatively regulates IL-6 transcription and that inactivation of BCL-6 leads to spontaneous IL-6 secretion and STAT3 activation, which in turn results in a hyperproliferation phenotype characterized by an accelerated G1/S transition. Our work suggests that this autocrine IL-6/STAT3 signaling pathway is normally repressed by BCL-6 in macrophages and that increased IL-6 production by macrophages provides the missing link between APCs and the hyper-Th2 inflammatory disease in BCL-6-/- mice.
Materials and methods
Mice, BM-derived macrophages, and retroviral infection
The generation of BCL-6-/- mice has been described.5,6 Mice used in this study were adults 6 to 12 weeks of age and maintained in pathogen-free barrier facilities. BM-derived macrophages (BMMs) were prepared as previously described14 and maintained in colony-stimulating factor 1 (CSF-1)-containing medium (α-modified Eagle medium [α-MEM] with 15% fetal bovine serum [FBS] and 120 ng/mL recombinant human CSF-1 [gift of Chiron, Emeryville, CA]). Day 7 to 10 macrophages were used for all experiments except when indicated otherwise. A Flag-tagged BCL-6 cDNA fragment was inserted into the EcoRI site of the pMSCV-IRES-GFP vector15 to make pMSCV-BCL-6-IRES-GFP. The pMSCV-based double-negative (DN) STAT3 construct has been described before.16 These and the control pMSCV virus were packaged in the Phoenix-Eco packaging cell line via calcium phosphate-mediated transfection. Viral supernatants were collected 48 hours after, filtered through 0.45-μm filters, and used to infect macrophages. Briefly, macrophages were incubated with fresh viral supernatants in the presence of 120 ng/mL CSF-1 and 4 μg/mL polybrene, centrifuged for 2 hours at 290 ref at 25°C, and incubated at 37°C overnight. Following 2 to 3 additional days of culturing in the CSF-1 medium, green fluorescent protein-positive (GFP+) cells were sorted by fluorescence-activated cell sorting (FACS) and used for either 3H-thymidine incorporation assays or reverse transcription-polymerase chain reactions (RT-PCRs).
Cell-cycle profile and proliferation assays
Cell-cycle profiling was performed by propidium iodide (PI) staining and flow cytometry analysis. To analyze the kinetics of the G1-to-S phase transition, macrophages were first starved of CSF-1 for 16 hours. At various time points after CSF-1 readdition, cells were fixed for PI staining. For 3H-thymidine incorporation assays, cells were cultured at a density of 3 × 103 cells/well in triplicate wells (96-well plates). Cultures were pulsed with 1.0 μCi (0.037 MBq) 3H-thymidine for 12 to 16 hours before harvesting the cells onto glass fiber filters and measuring the incorporated radioactivity by scintillation counting. Where indicated in Figure 5B, anti-mIL-6 antibody (PeproTech, Rocky Hill, NJ) was added to the culture at 4 μg/mL and anti-mMCP-1 and anti-murine macrophage inflammatory protein 1α (anti-mMIP-1α) antibodies (PeproTech) were added at 0.5 μg/mL each. MTT assays were performed in triplicate by staining the cells with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-tetrazolim bromide (MTT) as described before.17 AG490 (AG Scientific, San Diego, CA) was added on day 0 to the medium at the concentrations indicated in Figure 3D.
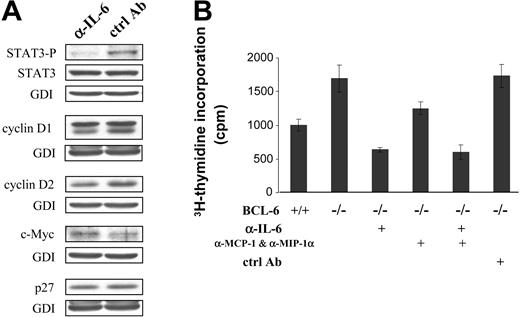
Role of spontaneous IL-6 production in activating STAT3 and promoting cell proliferation. (A) Western blot analysis of STAT3 and selected cell cycle regulators in BCL-6-/- macrophages that had been treated with either IL-6-neutralizing antibody (α-IL-6) or control IgG (ctrl Ab). (B) Cell proliferation measured by 3H-thymidine incorporation assays after antibody-mediated neutralization of IL-6 or chemokines MCP-1 and MIP-1α. Error bars indicate SD. Results are representative of 2 independent experiments.
Role of spontaneous IL-6 production in activating STAT3 and promoting cell proliferation. (A) Western blot analysis of STAT3 and selected cell cycle regulators in BCL-6-/- macrophages that had been treated with either IL-6-neutralizing antibody (α-IL-6) or control IgG (ctrl Ab). (B) Cell proliferation measured by 3H-thymidine incorporation assays after antibody-mediated neutralization of IL-6 or chemokines MCP-1 and MIP-1α. Error bars indicate SD. Results are representative of 2 independent experiments.
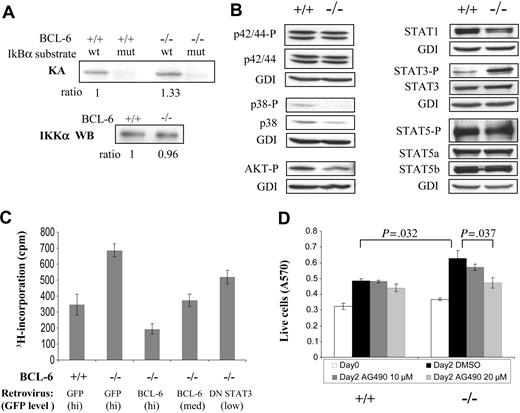
Increased STAT3 activation and its role in proliferation ofBCL-6-/-macrophages. (A) IKK kinase assay. KA indicates kinase assay; IKKα WB, IKKα Western blot for loading control. (B) Activation status of several cell-signaling pathways assessed by Western blotting using antibodies to phosphorylated/active forms of signal transducers. We did not detect any signal for activated STAT1 (phospho-Tyr701). (C) Growth behavior of BCL-6-/- macrophages can be corrected by retroviral-mediated expression of either a wild-type BCL-6 or a DN STAT3 gene. 3H-thymidine incorporation assay of sorted GFP+ cells was performed 30 hours after retroviral infection. (D) Proliferation of BCL-6-/- macrophages as measured by MTT assays was decreased in a dose-dependent manner by AG490. Numbers on top of the graph are the P values of 2-tailed Student t tests performed on the corresponding pair of triplicate tests. Error bars represent SD. Results are representative of 2 independent experiments.
