Abstract
Early-onset sarcoidosis (EOS) and inheritable Blau syndrome (BS) share characteristic clinical features of juvenile-onset systemic granulomatosis syndrome that mainly affects skin, joints, and eyes. However, no direct evidence has been shown for the possible common origin of these 2 diseases. Recent discovery of CARD15 mutations in BS families encouraged us to investigate similar CARD15 mutations in EOS patients. Among 10 EOS cases retrospectively collected in Japan, heterozygous missense mutations were found in 9 cases; 4 showed a 1000C>T (R334W in amino acid change) that has been reported in BS, 4 showed novel 1487A>T (H496L), 1538T>C (M513T), 1813A>C (T605P), and 2010C>A (N670K), and 1 case showed double 1146C>G (D382E)/1834G>A (A612T) mutations on different alleles. All 6 of these variants of CARD15 showed increased basal nuclear factor (NF)–κB activity. These findings indicate that the majority of EOS and BS cases share the common genetic etiology of CARD15 mutations that cause constitutive NF-κB activation.
Introduction
Sarcoidosis is a multiorganic inflammatory disease with unknown etiology, characterized by the histologic features of noncaseating epithelioid granulomas. In childhood, 2 distinct types of sarcoidosis have been described.1 Usually the disease is detected in older children by chest radiography and the clinical manifestations are characterized by a classical triad of lung, lymph node, and eye involvement, similar to those in adults. In contrast, early-onset sarcoidosis (EOS), which usually appears in those younger than 4 years of age, is quite rare and has a distinct triad of skin, joint, and eye disorders, without apparent pulmonary involvement. Compared with an asymptomatic and sometimes naturally disappearing course of the disease in older children, EOS is progressive and in many cases causes severe complications, such as blindness, joint destruction, and visceral involvement.2
Blau syndrome (BS), also showing early-onset granulomatous arthritis, uveitis, and skin rash, is a rare familial disease transmitted in an autosomal dominant manner.3 By linkage analysis, the responsible locus for BS was mapped to chromosome 16, 4 in which CARD15 has recently been identified as the susceptibility gene.5 CARD15 (NOD2) is a member of the growing family of nucleotide-binding oligomerization domain (NOD) proteins and composed of 2 amino-terminal caspase recruitment domains (CARDs), one NOD, and carboxy-terminal leucine-rich repeats (LRRs).6,7 While mutations in LRRs are reportedly associated with Crohn disease (CD) and psoriatic arthritis, 8-10 3 types of missense point mutations in the NOD, 1000C>T (R334W in amino acid change), 1001G>A (R334Q), and 1405C>T (L469F), have been discovered in BS families.5,11,12
It has been discussed since the first report of BS whether EOS and BS are the same diseases.13 However, no direct evidence of their common origin has been shown and confusion still remains.14 In the first paper describing genetic abnormalities in BS, the authors recognized no CARD15 mutation in 2 EOS patients and therefore proposed a different etiology of BS and EOS.5 However, we have recently described a sporadic case of systemic granulomatosis syndrome with clinical features of EOS that showed the same CARD15 mutation as detected in BS.15 In this report, therefore, we retrospectively collected Japanese EOS cases and searched for CARD15 mutations, to further evaluate the relationship between EOS and CARD15 mutations.
Study design
Patients and genetic analysis
The diagnosis of EOS was confirmed by the absence of family history of granuloma-forming diseases, as well as the typical clinical and histologic features. The agreement for genetic analysis was obtained from 10 Japanese EOS patients, whose clinical information is summarized in Table 1.15-20 Informed consent was provided according to the Declaration of Helsinki. The study was approved by the ethics committees of Kyoto University and the organizations where the patients were under medical treatment. Genomic DNA was extracted from peripheral blood of the patients, and all 12 exons of the CARD15 gene including exon-intron boundaries were amplified by polymerase chain reaction and sequenced. Genomic DNA of 100 healthy volunteers was examined for the mutations discovered in our patients.
