Abstract
Inhibitory killer immunoglobulin (Ig)-like receptors (KIRs) recognize HLA-C and -B epitopes on target cells, thereby regulating natural killer (NK) cell activity. In 178 patients receiving T-cell-depleted HLA-identical sibling transplants for acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), acute lymphoblastic leukemia (ALL), or myelodysplastic syndrome (MDS), analysis of donor KIR genotype with HLA genotype demonstrated that 62.9% of the patients lacked an HLA ligand for donor-inhibitory KIR. Lack of HLA ligand for donor-inhibitory KIR (missing KIR ligand) had no effect on disease-free survival (DFS), overall survival (OS), or relapse in patients receiving transplants for CML and ALL. In patients with AML and MDS, however, there was a significant missing KIR ligand effect on DFS (P = .014; hazard ratio [HR], 0.53; 95% confidence interval [95% CI], 0.28-0.88) and OS (P = .03; HR, 0.53; 95% CI, 0.3-0.93). Incidence of relapse was also lower in patients with AML and MDS who lacked the HLA ligand for donor-inhibitory KIR (P = .04; HR, 0.41; 95% CI, 0.18-0.97). AML and MDS patients lacking 2 HLA ligands for donor-inhibitory KIR had the highest DFS (P = .002) and OS (P = .003). There was no significant contribution of donor-activating KIR to transplantation outcome in these patients. These data indicate that the absence of class I ligand in the recipient for donor-inhibitory KIR can be a prognostic factor for transplantation outcome in HLA-identical sibling transplantation and that the lack of HLA-C or -B ligands for donor-inhibitory KIR can contribute to improved outcomes for patients with AML and MDS. (Blood. 2005;105:4878-4884)
Introduction
Allogeneic hematopoietic stem-cell transplantation (HCT) is a valuable therapy for refractory acute leukemias, leukemias with a high risk for relapse, myelodysplastic syndromes, and chronic myelogenous leukemia. HCT outcome is dependent on several factors, including the stage of disease, degree of human leukocyte antigen (HLA) identity between donor and recipient, conditioning regimen, and development of graft-versus-host disease (GVHD). Recent studies have indicated that another potential factor influencing transplantation outcome is the presence of donor-derived alloreactive natural killer (NK) cells.1,2
NK cells are bone marrow-derived lymphocytes capable of mediating early innate immune responses to virally infected cells and transformed malignant cells.3,4 As the first lymphocyte subset to reconstitute the peripheral blood after allogeneic HCT,5-7 NK cells have been implicated in the suppression of GVHD, the promotion of bone marrow engraftment,2,8-11 and the mediation of a graft-versus-leukemia effect.1,2,12,13 It is thought that NK cells are regulated by quantitative differences in cumulative activating and inhibitory signals. Mediators of the inhibitory signals include the inhibitory killer immunoglobulin (Ig)-like receptors (KIRs), many of which are known to have differential ligand specificities for HLA class I molecules. Upon interaction with target cells lacking HLA ligand, inhibitory KIR-expressing NK cells may have a lowered threshold for activation, particularly if target cells express additional activating NK receptor ligands.