Increased STAT3 activation and its role in proliferation ofBCL-6-/-macrophages. (A) IKK kinase assay. KA indicates kinase assay; IKKα WB, IKKα Western blot for loading control. (B) Activation status of several cell-signaling pathways assessed by Western blotting using antibodies to phosphorylated/active forms of signal transducers. We did not detect any signal for activated STAT1 (phospho-Tyr701). (C) Growth behavior of BCL-6-/- macrophages can be corrected by retroviral-mediated expression of either a wild-type BCL-6 or a DN STAT3 gene. 3H-thymidine incorporation assay of sorted GFP+ cells was performed 30 hours after retroviral infection. (D) Proliferation of BCL-6-/- macrophages as measured by MTT assays was decreased in a dose-dependent manner by AG490. Numbers on top of the graph are the P values of 2-tailed Student t tests performed on the corresponding pair of triplicate tests. Error bars represent SD. Results are representative of 2 independent experiments.
Western blot and RT-PCR analyses
Whole-cell lysates were prepared using RIPA buffer supplemented with protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). Western blotting was performed using standard procedures with antibodies reactive to proteins of interest. The results were visualized by the enhanced chemiluminescence system (ECL; Amersham Biosciences, Piscataway, NJ). Antibodies against cyclins D1, D2, D3, and E, CDK2 and 4, p16, p21, p27, Pim-1, AKT, STAT5a and b were from Santa Cruz Biotechnology (Santa Cruz, CA); antibodies against total Erk1/2, phospho-Erk1/2 (Thr202/Tyr204), p38 mitogen-activated protein kinase (MAPK), phospho-p38 (Thr180/Tyr182), phospho-AKT (Ser473), phospho-STAT1 (Tyr701), phospho-STAT3 (Tyr705), and phospho-STAT5 (Tyr694) were purchased from Cell Signaling Technology (Beverly, MA); antibody against c-Myc was from Upstate Biotechnology (Waltham, MA); antibodies against STAT1 and STAT3 were purchased from Transduction Laboratories (San Diego, CA). Antibody to guanine nucleotide dissociation inhibitor (GDI) was used as an internal control for protein loading. For semiquantitative RT-PCR, total RNA samples were prepared with the TriZol reagent (Invitrogen, Carlsbad, CA). RNA (3 μg) was used for cDNA synthesis with the Superscript Reverse Transcriptase (Invitrogen). PCRs were performed with serially diluted cDNA input (1:5 and 1:25) and the products were separated on agarose gels. The digitalized signals were normalized to those of the internal control, β-actin. Sequences of the primers used in the PCRs can be found in Document S1, available on the Blood website (see the Supplemental Materials link at the top of the online article).
IKK kinase assay
The IKK kinase assay was performed as previously described.18 Briefly, 600 μg of whole-cell lysate proteins were immunoprecipitated with 3 μg anti-IKKγ antibody (Santa Cruz Biotechnology) and 100 μL protein A-Sepharose (Amersham Biosciences) according to standard procedures. The kinase reaction was performed using GST-IκBα-(1-54) or the mutant GST-IκBα-(1-54)32A/36A as substrate (constructs from G. Bren, Mayo Clinic College of Medicine). Protein bands were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and dried gels exposed to phospho-imaging screens. The results were analyzed by the ImageQuant software (Amersham Biosciences). The amount of IKK proteins in the immunoprecipitates was measured by Western blotting with an anti-IKKα antibody (Santa Cruz Biotechnology).
Gel shift, construction of IL-6 reporters, and reporter assays
Procedures for nuclear extract preparation and gel shift analysis have been described before.19 For reporter assays in 293T cells, 0.125 μg of the reporter, 0.04 μg of a CMV-β-gal plasmid plus various amount of pMT2T-BCL-6 were used to transfect cells in each well of 24-well plates using the Superfect transfection reagent (Qiagen, Valencia, CA). All transfections were performed in duplicate and harvested 48 hours later. Luciferase activities were analyzed with the Luciferase Assay System (Promega, Madison, WI) and normalized by the control readings from the β-gal enzyme. To construct the 1.8-kb mIL-6S and 9-kb mIL-6L luciferase reporter constructs, the corresponding regions of the mouse IL-6 locus were amplified by PCR from mouse genomic DNA (gDNA) and subcloned into the pGL3-basic plasmid (Promega). Detailed cloning procedures and PCR primer sequences can be found in Document S1.
Assays of BM progenitors
Total BM cells were plated at 5 × 104/mL in Iscove modified Dulbecco medium (IMDM) containing 1% methylcellulose and 30% vol/vol fetal bovine serum (FBS) either without addition (control) or with the addition of 10 ng/mL recombinant mouse CSF-1 (R&D Systems, Minneapolis, MN) or CSF-1 and 50 ng/mL murine SCF (Immunex, Seattle, WA), 1 μg/mL anti-mIL-6 antibodies (monoclonal antibody [mAb] from R&D Systems [no. 1 in Table 2] and rabbit polyclonal antibody from PeproTech [no. 2 in Table 2]) or control antibodies (isotype-matched control IgGs from the corresponding sources), or 10 ng/mL recombinant mIL-6 (PharMingen, San Diego, CA). Colonies were scored after 7 days of incubation as previously described.20 For analysis of secondary colonies, 4 individual primary colonies per mouse were picked from 7-day CSF-1 or CSF-1 plus stem cell factor (SCF)-stimulated methyl cellulose culture plates and each colony individually dispersed in phosphate-buffered saline (PBS) and replated in CSF-1-containing agar medium in 3 secondary plates. Secondary colonies and clusters were scored on day 7.
