The NOTCH ligand, JAG2, was found to be overexpressed in malignant plasma cells from multiple myeloma (MM) patients and cell lines but not in nonmalignant plasma cells from tonsils, bone marrow from healthy individuals, or patients with other malignancies. In addition, JAG2 overexpression was detected in 5 of 5 patients with monoclonal gammopathy of undetermined significance (MGUS), an early phase of myeloma disease progression. This overexpression appears to be a consequence of hypomethylation of the JAG2 promoter in malignant plasma cells. An in vitro coculture assay was used to demonstrate that JAG2 induced the secretion of interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), and insulin-like growth factor-1 (IGF-1) in stromal cells. Further, the induction of IL-6 secretion was blocked in vitro by interference with anti–Notch-1 monoclonal antibodies raised against the binding sequence of Notch-1 with JAG2. Taken together, these results indicate that JAG2 overexpression may be an early event in the pathogenesis of multiple myeloma involving IL-6 production.
Introduction
Multiple myeloma (MM) is the second most common hematologic malignancy in the United States, with some 13 000 new cases diagnosed each year.1,2 MM is a clonal B-cell malignancy that affects terminally differentiated B cells (ie, plasma cells). Asymptomatic patients present with a low level monoclonal paraproteinemia, no bone lesions, and with less than 10% plasma cells in the bone marrow; by definition they have a monoclonal gammopathy of undetermined significance (MGUS), and they have a relatively good long-term prognosis. Most such patients never have any symptoms related to their monoclonal protein. Approximately 1% of MGUS patients per year develop MM. Some of the remaining patients present with smoldering MM, where 10% to 30% plasma cells are seen in the bone marrow but bone lesions are absent. Patients presenting with de novo active phase may progress to a fulminant phase with the frequent occurrence of extramedullary proliferation and an increase in plasmablastic cells.1,2 These plasmablastic cells present a highly rearranged immunoglobulin Heavy chain (IgH) region due to numerous physiological switch recombinations.3
In the bone marrow (BM), the myeloma cells and stromal cells secrete cytokines and interact through adhesion molecules. Activation of the stromal cells (including osteoclasts) further supports the growth and survival of the myeloma cells and leads to the osteolytic complications associated with MM.4
It is now well recognized that the cytokine interleukin-6 (IL-6) is a major growth factor that promotes the proliferation of malignant plasma cells in MM. Although myeloma cell lines are notoriously difficult to establish, this task has become easier with the demonstration that IL-6 promotes the growth of myeloma cells in vitro.5 Elevated IL-6 levels are directly correlated with tumor burden, bone destruction, and other manifestations of the disease in myeloma patients,6 suggesting that IL-6 plays a pivotal role in MM. Moreover, some studies have shown that myeloma cells induce IL-6 expression in stromal cells in a largely cell contact–dependent manner.7 Therefore, the increased levels of IL-6 production likely reflect disease-associated alteration of IL-6 regulation.
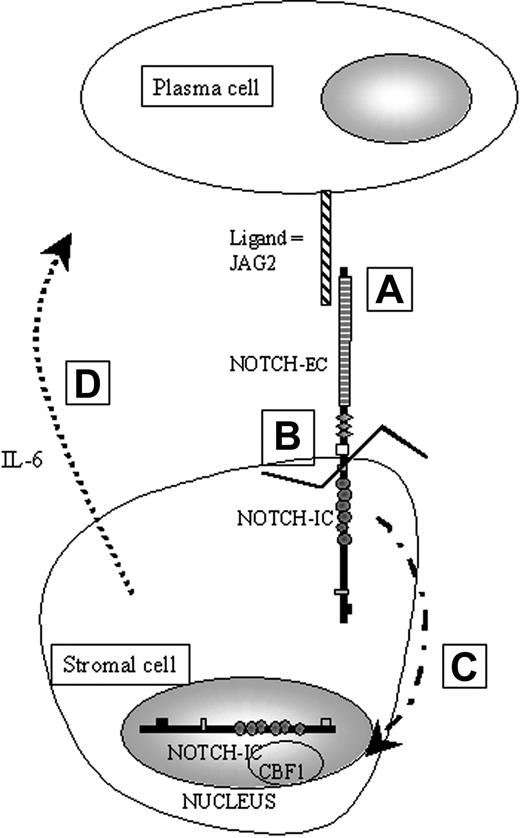