In transplantations between HLA nonidentical donors and recipients, it has been observed that donor NK cells encountering recipient target cells lacking an HLA class I allele present in the donor HLA genotype can mediate antileukemic effects in myeloid leukemias if the class I mismatch predicts lack of ligand for donor-inhibitory KIR. These antileukemic effects include lower rates of relapse, graft failure, and GVHD, ultimately translating into higher overall survival (OS).1,2 Invoking donor-recipient KIR ligand incompatibility as the basis for NK alloreactivity in HLA nonidentical transplants, several studies have supported the clinical concept of selecting donors for certain myeloid malignancies based on KIR ligand incompatibility in a graft-versus-host direction.1,2,14 Other studies examining the KIR ligand incompatibility concept in transplantation, however, have provided conflicting results.15-18
Central to the KIR ligand incompatibility model is the principle that NK-cell alloreactivity occurs when the HLA genotype of the donor includes a KIR epitope that is not part of the HLA genotype of the allogeneic target cell. The implication of the KIR ligand incompatibility model is that a donor's NK cells exhibit the relevant inhibitory KIR for the donor's HLA ligands. It is known, however, that the KIR genes, which are located on human chromosome 19q13.4, and the HLA genes, which are located on human chromosome 6, segregate independently of each other in normal Mendelian fashion. This allows for the possibility of persons who lack KIR receptors for their HLA ligands and, vice versa, persons who lack HLA ligands for their KIR receptors.19 Furthermore, the expressed KIR repertoire of a person has been shown to be primarily regulated by KIR genotype and not by HLA genotype.20-25
The inhibitory KIRs with identified HLA ligands are KIR2DL2 and KIR2DL3, which recognize the HLA-C group 1-related alleles characterized by an asparagine residue at position 80 of the α-1 helix (HLA-CAsn80), KIR2DL1, which recognizes the HLA-C group 2-related alleles characterized by a lysine residue at position 80 (HLA-CLys80), and KIR3DL1, which recognizes the HLA-Bw4 alleles. Population KIR genotyping studies have revealed that there is significant KIR diversity between persons, such that one or more inhibitory KIR may be lacking in a significant number of persons. Indeed, in a genotype analysis of white persons, 4.8% lack KIR3DL1, and 8.4% lack KIR2DL1.26 Similar to HLA, there are significant differences in KIR genotypes between ethnicities.26-37
Studies investigating the KIR-HLA relationship and susceptibility to viral disease progression and autoimmunity have recognized that the lack of HLA ligand for inhibitory KIR receptors within a patient can have significant effects on clinical outcome in these disease states.38,39 Furthermore, a recent study examining the KIR genotype of the mother and the HLA genotype of the fetus has demonstrated that certain KIR-HLA combinations predispose to the development of preeclampsia.40 Taken together, these clinical studies and KIR immunogenetics studies2,14,19-23,41 suggest that KIR-driven alloreactivity in the transplantation setting might be better predicted if the donor KIR genotype is considered in addition to the HLA genotype of the recipient. A recent study in HLA-haplotype nonidentical transplantations has examined the effect of lack of HLA ligand in the recipient for the donor-inhibitory KIR, finding that this analysis may be more predictive for outcome than the KIR ligand incompatibility approach.25 A logical extension of these findings is that the prediction of NK alloreactivity need not rely on HLA nonidentity between donor and recipient, as is required for the KIR ligand incompatibility studies, but instead on the missing ligand in the recipient, a scenario that can be readily found even in HLA-identical allotransplantations.19
The goal of this study was to test the hypothesis that HLA-identical sibling transplant recipients who lack HLA ligands for their donor-inhibitory KIR have differences in transplantation outcome. Because T-cell depletion has been suggested as facilitating NK-cell recovery and perhaps NK alloreactivity, patients receiving T-cell-depleted transplants were specifically selected for this study.14 In this retrospective single-center study, we analyzed the outcomes of patients with various hematologic malignancies who received T-cell-depleted transplants from HLA-identical sibling donors, grouped according to lack or presence of recipient HLA ligand for donor-inhibitory KIR.
Patients, materials, and methods
Patients
The institutional review board of the Medical College of Wisconsin approved the use of patient samples for this study. All samples were collected with the written consent of the patients or of their legal guardians. One hundred seventy-eight patients with acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), acute lymphoblastic leukemia (ALL), or myelodysplastic syndrome (MDS) receiving T-cell-depleted (TCD) HCT from HLA-identical siblings between 1981 and 1998 were included in this study. Sibling donors were selected prospectively based on low-resolution typing for HLA class I and oligonucleotide typing for class II loci. High-resolution typing was performed retrospectively on archived peripheral blood and bone marrow samples for the purposes of this study. All donor-recipient pairs were confirmed to be HLA-identical. Patient and transplant characteristics are detailed in Table 1.