Acidic β-gal staining
A Senescence β-Galactosidase Staining Kit (Cell Signaling Technology) was used for histochemical staining of the acidic β-galactosidase.
IL-6 enzyme-linked immunosorbent assay
A mouse IL-6 kit from BD Biosciences (San Jose, CA) was used to measure IL-6 secretion by cultured macrophages.
Results
BCL-6-/- macrophages hyperproliferate due to accelerated G1/S transition
Using a 3-step procedure, we isolated undifferentiated BM progenitor cells from both wild-type and BCL-6-/- mice and differentiated them first to nonadherent macrophage progenitors in low CSF-1 medium. Their further differentiation to adherent macrophages was accomplished in high CSF-1 medium.21 No difference between the BCL-6-/- and wild-type cells were observed in the first 2 steps of this procedure. However, in the high CSF-1 medium, BCL-6-/- macrophages displayed a remarkable hyperproliferation phenotype. Compared to wild-type controls, they formed much bigger colonies (Figure 1A) and accumulated 1.6-fold more cells during the first week in the high CSF-1 medium (Figure 1B). To pinpoint the cell cycle defect in these cells, we analyzed cell cycle profiles of these macrophages by PI staining using a cohort of 8 mice per genotype. As shown in Figure 1C, on average, only 59% of unsynchronized BCL-6-/- macrophages were in G1 compared to 72% of wild-type cells in the same cell cycle phase. This statistically significant difference suggests an accelerated G1/S transition in the absence of BCL-6. Despite their accelerated rate of proliferation, BCL-6-/- macrophages were still strictly dependent on CSF-1 for proliferation and survival just like their wild-type counterparts (Figure 1D and data not shown). This allowed us to determine the duration of G1 phase in a CSF-1 withdrawal and readdition experiment. As shown in Figure 1D, CSF-1 starvation synchronized the majority of macrophages in G1 with a minute fraction in G2/M. Approximately 8 hours after CSF-1 readdition, the G2/M peak in both wild-type and BCL-6-/- cells was much reduced, suggesting BCL-6 does not play a major role in G2/M phases (Figure 1D and data not shown). In contrast, whereas the G1 phase BCL-6-/- macrophages took only 8 hours to complete G1 and enter S phase, the wild-type cells required at least 12 hours to do so (Figure 1D). Clearly, inactivation of BCL-6 resulted in an accelerated G1/S transition and shortened cell doubling time.
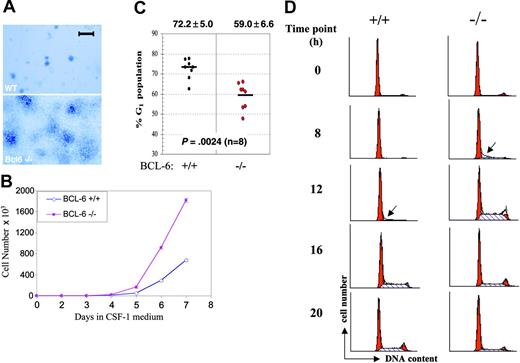
BM-derivedBCL-6-/- macrophages hyperproliferate due to accelerated G1-to-S phase transition. (A) Morphology of macrophage colonies from liquid culture stained with methylene blue. A Zeiss Stemi microscope (Carl Zeiss Microimaging, Thornwood, NY) was used, and was equipped with a 4 ×/0.65 objective lens. The image was acquired with QImaging Retiga (QImaging, Burnaby, BC, Canada). Scale bar is equal to 1 mm. (B) Growth curves of wild-type (⋄) and BCL-6-/- macrophages (▪) in the high CSF-1 medium. (Error bars representing SD are too small to be visible in this case.) (C) Cell cycle analysis based on PI staining and flow cytometry. Percentages of G1 cells from 8 independent primary macrophage preparations are plotted for each genotype. Bars in the graph represent group means, shown with SD at the top and the P from a 2-tailed Student t test below. (D) Cell cycle progression of CSF-1-starved cells at 0, 8, 12, 16, and 20 hours after CSF-1 readdition was monitored by PI staining followed by flow cytometry analysis. Red peaks indicate G1 and G2/M phases and dashed areas represent S phase fractions. Arrows point to the first appearance of S phase cells. Results in panels A, B, and D are representative of 2 independent experiments.
BM-derivedBCL-6-/- macrophages hyperproliferate due to accelerated G1-to-S phase transition. (A) Morphology of macrophage colonies from liquid culture stained with methylene blue. A Zeiss Stemi microscope (Carl Zeiss Microimaging, Thornwood, NY) was used, and was equipped with a 4 ×/0.65 objective lens. The image was acquired with QImaging Retiga (QImaging, Burnaby, BC, Canada). Scale bar is equal to 1 mm. (B) Growth curves of wild-type (⋄) and BCL-6-/- macrophages (▪) in the high CSF-1 medium. (Error bars representing SD are too small to be visible in this case.) (C) Cell cycle analysis based on PI staining and flow cytometry. Percentages of G1 cells from 8 independent primary macrophage preparations are plotted for each genotype. Bars in the graph represent group means, shown with SD at the top and the P from a 2-tailed Student t test below. (D) Cell cycle progression of CSF-1-starved cells at 0, 8, 12, 16, and 20 hours after CSF-1 readdition was monitored by PI staining followed by flow cytometry analysis. Red peaks indicate G1 and G2/M phases and dashed areas represent S phase fractions. Arrows point to the first appearance of S phase cells. Results in panels A, B, and D are representative of 2 independent experiments.