Generation of CARD15 mutants and NF-κB luciferase assay
The wild-type CARD15 cDNA was generated from a healthy volunteer by reverse transcription–polymerase chain reaction. Each CARD15 mutant cDNA was generated using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and subcloned into p3xFLAG-CMV-14 vector (Sigma, St Louis, MO). HEK293T cells (1 × 105) were transfected with 1000 ng plasmids, containing 100 ng nuclear factor (NF)–κB reporter plasmid (pNF-κB-Luc; BD Biosciences Clontech, Palo Alto, CA), 30 ng expression construct of a CARD15 variant, 10 ng internal control for normalization of transfection efficiency (pRL-TK; Toyo Ink, Tokyo, Japan), and the corresponding mock vector, using TransIT-293 Transfection Reagent (Mirus Bio, Madison, WI). The cells were cultured with or without 5 μg/mL muramyl dipeptide (MDP; Sigma) for 12 hours after transfection and measured for NF-κB activity using PicaGene Dual Luciferase Kit (Toyo Ink). Protein expression of each CARD15 variant was examined by Western blotting using anti-FLAG (8 amino acids; DYKDDDDK) M2 monoclonal antibody (Sigma).
Results and discussion
The genetic analysis of 10 EOS patients revealed 9 cases with heterozygous missense mutations in the NOD of the CARD15 gene. As shown in Table 1, 4 cases showed a 1000C>T (R334W), the same mutation as reported in BS, and 4 showed novel mutations, 1487A>T (H496L), 1813A>C (T605P), 2010C>A (N670K), and 1538T>C (M513T). Case 919 showed double mutations, a novel 1146C>G (D382E) and a known 1834G>A (A612T), which were present on different alleles as indicated by sequencing after cloning. None of these 7 mutations is identical to a reported single nucleotide polymorphism, nor was it detected by the analysis of 100 Japanese healthy volunteers except for 1834G>A of case 9. 1834G>A, which had already been reported in one CD patient, 21 was identified in 1 of 200 alleles. Only case 10, who developed huge hepatosplenomegaly since the disease onset, 20 showed no detectable mutation in CARD15. These results indicate that the majority of EOS cases are related to CARD15 mutations, especially in the NOD.
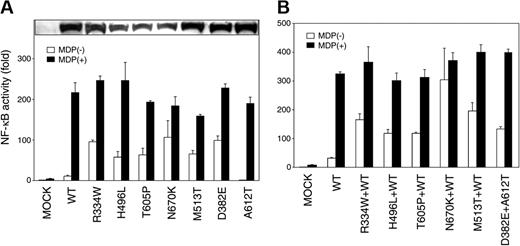
CARD15 is expressed intracellularly in phagocytic cells and recognizes MDP, a component of bacterial peptidoglycan, to induce immune responses through NF-κB activation.6,7,22,23 The BS-related CARD15 variants reportedly show increased basal MDP-independent NF-κB activity.24 Accordingly, the MDP-independent and -dependent NF-κB transactivation by the novel CARD15 mutations discovered in our EOS cases was examined in vitro to define their biologic effects. At equivalent protein expression levels, 5 novel CARD15 variants (H496L, T605P, N670K, M513T, and D382E) significantly increased the basal NF-κB activity compared with the wild-type CARD15, similar to the R334W found in both BS families and our EOS cases (Figure 1A, open bars). Although the A612T showed reduced basal NF-κB activity as reported previously, 24 cotransfection of D382E and A612T increased the basal NF-κB activity (Figure 1B, open bars), indicating the dominant positive effect of the D382E in case 9. Similar dominant positive effects of other 5 mutations were observed when cotransfected with the wild-type CARD15. In contrast, addition of a maximum dose of MDP (5 μg/mL) to each CARD15 mutant further elevated the NF-κB activity up to almost the same level as the case of the wild-type CARD15 (Figure 1A-B, filled bars). Collectively, all 6 combinations of CARD15 variants mimicking the genotype of our EOS patients showed increased basal NF-κB activity (Figure 1B, open bars) and shared the common biologic effect with the BS-related CARD15 variants. Although the basal NF-κB activation levels of these 6 CARD15 variants were divergent, no remarkable correlation could be observed between the basal NF-κB activity and the disease severity (Table 1).