HLA and KIR genotyping
HLA typing of patients and donors was initially performed by HLA serology and intermediate-level resolution of HLA specificities by a combination of sequence-based amplification (polymerase chain reaction-sequence-specific primer [PCR-SSP])42 and oligonucleotide probing of genomic DNA (PCR-sequence-specific oligonucleotide probe [PCR-SSOP]).43 When needed for unambiguous determination of HLA identity between donor and recipient, additional HLA allele-level typing was performed with direct, sequenced-based typing.44 KIR genotyping was performed according to methods previously described.26
Missing ligand algorithm
Donor-recipient pairs were divided into 2 categories depending on the presence or absence of recipient HLA ligand for donor-inhibitory KIR. KIR genotyping for donors identified the presence or absence of inhibitory KIR2DL1, KIR2DL2, KIR2DL3, and KIR3DL1. Recipient HLA-C and -B alleles were identified by high-resolution DNA-based HLA typing and were segregated, where appropriate, into the epitope groups HLA-C group 1 (HLA-CAsn80), HLA-C group 2 (HLALys80), and HLA-Bw4. KIR ligand presence is defined as the presence of recipient HLA epitopes for the identified donor-inhibitory KIR, and KIR ligand absence (missing KIR ligand) is defined as the absence of one or more recipient HLA epitopes for the identified donor-inhibitory KIR. In this way, patients lacking HLA-Bw4 alleles would be considered as lacking ligand for KIR3DL1 if the donor genotype contains KIR3DL1. If the donor genotype does not include KIR3DL1, the patient would not be considered to be lacking HLA-Bw4 ligand, regardless of HLA genotype. Similar analysis was performed for the patient HLA-C epitopes and their respective donor-inhibitory KIR.
T-cell-depleted transplantation
All patients in this study underwent myeloablative therapy. Patients received total body irradiation (TBI) in addition to a standard conditioning regimen of intravenous cytarabine (3 g/m2 every 12 hours for 6 doses), cyclophosphamide (45 mg/kg given 6 hours after the second and fourth doses of cytarabine), and methylprednisolone (1 g/m2 at 12-hour intervals on days -2 and -1). TBI was begun 48 hours after the last dose of cytarabine and was delivered at dose rates of 8 to 25 cGy/min in 9 fractions over 3 days to a total of 1400 cGy or 6 fractions over 3 days to a total of 1320 cGy.
Bone marrow served as the source of hematopoietic stem cells in all 178 patients, and each patient received a hematopoietic stem-cell product depleted of T cells using the T10B9 (n = 144) or the OKT3 (n = 34) monoclonal antibody/complement depletion method.45 Reliable data of infused doses of T cells were available for 100 of the 178 patients. The mean number of residual T cells for patients receiving allografts depleted according to the T10B9 antibody/complement depletion method (n = 66) was 8.16 × 105/kg; for patients receiving allografts depleted according to the OKT3 antibody method (n = 34), the mean number of residual T cells was 9.26 × 105/kg. There was no statistically significant difference in residual T cells between the 2 methods (P = .75). All patients received GVHD prophylaxis with cyclosporin A after transplantation.
Definitions
Patients with AML in first or second complete remission (CR) were considered to be at standard risk for relapse. Patients with AML who underwent transplantation in CR3 or greater, those with primary refractory or relapsed disease, and those with secondary AML were considered to be at high risk for relapse. Patients with ALL in first CR were considered to be standard risk, and patients with ALL in second CR or with relapsed or refractory disease were considered to be at high risk for relapse. Patients with CML in first chronic phase were considered to be at standard risk, and patients with CML in second chronic phase or in accelerated or blastic phase were considered to be at high risk for relapse. MDS patients with refractory anemia were felt to be at standard risk, whereas all other MDS subgroups were felt to be at high risk for relapse after transplantation. Overall survival (OS) was calculated for the interval between the date of transplantation and death or last follow-up visit. Disease-free survival (DFS) was calculated for the interval between the date of transplantation and either first relapse or death in CR. Relapsed disease for AML, ALL, and MDS was defined by morphologic or cytogenetic evidence either in peripheral blood or in bone marrow. Relapsed disease for CML was defined by cytogenetic evidence in the blood or bone marrow. Of the 61 patients with CML, 26 received donor lymphocyte infusion (DLI) at the time of hematologic or cytogenetic relapse.46 However, for DFS or OS analyses, reinduction of remission was not considered.