Altered expression of G1 checkpoint regulators
To gain mechanistic insight into this cell cycle phenotype, we examined expression of a number of proteins known to be involved in G1 phase progression. Consistent with a faster G1/S transition, BCL-6-/- macrophages showed a decrease in p27, a moderate increase in both cyclin D1 and D2, and a remarkable increase in c-Myc (Figure 2A). Levels of other regulators examined, including cyclin D3, CDK2, CDK4, and p21, were similar between the 2 genotypes (Figure 2A). Moderately reduced cyclin E and Pim-1 were also seen in BCL-6-/- cells although the significance of such reduction is unclear (Figure 2A). Comparing protein levels of cyclin D1 and D2 and c-Myc with their relative mRNA levels assessed by semiquantitative RT-PCR (Figure 2B), it seems that increased expression of cyclin D2 and c-Myc is the result of enhanced transcription, whereas a posttranscriptional mechanism may be responsible for elevated cyclin D1 protein in BCL-6-/- macrophages. Hyperproliferation ofBCL-6-/-macrophages is dependent upon activation of STAT3
Expression of various cell cycle regulators inBCL-6-/-macrophages. (A) Western blot analysis of various cell cycle regulators involved in G1 progression. (B) Semiquantitative RT-PCR analysis of cyclin D1 and D2 and c-Myc. Template cDNAs were diluted 1:5 as well as 1:25 for each sample. Panels on the right are bar graphs of the data normalized with respect to the β-actin control. Results are representative of 2 to 3 independent experiments.
Expression of various cell cycle regulators inBCL-6-/-macrophages. (A) Western blot analysis of various cell cycle regulators involved in G1 progression. (B) Semiquantitative RT-PCR analysis of cyclin D1 and D2 and c-Myc. Template cDNAs were diluted 1:5 as well as 1:25 for each sample. Panels on the right are bar graphs of the data normalized with respect to the β-actin control. Results are representative of 2 to 3 independent experiments.
Increased cyclin D2 expression in BCL-6-/- macrophages is in line with published work describing it as a BCL-6 target gene.2 Increased cyclin D1 and decreased p27 expression, on the other hand, are inconsistent with previous studies showing overexpressed BCL-6 increases cyclin D1 expression in fibroblasts and B cells,22,23 and that BCL-6 can repress p27 under certain experimental conditions.2 Furthermore, at the time this study was conducted, c-myc had not been shown to be a BCL-6 target gene. Thus, we explored the possibility that BCL-6 might affect these G1 phase regulators indirectly via cell signaling pathways. A well-known growth-promoting pathway, nuclear factor κB (NF-κB), was examined first. Because the IKK kinase assay only revealed a slight increase in the BCL-6-/- macrophages (Figure 3A) and cell proliferation of both genotypes were equally sensitive to a NF-κB inhibitor, Bay 11-7082 (not shown), we concluded that NF-κB signaling is not notably affected in the absence of BCL-6. Using antibodies against phosphorylated/activated forms of signaling molecules, several other signal transduction pathways were then examined. Among these, levels of activated Erk1/2 and STAT5 were similar between wild-type and BCL-6-/- cells; activated forms of p38 MAPK and AKT were reduced in the absence of BCL-6, whereas the STAT3 pathway was uniquely and significantly activated (Figure 3B). A number of published studies have shown that STAT3 plays a critical role in gp130-mediated G1-to-S phase transition. In particular, in BAF/B03 cells, STAT3 activation is essential for up-regulation of cyclins D2, D3, cdc25A, and down-regulation of p21 and p27.24 c-myc has also been shown to be a direct STAT3 target gene.25 Thus, we evaluated the role of STAT3 activation in the hyperproliferation phenotype of BCL-6-/- macrophages. As expected, retroviral infection of BCL-6-/- macrophages with DN STAT3 reduced proliferation of these cells, although the degree of reduction is not as substantial as cells reconstituted with the wild-type BCL-6 protein (Figure 3C). Because we consistently observed lower infection efficiency and lower levels of GFP expression in DN STAT3-infected cells compared to either GFP- or BCL-6-infected cells, we suspect that significant reduction of STAT3 activity may interfere with cell survival. Thus, we turned to a JAK-selective inhibitor, tyrphostin AG490, a compound known to block STAT3 activation in cancer cells.26 When used within a 10- to 20-μM range, AG490 had minimum effect on wild-type macrophages, yet it reduced proliferation of BCL-6-/- cells in a dose-dependent manner to a level comparable to the wild-type controls (Figure 3D). Therefore, we conclude that STAT3 activation is essential for the hyperproliferation phenotype of BCL-6-/- macrophages.