Biologic effects of CARD15 variants discovered in EOS patients. (A) MDP-independent and -dependent NF-κB transactivation by CARD15 variants discovered in EOS patients. HEK293T cells were cotransfected with 30 ng expression construct of a CARD15 variant together with the NF-κB reporter plasmid and measured for NF-κB activity after 12 hours' incubation with (▪) or without (□) 5 μg/mL MDP. Mock vector and the wild-type (WT) CARD15 were used as controls. Values represent the mean of normalized data (mock without MDP = 1) of triplicate cultures, and error bars indicate SD. Shown in 1 representative result of 3 independent experiments. Protein expression levels of CARD15 variants analyzed by Western blotting are shown in the top column. (B) MDP-independent and -dependent NF-κB transactivation by combinations of CARD15 variants mimicking the genotype of EOS patients. To reach a phenotype closer to the heterozygous CARD15 gene expression in case 1 to 8, 15 ng of each CARD15 mutant (R334W, H496L, T605P, N670K, and M513T) was cotransfected with the same amount of the wild-type CARD15 with (▪) or without (□)5 μg/mL MDP. For case 9, 15 ng of each D382E and A612T were cotransfected. Mock vector alone and the wild-type CARD15 alone were added as controls.
Biologic effects of CARD15 variants discovered in EOS patients. (A) MDP-independent and -dependent NF-κB transactivation by CARD15 variants discovered in EOS patients. HEK293T cells were cotransfected with 30 ng expression construct of a CARD15 variant together with the NF-κB reporter plasmid and measured for NF-κB activity after 12 hours' incubation with (▪) or without (□) 5 μg/mL MDP. Mock vector and the wild-type (WT) CARD15 were used as controls. Values represent the mean of normalized data (mock without MDP = 1) of triplicate cultures, and error bars indicate SD. Shown in 1 representative result of 3 independent experiments. Protein expression levels of CARD15 variants analyzed by Western blotting are shown in the top column. (B) MDP-independent and -dependent NF-κB transactivation by combinations of CARD15 variants mimicking the genotype of EOS patients. To reach a phenotype closer to the heterozygous CARD15 gene expression in case 1 to 8, 15 ng of each CARD15 mutant (R334W, H496L, T605P, N670K, and M513T) was cotransfected with the same amount of the wild-type CARD15 with (▪) or without (□)5 μg/mL MDP. For case 9, 15 ng of each D382E and A612T were cotransfected. Mock vector alone and the wild-type CARD15 alone were added as controls.
Recently, an extensive in vitro CARD15 mutation study has revealed that P668H and I673P mutations in the C-terminal NOD region show the increased basal NF-κB activity and minimal elevation of the activity by addition of MDP.25 Here, we found 5 novel mutations with increased basal NF-κB activity after systematic analysis of Japanese EOS cases. Indeed, location and a biologic effect of the N670K are similar to those of the P668H and I673P, but the mutations we found were distributed through the whole NOD and showed no significant difference in NF-κB activity induced by the maximum dose of MDP.
In conclusion, our results clearly show that EOS is closely related with CARD15 mutations causing constitutive NF-κB activation and shares the common genetic etiology with BS. These findings strongly support the long-standing hypothesis that sporadic EOS and familial BS represent different types of the same juvenile systemic granulomatosis syndrome.13
Prepublished online as Blood First Edition Paper, September 30, 2004; DOI 10.1182/blood-2004-07-2972.
N. Kanazawa and I.O. contributed equally to this work.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Drs M. B. Lutz and T. Berger (University of Erlangen, Erlangen, Germany) for critically reviewing the manuscript.