Statistical analysis
The primary end points of this study were OS, DFS, and relapse. The Kaplan-Meier method was used to estimate OS and DFS, whereas the cumulative incidence function was used to estimate the probability of relapse. The log-rank statistic was used to evaluate the univariate effects on OS and DFS of missing KIR ligand and of other prognostic factors such as patient age (younger than 18 years vs older than 18 years), cytomegalovirus (CMV) status (CMV-seropositive donor or patient vs CMV-seronegative donor and patient), diagnosis, time to treatment from diagnosis, GVHD, total nucleated cell count per kilogram, T-cell depletion method, and year of transplantation. Gray statistic was used to evaluate the KIR effect on relapse in the presence of competing causes of failure. For our data, death was the competing cause of failure. A proportional hazards model for the end points of OS and DFS and a competing risk model for relapse were developed to examine potential effects of KIR ligand absence or presence on outcome after adjustments for patient age, CMV status, diagnosis, time to treatment for diagnosis, GVHD (treated as a time-dependent covariate), total nucleated cell count per kilogram, and T-cell depletion method. Data were analyzed as of December 1, 2004.
Results
Donor KIR and recipient HLA relationship
Patient and donor characteristics are listed in Table 1. Peripheral blood and bone marrow samples from donor-recipient sibling pairs undergoing TCD HLA-matched HCT were evaluated for KIR and HLA genes. KIR genotyping of 178 donors revealed 92.7% of them to be positive for KIR2DL1 and 92.1% to be positive for KIR3DL1. In addition, 49% were positive for KIR2DL2 and 85.4% were positive for KIR2DL3, with all but 2 donors positive for 1 or both receptors (data not shown). We observed that only 84.8% of donors could be characterized as having an inhibitory KIR for each of the 3 known class I ligands. When analyzed with HLA genotyping, it could be determined that 112 of 178 (62.9%) donor-recipient pairs could be characterized by lack of recipient HLA ligand for donor KIR. The distribution of HLA ligands missing for donor KIR is detailed in Table 2.
Missing ligand and HCT survival
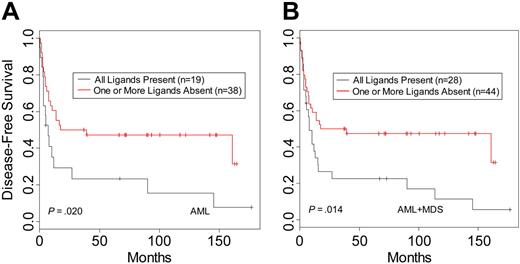
When donor-recipient pairs were segregated based on the presence or absence of recipient HLA ligand for donor KIR, there were no differences in DFS for CML (n = 61; P = .73) or ALL patients (n = 45; P = .55). In contrast, analysis of AML patients revealed a beneficial effect on DFS within the pairs characterized by lack of KIR ligand in the recipient (n = 57; P = .02) (Table 3; Figure 1A). This effect was more apparent when MDS patients were included with the AML patients (n = 72; P = .014) (Table 3; Figure 1B). Similar results were seen for OS (Table 3). The MDS group was too small (n = 15) for reliable analysis as a separate disease category; however, the hazard ratios for DFS and OS suggest a benefit in survival in the group lacking HLA ligand for donor-inhibitory KIR (Table 3). Multivariate analysis was performed to examine the effects of KIR status on outcome after adjustment for patient age, CMV status, time to treatment from diagnosis, GVHD, total nucleated cell count per kilogram, and T-cell depletion method. The results confirmed that missing KIR ligand is an independent predictor of outcome within the AML and MDS groups for relapse and trended to significance as a predictor for DFS. In addition to missing KIR ligand, other important prognostic factors were the time to treatment from diagnosis and total nucleated cell count per kilogram for the DFS and OS end points and GVHD when the end point was relapse (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article).
Disease-free survival of leukemia patients with HLA ligands present or missing for donor-inhibitory KIR. Kaplan-Meier estimates for the probability of disease-free survival in (A) AML patients and (B) AML and MDS patients.