BCL-6 is a potent inhibitor of IL-6 transcription
STAT3 is primarily activated in response to several IL-6 family cytokines using gp130 as the shared signal-transducing component. Enzyme-linked immunosorbent assay (ELISA) performed with culture supernatants showed that BCL-6-/- macrophages constitutively produced IL-6 either in the presence or absence of CSF-1, whereas control wild-type cells did not secrete any detectable IL-6 (Table 1). In response to lipopolysaccharide (LPS) treatment, wild-type macrophages did produce a large amount of IL-6 as expected, but 20-fold more IL-6 was produced by BCL-6-/- macrophages (Table 1). Thus, BCL-6 is a potent inhibitor of IL-6 production under both basal and LPS-stimulated conditions. Semiquantitative RT-PCR analysis confirmed that IL-6 was expressed only in BCL-6-/- macrophages but not the wild-type controls (Figure 4A). Furthermore, the aberrant IL-6 mRNA expression in BCL-6-/- cells was completely shut off by retroviral-mediated BCL-6 reconstitution (Figure 4A). These results strongly suggest that IL-6 is a transcriptional target of BCL-6. To further investigate this possibility, a luciferase reporter construct was made that contains a 1.8-kb genomic fragment from the 5′ flanking region of mouse IL-6 gene (IL-6S, Figure 4B). To our surprise, although this region contains all of the important IL-6 regulatory elements that have been studied by others, it was only marginally responsive to BCL-6 repression in reporter assays (Figure 4D). The entire 9-kb IL-6 locus was then analyzed by the MacInspector program,27 which revealed 3 other BCL-6 motifs scattered in the second and third intron of the IL-6 gene; in particular, site 4 in intron 3 has a perfect match to the 9-bp core of BCL-6 consensus site (Figure 4C). BCL-6 specifically recognized site 1, 2, and 4, whereas weaker binding to site 3 was also detected in gel shift assays (Figure 4C). As a result, a luciferase reporter construct (IL-6L, Figure 4B) was made that includes the entire 9-kb IL-6 locus with all 5 exons. Because of the built-in internal ribosome entry site (IRES) feature, this construct also allows measurement of secreted IL-6. In agreement with the gel shift results, transcription from this construct was repressed by BCL-6 in a dose-dependent manner in reporter assays. When 1 pmol BCL-6 plasmid was cotransfected with the reporter, transcription from the IL-6 locus was suppressed 8-fold, whereas the 1.8-kb IL-6 reporter was repressed only 2-fold and the negative control reporter, pGL3-control, was completely resistant to BCL-6 (Figure 4D). ELISA results also showed that the cloned IL-6 locus can produce secreted IL-6 as expected and that the amount of IL-6 released into the medium was similarly inhibited by BCL-6 in a dose-dependent manner (Figure 4E). These results strongly suggest that BCL-6 negatively regulates IL-6 production in macrophages due to its ability to inhibit IL-6 transcription. To examine in vivo association of BCL-6 with the IL-6 locus, we also performed chromatin immunoprecipitation (ChIP) experiments (see “Discussion”).
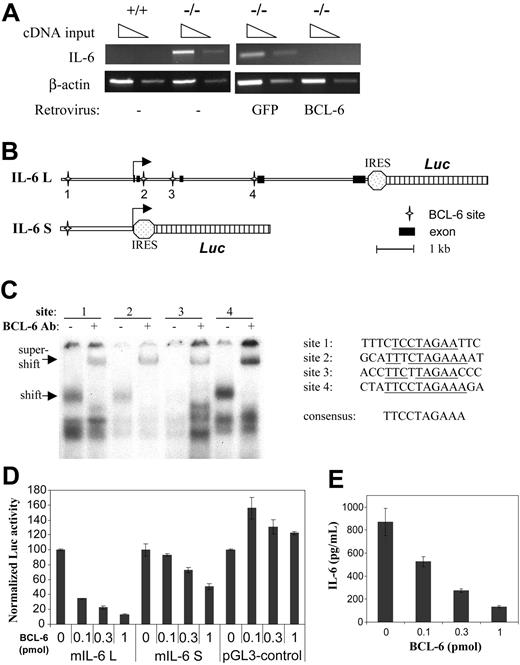
BCL-6 is a repressor of IL-6 transcription. (A) RT-PCR analyses of IL-6 mRNA expression in wild-type and BCL-6-/- macrophages, as well as in BCL-6-/- macrophages in which either BCL-6 or control GFP was retrovirally expressed. (B) Schematic representation of the 2 IL-6 reporter constructs used in the reporter assay. Locations of the 4 BCL-6 binding sites are also shown. (C) Gel shift analysis using synthetic oligos corresponding to the 4 BCL-6 motifs and nuclear extract from a BCL-6+ lymphoma cell line, Ly1. Specificity of the BCL-6/DNA complexes was shown in super-shift assays using a BCL-6 antibody. Sequences of the 4 BCL-6-binding sites studied are given in comparison with the 9-bp consensus BCL-6-binding site. (D) Reporter assays performed in 293T cells with the 2 IL-6 reporters and the indicated amounts of BCL-6 expression plasmids (pmol/plate). The experiments were performed in duplicate, and luciferase activities in all transfections were normalized to that in the absence of BCL-6, which is set as 100 in the graph. (E) Culture supernatants from the reporter assays in panel D were measured for IL-6 secretion by ELISA.
BCL-6 is a repressor of IL-6 transcription. (A) RT-PCR analyses of IL-6 mRNA expression in wild-type and BCL-6-/- macrophages, as well as in BCL-6-/- macrophages in which either BCL-6 or control GFP was retrovirally expressed. (B) Schematic representation of the 2 IL-6 reporter constructs used in the reporter assay. Locations of the 4 BCL-6 binding sites are also shown. (C) Gel shift analysis using synthetic oligos corresponding to the 4 BCL-6 motifs and nuclear extract from a BCL-6+ lymphoma cell line, Ly1. Specificity of the BCL-6/DNA complexes was shown in super-shift assays using a BCL-6 antibody. Sequences of the 4 BCL-6-binding sites studied are given in comparison with the 9-bp consensus BCL-6-binding site. (D) Reporter assays performed in 293T cells with the 2 IL-6 reporters and the indicated amounts of BCL-6 expression plasmids (pmol/plate). The experiments were performed in duplicate, and luciferase activities in all transfections were normalized to that in the absence of BCL-6, which is set as 100 in the graph. (E) Culture supernatants from the reporter assays in panel D were measured for IL-6 secretion by ELISA.
Hyperproliferation is dependent on spontaneous IL-6 production by BCL-6-/- macrophages
To evaluate the functional role of spontaneous IL-6 production in STAT3 activation and hyperproliferation of BCL-6-/- macrophages, we treated these cells with a neutralizing antibody against IL-6. Compared to cells treated with control IgG, anti-IL-6 treatment dramatically reduced STAT3 activation, moderately decreased cyclin D2, and slightly decreased cyclin D1 (Figure 5A). Interestingly, p27 was also reduced somewhat, whereas c-Myc was even further increased (Figure 5A; see “Discussion”). 3H-thymidine incorporation assays showed that IL-6 neutralization completely reversed the hyperproliferation phenotype of BCL-6-/- macrophages (Figure 5B). Because these macrophages also overproduce chemokines MCP-1 and MIP-1α,12 their potential role in cell proliferation was evaluated with the use of neutralizing antibodies as well. As shown in Figure 5B, combined use of antibodies to MCP-1 and MIP-1α had only a moderate effect that was not additive with the anti-IL-6 effect. It is possible that in our experimental system, a minor proliferation effect of MCP-1 and MIP-1α works through gp130/STAT3.28,29
Formation of IL-6-dependent, spontaneous macrophage colonies by BCL-6-/- BM progenitors
Our results so far demonstrate that spontaneous IL-6 production and STAT3 activation are essential components of a mechanism responsible for hyperproliferation of in vitro differentiated BCL-6-/- macrophages. To extend our findings to cells of the monocyte/macrophage lineage in vivo, we studied the macrophage colony-forming ability of BCL-6-/- BM progenitors. In control medium devoid of any hematopoietic cytokines, no colonies formed from wild-type BM cells as expected, whereas low numbers of macrophage-type colonies were consistently obtained from BCL-6-/-BM (Table 2). These colonies are IL-6 dependent because they were almost completely eliminated by 2 independent IL-6-neutralizing antibodies and significantly increased by addition of exogenous IL-6 (Table 2). When stimulated with CSF-1, BCL-6-/- BM progenitors produced nearly 3 times more colonies than their wild-type counterparts (Table 2). Interestingly, whereas stimulation with CSF-1 plus SCF increased the number of wild-type colonies by almost 2-fold, there was no effect of adding SCF to the CSF-1-stimulated BCL-6-/- colonies. Still, the colony numbers from SCF plus CSF-1-stimulated BCL-6-/- cells were higher than that from the wild-type controls (Table 2). Furthermore, BCL-6-/- colonies were considerably larger than their wild-type counterparts (Figure S1). These differences in colony number and size parallel that exhibited by cultured primary macrophages (Figure 1A-B). Thus, an IL-6-driven growth advantage is also manifested by cells committed to the monocyte/macrophage lineage in vivo. Interestingly, in the absence of other hematopoietic cytokines, exogenous IL-6 failed to stimulate colony formation from wild-type BM cells (Table 2), suggesting that IL-6 production per se is necessary but not sufficient for the observed growth advantage associated with BCL-6 inactivation, an interpretation that is consistent with our results from in vitro differentiated macrophage cultures (Figure S2).
Late-passage BCL-6-/- macrophages enter senescence prematurely
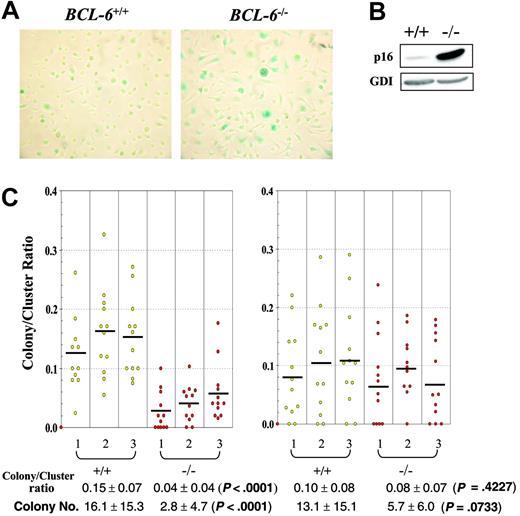
Although in vitro differentiated BCL-6-/- macrophages proliferated notably faster than their wild-type counterparts during the first 10 days in high CSF-1 medium, beginning on day 11 to 12, their population growth rate started to drop rapidly, resulting in permanent growth arrest around day 13, at 2 to 3 days prior to growth arrest of the wild-type cells. Throughout 8 to 14 days of culture and compared to wild-type controls, more BCL-6-/- macrophages increased in size and stained positive for the senescence histochemical marker, acidic β-galactosidase; furthermore, the staining was much stronger in BCL-6-/- macrophages than in wild-type cells (Figure 6A). The large increase in p16 expression in BCL-6-/- macrophages is also consistent with this senescence phenotype (Figure 6B). A similar premature senescence phenotype was also observed in BCL-6-/- BM progenitors when their proliferative potential was evaluated in secondary plating experiments in which colonies were picked from primary plates, dissociated, and the cells from single isolated colonies tested for their colony-forming ability by replating in secondary plates. In such assays, the ratio of colony numbers to cluster numbers from secondary plate count is higher for primary colony cells that have greater proliferative potential. As shown in Figure 6C, when colonies were picked from primary plates stimulated with CSF-1 alone, significantly fewer BCL-6-/- cells were able to form colonies in secondary plates than the wild-type cells. On the other hand, when colonies were picked from primary plates stimulated with the combination of CSF-1 and SCF, there was no significant difference in the secondary colonies between the 2 genotypes, suggesting that the role of BCL-6 in delaying senescence is specific to BM progenitors responsive to CSF-1 alone that have fully committed to the monocyte/macrophage lineage, but not the more immature progenitors responsive to CSF-1 plus SCF. A similar conclusion is reached when colony numbers are used as the indicator rather than the colony-to-cluster ratios (Figure 6C bottom). Although determination of the exact mechanism responsible for this premature senescence phenotype requires future studies, the complete agreement between the senescence behavior of the in vitro differentiated macrophages and the in vivo committed monocyte/macrophage progenitors once again validates the physiologic relevance of the macrophage culture system we have used to uncover the IL-6/STAT3-based hyperproliferation mechanism.