Disease-free survival of leukemia patients with HLA ligands present or missing for donor-inhibitory KIR. Kaplan-Meier estimates for the probability of disease-free survival in (A) AML patients and (B) AML and MDS patients.
To determine whether disease stage contributed to the missing KIR ligand effect, AML patients were categorized as at standard risk (defined as patients who underwent transplantation while in first or second CR) or high risk (defined as patients with secondary AML, patients who underwent transplantation while in remission beyond second CR, and patients with relapsed or refractory disease). Although none of the values reached statistical significance, it appeared that most of the benefit of missing KIR ligand was in the AML and MDS patients at high risk in terms of improved survival. Hazard ratios for DFS and OS in the high-risk AML group were 0.49 (95% CI, 0.2-1.2) and 0.52 (95% CI, 0.2-1.3), respectively. Similarly, hazard ratios for DFS and OS in MDS patients were 0.6 (95% CI, 0.2-2.3) and 0.55 (95% CI, 0.1-2.1), respectively (Table 3). Because MDS is composed of multiple subtypes, some of which can be considered to involve higher risk for transplantation failure or disease relapse, patients with refractory anemia were examined separately from patients with other MDS subtypes. Because of limitations of sample size, however, it could not be demonstrated that MDS subtype conferred greater susceptibility to a missing KIR ligand effect (data not shown).
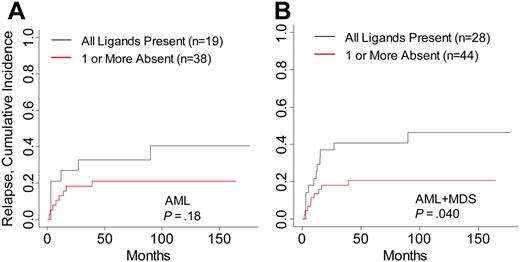
Differences in relapse rates contributed to the increased survival in AML and MDS patients in this study. Cumulative incidence analysis of relapse within AML and MDS patients demonstrated a decreased incidence of relapsed disease in patients lacking HLA ligand for donor-inhibitory KIR (P = .04; Table 3; Figure 2). The number of relapse events within separate AML and MDS disease stages was small, attenuating the comparative analysis of relapse within each disease risk category. Risk for GVHD was not modified by the absence of HLA ligand for donor-inhibitory KIR in any disease group (data not shown). To identify whether high-risk features in ALL and CML were associated with higher survival in patients lacking KIR ligand, patients were stratified into high- and low-risk groups. No significant differences in DFS, OS, or relapse were noted in patients lacking KIR ligand in the high-risk CML and ALL groups, though the favorable hazard ratios in high-risk CML suggest that patients with accelerated or blastic-phase CML who lacked HLA ligand for donor-inhibitory KIR might have had higher DFS and OS rates (Table 3).
Missing KIR ligand specificity
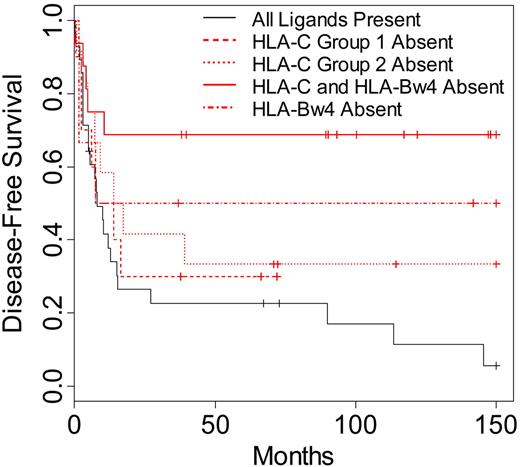
To identify whether a specific missing KIR ligand might have contributed to the increased survival and lower relapse rates seen in the AML and MDS patients, donor-recipient pairs were segregated into specific missing ligand groups. These groups were defined respectively by the lack of recipient HLA-C group 1, HLA-C group 2, and HLA-Bw4 for known donor KIR. A group representing patients lacking 2 ligands (HLA-C ligand in addition to HLA-Bw4) for donor KIR was also included in the analysis. When these groups were compared with patients exhibiting all ligands for known donor-inhibitory KIR, all groups characterized by missing ligand displayed higher DFS and OS (Figure 3). Lack of one class I ligand (HLA-C group 1, HLA-C group 2, or HLA-Bw4 absent) for donor-inhibitory KIR resulted in higher DFS and OS (DFS P = .16; OS P = .22). More strikingly, patients lacking 2 ligands (HLA-C and HLA-Bw4) for donor-inhibitory KIR displayed the highest differences in DFS and OS when compared with patients exhibiting all class I ligands for donor KIR (DFS P = .002, OS P = .003).