BCL-6-/- macrophages and determined macrophage progenitor cells enter senescence prematurely compared to wild-type controls. (A) Acidic β-galactosidase staining of in vitro differentiated macrophages on day 10 in CSF-1 medium. Representative images (original magnification ×10) from 6 independent experiments are shown. Wild-type macrophages in left panel; BCL-6-/- macrophages, right panel. (B) Western blot analysis of p16 expression in in vitro differentiated wild-type and BCL-6-/- macrophages. The result shown was obtained from day 10 macrophages, yet day 6 macrophages gave essentially the same result. (C) Colony-to-cluster ratios and colony numbers for cells of individual primary colonies picked from plates stimulated with either CSF-1 alone (left) or CSF-1 plus SCF (right). The primary colony cells were plated with CSF-1 on secondary plates for determination of their colony-forming (> 50 cells) and cluster-forming (< 50 cells) ability. Bars indicate the mean value for each mouse. Mean colony-to-cluster ratios and mean colony numbers ± SD for each genotype are presented under the graph. Because not all data sets conform to a gaussian distribution, the P values are from 2-sided Wilcoxon rank-sum tests performed to compare the results between BCL-6+/+ and BCL-6-/- cells.
BCL-6-/- macrophages and determined macrophage progenitor cells enter senescence prematurely compared to wild-type controls. (A) Acidic β-galactosidase staining of in vitro differentiated macrophages on day 10 in CSF-1 medium. Representative images (original magnification ×10) from 6 independent experiments are shown. Wild-type macrophages in left panel; BCL-6-/- macrophages, right panel. (B) Western blot analysis of p16 expression in in vitro differentiated wild-type and BCL-6-/- macrophages. The result shown was obtained from day 10 macrophages, yet day 6 macrophages gave essentially the same result. (C) Colony-to-cluster ratios and colony numbers for cells of individual primary colonies picked from plates stimulated with either CSF-1 alone (left) or CSF-1 plus SCF (right). The primary colony cells were plated with CSF-1 on secondary plates for determination of their colony-forming (> 50 cells) and cluster-forming (< 50 cells) ability. Bars indicate the mean value for each mouse. Mean colony-to-cluster ratios and mean colony numbers ± SD for each genotype are presented under the graph. Because not all data sets conform to a gaussian distribution, the P values are from 2-sided Wilcoxon rank-sum tests performed to compare the results between BCL-6+/+ and BCL-6-/- cells.
Discussion
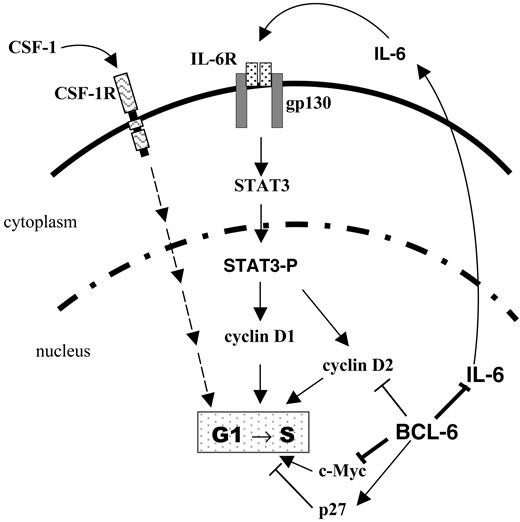
Our results show that BCL-6 is a potent inhibitor of IL-6 production that maintains IL-6 expression in an off state in the absence of inflammatory stimuli such as LPS. They also demonstrate that BCL-6 inactivation in macrophages results in aberrant IL-6 production and STAT3 activation leading to an accelerated G1/S transition and a hyperproliferation phenotype (Figure 7). Although many of our experiments are conducted using in vitro differentiated macrophages, a similar, IL-6-dependent hyperproliferation phenotype is also exhibited by BCL-6-/- BM progenitors that have committed to the monocyte/macrophage lineage in vivo.
A model for regulation of macrophage cell cycle by BCL-6 via IL-6 and STAT3. Solid lines indicate regulatory relationships (direct or indirect) studied in this work; dashed lines, regulatory relationships not directly examined in this study. CSF-1R indicates CSF-1 receptor tyrosine kinase.
A model for regulation of macrophage cell cycle by BCL-6 via IL-6 and STAT3. Solid lines indicate regulatory relationships (direct or indirect) studied in this work; dashed lines, regulatory relationships not directly examined in this study. CSF-1R indicates CSF-1 receptor tyrosine kinase.
Results from our gene expression studies indicate that BCL-6 is a potent inhibitor of IL-6 production due to its ability to suppress IL-6 transcription. Our results in Figure 4D further suggest that the BCL-6-binding sites scattered in the introns of IL-6 are most important for mediating BCL-6 repression. This is a unique situation because previously reported BCL-6 target genes all contain either a single or closely spaced BCL-6 sites. Nevertheless, as a POZ/BTB domain-containing transcription factor, repression through multiple distant sites is quite possible and this has been shown for PLZF, another POZ/BTB-containing zinc finger protein, in its repression of the Hox genes.30 We have been unsuccessful in our attempt to demonstrate in vivo occupancy of the 4 binding sites by BCL-6 using the ChIP technique. The reason for this remains unclear, but because the level of BCL-6 protein in macrophages is very low and there are several scattered binding sites in the IL-6 locus, one possibility is that at any given time, BCL-6 may be associated with only one of these sites, and thus the ChIP signal may be much weaker compared to other target gene loci.
Prior to this study, limited information existed on the role of BCL-6 in cell proliferation. A study based on lymphoma cell lines suggested a growth-promoting effect although the detailed mechanism and target genes responsible for such a phenotype were unclear.2 Our work demonstrates an opposite and potent growth-inhibitory role of BCL-6 in BM-derived macrophages. Although our results show that spontaneous IL-6 production and STAT3 activation are required for the hyperproliferation phenotype of BCL-6-/- macrophages, BCL-6 is likely to control other aspects of the cell cycle independently of IL-6/STAT3 signaling (Figure 7). This is because although the wild-type and BCL-6-/- macrophages express similar levels of the IL-6 receptor (not shown), IL-6 treatment alone does not enhance proliferation of wild-type macrophages (Figure S2) nor does it stimulate colony formation by wild-type BM progenitors in the absence of other hematopoietic cytokines (Table 2). Part of the explanation may lie in the fact that the increased c-Myc and decreased p27 expression in BCL-6-/- macrophages cannot be corrected by blocking IL-6 (Figure 5A). In fact, c-Myc protein was even further increased following IL-6 neutralization and STAT3 inactivation (Figure 5A). This negative effect of IL-6/STAT3 on c-Myc expression has been previously observed in IL-6-treated M1 cells.31 Thus, although STAT3 can positively regulate c-Myc expression in certain cell types such as pro-B cells,25 it appears to be a negative regulator of c-myc in the monocytic/macrophage lineage. The observation that BCL-6-/- macrophages express elevated levels of c-Myc mRNA that can be down-regulated by BCL-6-expressing retrovirus (Figure 2B and data not shown) suggests that c-myc is likely to be a target gene of BCL-6. Although p27 is thought to be a BCL-6 target gene in B cells,2 its protein level actually decreased in BCL-6-/- macrophages. Thus, p27 may be regulated by BCL-6 in an indirect manner. Regardless of the underlying explanations linking BCL-6 to these cell cycle regulators, our results demonstrate a negative effect of BCL-6 on macrophage proliferation and raise the important question as to whether such an effect can be extended to other cell types including normal B cells.
BCL-6 was previously described to suppress plasma cell differentiation by inhibiting STAT3-mediated B-lymphocyte-induced maturation protein 1 (Blimp-1) expression.32 It was suggested that BCL-6 may inhibit Blimp-1 expression by competitively binding to STAT3 sites. However, not all STAT3 sites are recognized by BCL-6; in addition, 2 later reports on Blimp-1 regulation suggest that BCL-6 inhibits Blimp-1 transcription either through an AP-1 site in the 5′ regulatory region or a BCL-6 site in intron 5.33,34 Our study reveals a completely different mechanism by which BCL-6 inhibits STAT3 signaling. Because this mechanism involves regulation of IL-6 production instead of competitive binding of BCL-6 to STAT3 sites on DNA, it has the potential to affect many IL-6/STAT3-responsive genes. A senescence inhibitory role has previously been described for BCL-6 based on overexpression of BCL-6 in fibroblasts and in vitro activated mature B cells, where BCL-6 was found to antagonize the p53-mediated senescence program at least in part by up-regulating cyclin D1.22,23 Although BCL-6-/- macrophages enter senescence prematurely, p21 expression is not significantly altered and cyclin D1 is moderately increased in these cells (Figure 2A). Nevertheless, p16, which is often up-regulated in cells entering replicative senescence, is dramatically elevated in BCL-6-/- macrophages (Figure 6B). Thus, it is possible that BCL-6 interacts with the cell cycle and senescence programs in a cell type-specific manner. In the B-cell lineage, high-level BCL-6 expression is restricted to GC B cells. During the period of rapid cell division prior to their differentiation to memory or plasma cells, GC B cells are prevented from entering replicative senescence due to reactivation of telomerase.35 Our observation that BCL-6 inhibits macrophage senescence is consistent with a direct role of BCL-6 in delaying the senescence program in GC B cells.
A prominent phenotype exhibited by BCL-6-/- mice is the hyper-Th2 inflammatory response involving both T cells and APCs.12 On the T-cell side, our previous studies indicate that BCL-6 inactivation alters the properties of naïve CD4+ cells at the initial priming phase of Th2 differentiation at least partly through its ability to negatively regulate GATA3, a well known regulator of Th2 differentiation.8,9 In contrast, the role played by BCL-6-/- APCs in this process was poorly understood. Our findings in this work strongly suggest that in the absence of BCL-6, macrophages, and, perhaps, other cell types as well, provide a strong, Th2-skewing cytokine environment due to their ability to spontaneously produce IL-6, a cytokine that is well known to promote Th2 differentiation at the expense of Th1 response.11 A major manifestation of the hyper-Th2 disease in BCL-6-/- mice is acidophilic macrophage pneumonia characterized by heavy infiltration of the lung by activated macrophages.5 The observation that IL-6 is abundantly produced in the lungs of these mice6 supports our hypothesis that IL-6 plays an important role in this hyper-Th2 phenotype. This IL-6-based hyper-Th2 model also explains, at least in part, the observation that the hyper-Th2 disease in BCL-6-/- mice can develop independently of STAT68 and that BCL-6-/- T cells only undergo abnormal Th2 differentiation when stimulated with IL-6 but not IL-4.9 In conclusion, we propose that the inhibitory effect of BCL-6 on IL-6/STAT3 signaling represents an important mechanism by which BCL-6 regulates a variety of immune responses including Th1/Th2 differentiation.
Prepublished online as Blood First Edition Paper, October 26, 2004; DOI 10.1182/blood-2004-08-3171.
Supported by National Institute of Health grants RO1 CA85573 (B.H.Y.); RO1 CA32551 and RO1 CA25604 (E.R.S.); RO1 HL56416, RO1 HL67384, and RO1 DK53674 (H.E.B.); RO1 AI46410 (A.L.D.); and KO8 CA097348 (F.J.P.); and the Albert Einstein College of Medicine (AECOM) Cancer Center (5P30-CA13330). B.H.Y. is the recipient of a grant from the G & P Foundation for Cancer Research, R.Y.-L.Y. is supported by the Harry Eagle Postdoctoral Scholarship from the Department of Cell Biology, AECOM.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Connie J. Eaves for generously providing the DN STAT3 retrovirus; Gary Bren for sharing the GST-IκBα construct; and Liang Zhu, Arthur Skoultchi, and Philipp Scherer for help with other reagents. We are grateful to B. Belinda Ding, Lourdes M. Mendez, and Scott H. Cooper for expert technical assistance, and to Hong Qian for help with statistical analysis.