Risk for relapse in AML and MDS patients with HLA ligands present or missing for donor-inhibitory KIR. Cumulative incidence estimates for the probability of relapse in (A) AML patients and (B) AML and MDS patients.
Risk for relapse in AML and MDS patients with HLA ligands present or missing for donor-inhibitory KIR. Cumulative incidence estimates for the probability of relapse in (A) AML patients and (B) AML and MDS patients.
Donor-activating KIR
To examine the possibility of contribution of donor-activating KIR receptors to transplantation outcome, patient-donor pairs were separated according to the presence or absence of donor-activating KIR and were analyzed for differences in OS and relapse. When each activating KIR (KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, and KIR2DS5) was analyzed in a univariate analysis (eg, presence or absence of each specific donor-activating KIR gene), there was no effect on survival. Similarly, when each activating KIR was analyzed in combination with the absence of HLA ligand for donor-inhibitory KIR, there was no evidence of activating KIR effect on survival (data not shown).
Disease-free survival of AML and MDS patients according to specific KIR ligand absence. Kaplan-Meier estimates for the probability of disease-free survival in patients lacking the HLA-CAsn80 ligand for donor KIR2DL2/2DL3 (HLA-C group 1 absent; n = 10), patients lacking the HLA-CLys80 ligand for donor KIR2DL1 (HLA-C group 2 absent; n = 12), patients lacking the HLA-Bw4 ligand for donor KIR3DL1 (HLA-Bw4 absent; n = 6), patients lacking both an HLA-Bw4 ligand and an HLA-C ligand for donor KIR (HLA-C and HLA-Bw4 absent; n = 16), or patients with all ligands present for donor-inhibitory KIR (n = 28). Comparison of survival between patients lacking 1 KIR ligand and patients with all ligands present (P = .16), between patients lacking 2 ligands and patients lacking 1 (P = .06), and between patients lacking 2 ligands and patients with all ligands present (P = .002). Significance for heterogeneity between the study groups was tested by the log-rank statistic.
Disease-free survival of AML and MDS patients according to specific KIR ligand absence. Kaplan-Meier estimates for the probability of disease-free survival in patients lacking the HLA-CAsn80 ligand for donor KIR2DL2/2DL3 (HLA-C group 1 absent; n = 10), patients lacking the HLA-CLys80 ligand for donor KIR2DL1 (HLA-C group 2 absent; n = 12), patients lacking the HLA-Bw4 ligand for donor KIR3DL1 (HLA-Bw4 absent; n = 6), patients lacking both an HLA-Bw4 ligand and an HLA-C ligand for donor KIR (HLA-C and HLA-Bw4 absent; n = 16), or patients with all ligands present for donor-inhibitory KIR (n = 28). Comparison of survival between patients lacking 1 KIR ligand and patients with all ligands present (P = .16), between patients lacking 2 ligands and patients lacking 1 (P = .06), and between patients lacking 2 ligands and patients with all ligands present (P = .002). Significance for heterogeneity between the study groups was tested by the log-rank statistic.
Discussion
The “missing self hypothesis,” proposed by Karre et al47 more than a decade ago to explain the regulation of NK-cell activation through its inhibitory receptors, has been tested with mixed results in patients who have undergone allogeneic stem-cell transplantation.2,14-18,48 The KIR ligand incompatibility model predicts inhibitory KIR-driven donor NK alloreactivity in the clinical situation, where HLA disparity between donors and recipients fulfills the criterion of “missing self.”1,2,41 Because KIR and HLA genotypes segregate independently of each other, however, the possibility exists for NK cells to exhibit KIR for which they have no HLA ligand, and, conversely, for persons to exhibit HLA ligands for which they have no KIR. Our study was designed to test the hypothesis that the lack of HLA ligand in the recipient of donor KIR can be used as a genetic indicator of outcome in HLA-identical sibling transplantation. As the first study to examine the missing KIR ligand effect in HLA-identical transplantation and within separate disease categories, we demonstrate that more than 60% of HLA-identical donor-recipient pairs exhibit missing KIR ligand in the recipient, that missing KIR ligand in the recipient leads to increased DFS and OS in AML and MDS patients through a decrease in disease relapse, and that there may be a dose effect of missing KIR ligand such that the lack of both an HLA-B and an HLA-C ligand for donor-inhibitory KIR results in higher probability of OS and DFS than does the lack of either one alone.
The effect of lack of HLA ligand for inhibitory KIR has been the focus of investigation for other disease states, such as autoimmune disease and hepatitis C infection. In patients with psoriatic arthritis, there is a clear association between lack of HLA ligand for inhibitory KIR and development of disease, particularly when the activating KIR receptors KIR2DS1 and KIR2DS2 are also present, leading the authors to conclude that the combination of incoming activating signals with the lack of inhibition of NK cells in these patients tips the balance of NK regulation to favor the autoreactive state.38 Studies examining KIR-HLA in hepatitis C have also demonstrated a relationship between HLA genotype, inhibitory KIR genotype, and likelihood for viral clearance.39 With the exception of allogeneic transplantation, the only clinical situation in which NK alloreactivity might occur with predictable frequency is during pregnancy at the maternal-fetal interface. A recent study examining maternal KIR and fetal HLA has proposed that NK inhibition through fetal HLA engagement of the maternal inhibitory KIR2DL1 receptor favors the development of preeclampsia by reducing NK-assisted, trophoblast-mediated remodeling of uterine vasculature.40 These studies underscore the importance of examining HLA and KIR genotypes in studies on NK cells and disease.
HLA and KIR genotyping was sufficient in this study to demonstrate a genetic algorithm for the prediction of transplantation outcomes in AML and MDS patients according to lack of KIR ligand. In the present study of HLA-identical siblings, T-cell alloreactivity caused by HLA disparity is not a confounding issue, whereas it may be in KIR studies performed in HLA-mismatched and HLA-haploidentical transplantation.2,14,15,17,18,25,49,50 Furthermore, homogeneity in treatment procedure removes potential treatment-related variation, a factor that has been suggested to influence KIR effects.14,15 The higher probability of OS and DFS found in AML and MDS patients lacking HLA ligand for donor-inhibitory KIR support previous findings of antileukemic NK effects in AML but not in ALL in haploidentical transplantation.2,14 Our findings indicate that the higher probability of OS and DFS in AML and MDS patients lacking ligand for donor-inhibitory KIR is related to a lower relapse rate, presumably through NK-mediated antileukemic effects. There is a suggestion that advanced stages of CML, such as accelerated or blastic CML, may be more susceptible to an NK effect driven by lack of KIR ligand. This would be consistent with previous studies performed in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice engrafted with human blastic CML, in which the transfer of human alloreactive NK clones into the mice resulted in clearance of leukemia and longer survival.1
Our findings are in direct contrast to those of a previous report that examined the effect of donor KIR and HLA-C genes on transplantation outcomes between HLA-identical siblings and showed that patients with myeloid disease who are homozygous for HLA-C group 2 alleles had worse survival outcomes when their donors were positive for activating KIR2DS2 than did patients homozygous or heterozygous for HLA-C group 1 alleles.48 Our study, which examined KIR-HLA for each myeloid leukemia separately, did not find any association between donor activating KIR, recipient HLA, and outcome (survival or relapse) in patients with any of the leukemias analyzed, and we found that lack of any known ligands for donor-inhibitory KIR led to an improvement in survival outcome, not deterioration. One major difference between the present study and the previous report is our use of ex vivo T-cell depletion, a factor that may facilitate posttransplantation NK effects.14
Our findings indicate that the absence of HLA-C and HLA-B ligands for donor-inhibitory KIR can mediate an improvement in survival in patients with AML and MDS, and this effect is amplified when both ligands are absent. Previous studies in HLA-haplotype nonidentical transplantation have indicated that the HLA-C ligands are the predominant class I molecules involved in NK alloreactivity,1,2 though HLA-Bw4 effects may be variable because of allotypic variation in KIR3DL1-mediated inhibition.51,52 The current understanding of NK biology is that the regulation of NK function occurs as a result of cumulative, and occasionally opposing, signals from inhibitory and activating receptors.53 It is, therefore, not surprising that incremental release from inhibitory signals because of the lack of HLA-C and HLA-B ligands leads to increased NK activation, particularly if the target cell also exhibits ligands for NK activating receptors, as AML cells are known to do.54
The implication of the present study is twofold: (1) clonal distribution of KIR receptors may exist to give rise to KIR-driven alloreactive NK clones, and (2) these NK clones or their progenitors must also exist in the donor, who exhibits the same HLA genotype as the sibling recipient. There is clear evidence for the clonal distribution of KIR receptors. In addition to demonstrating that the KIR genotype determines KIR repertoire with minimal HLA influence,21,23,55 it has also been shown that individual NK clones can exhibit varying degrees of KIR expression,2,28,56,57 thereby supporting the likelihood that a subset of donor NK clones expressing one inhibitory KIR receptor can be triggered by the lack of engagement of this KIR with its HLA ligand.
It is common for a person to lack HLA ligand for his or her own KIR. As shown by this study, more than 60% of the population is missing one or more ligands for inhibitory KIR, suggesting the possibility of autoreactive NK clones. A recent study by Grau et al57 demonstrated autologous stimulation and cytotoxicity of KIR2DL2-expressing NK clones with ligand specificity for HLA-C group 1 alleles derived from a person who was homozygous for HLA-C group 2 alleles. The frequency of these clones was low in the peripheral blood of the person, leading the authors to speculate that the NK clones are quiescent and thus not functionally autoreactive. The study presented here opens the possibility that the NK-cell repertoire emerging after transplantation contains these clones, which, on expansion in an altered hematopoietic environment, may mediate higher survival through their antileukemic effects.
In summary, this work reveals that missing KIR ligand in the recipient can be commonly found during allotransplantation, occurring not only in HLA nonidentical transplants, but also in HLA-identical sibling transplantation. Using patient homozygosity for HLA-C group 1 alleles, HLA-C group 2 alleles, or HLA-Bw6 alleles as genetic markers predicting NK-mediated antileukemic effects can to some extent be applied in the absence of direct genomic identification of donor KIR genotype because 84.8% of the donors displayed inhibitory KIR receptors for both HLA-C groups and for HLA-Bw4 alleles. If, however, the KIR-HLA class I relationship between recipient and donor in the future becomes part of the donor evaluation and selection, it will be necessary to directly identify the presence or absence of a given KIR gene in the donor. A better understanding of the biologic mechanisms involved in the observed effects must be achieved. This study did not examine host KIR genotype with donor HLA because the likelihood that host NK cells would survive a fully myeloablative conditioning regimen is rare. Alloreactive host NK cells against donor stem cells might be manifested as a failure to engraft, an outcome that did not occur in any of the patients in this study. The issue of host KIR-donor HLA effects may be more relevant in the nonmyeloablative setting, where donor and host NK cells could be predicted to coexist. Finally, it should be noted that inhibitory KIR receptors are expressed not only on NK cells but also on a subpopulation of CD8+TCRαβ T cells.58,59 It will be important to determine the combined effects of missing KIR ligand on NK-cell alloreactivity and adaptive T-cell responses.
Prepublished online as Blood First Edition Paper, February 24, 2005; DOI 10.1182/blood-2004-12-4825.
Supported by grants from the National Institutes of Health (AI49213, CA23766, HL070053, CA08748) and by the Amy Strelzer Manasevits Scholar Program, The Marrow Foundation (K04-01).
K.C.H. and C.A.K. contributed equally to this study.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.