The IL-6 gene can be regulated by a variety of factors, including IL-1, tumor necrosis factor-α (TNF-α) and, as recently demonstrated, the NOTCH genes products. The NOTCH genes were originally identified in Drosophila melanogaster and are members of an evolutionarily conserved family of transmembrane receptors that help to determine cell fate during development.8 During fetal and adult development, expression of NOTCH continues in the proliferative layers of several mature tissues.9-11 These patterns of expression suggest that NOTCH proteins can in some cases maintain the proliferative capacity of immature cells. NOTCH intracellular subunit (NIC) can bind to the transcription factor CBF1/RBP-Jκ 12 (Figure 1), which then becomes a transcriptional activator13 of numerous genes, including nuclear factor–kappa B2 (NFκB2) and IL-6.14,15 This activation has been shown to involve the disruption of a corepressor complex containing silencing mediator of retinoic and thyroid hormone receptor (SMRT) and histone deacetylase-1 (HDAC1).16,17 Therefore, one can hypothesize that overexpression of a NOTCH ligand could activate one or more NOTCH receptors in bone marrow stromal cells and thus, through CBF1 regulation, lead to increased IL-6 production that may promote plasma cell survival and proliferation in MM.
Schematic representation of the physiological activation of NOTCH in our working model of paracrine activation of IL-6. A: JAG2 binds NOTCH via cell-to-cell contact. B: Binding of JAG2 induces a proteolytic cleavage of the intracellular part of NOTCH (NIC). C: When cleaved, NIC is translocated into the nucleus. D: Once in the nucleus, NIC will be able to bind to downstream effectors such as CBF1 to activate, for example, the IL-6 gene transcription.
Schematic representation of the physiological activation of NOTCH in our working model of paracrine activation of IL-6. A: JAG2 binds NOTCH via cell-to-cell contact. B: Binding of JAG2 induces a proteolytic cleavage of the intracellular part of NOTCH (NIC). C: When cleaved, NIC is translocated into the nucleus. D: Once in the nucleus, NIC will be able to bind to downstream effectors such as CBF1 to activate, for example, the IL-6 gene transcription.
Previously published work suggested that JAG2 might be the ligand of interest in the context of MM. Indeed, JAG2 is localized at 14q32.3, just upstream of the IgH switch regions.18 Moreover, mouse JAG2 promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact.19 JAG2 has also been shown to induce cell cycling in confluent fibroblasts susceptible to density-dependent inhibition of cell division and therefore may contribute to neoplastic transformation.20 In addition, JAG2 is down-regulated during normal bone marrow differentiation.19 Finally, because MM cells have been shown to induce IL-6 expression in stromal cells in a largely cell contact–dependent manner,7 we hypothesized that MM cells induce production of IL-6 in stromal cells21 through overexpression of JAG2. Once secreted, IL-6 enhances proliferation of myeloma cells in a paracrine fashion. On the basis of these data and the IL-6–mediated paracrine stimulation of MM cells by bone marrow stromal cells, we sought to assess the expression level of JAG2 in the malignant plasma cells of MM and MGUS patient samples and cell lines (as compared with controls) and to determine if alteration of JAG2 expression could play an oncogenic role in these malignancies.
Materials and methods
Samples and cell lines
RPMI 8226, U266 (MM cell lines), and MRC5 (immortalized fibroblasts) were obtained from the American Type Culture Collection ([ATCC] Manassas, VA); K620, an IgG-κ plasma cell leukemia cell line,22 and DoHH2 (transformed non-Hodgkin lymphoma [NHL])23 were a gift from Dr Elisabeth Nacheva (Royal Free Hospital, London, United Kingdom); KMSM-1 and -2 (2 unpublished cell lines established from plasma cell leukemia patients presenting an IgG-κ light chain expression) were a gift from Dr Masami Nagai (Kagawa University, Japan); MM1-S and -R (dexamethasone sensitive and resistant, respectively) were a gift from Dr Steve Rosen (Northwestern University, Chicago, IL); EBV-LIN (lymphoblastoid cell line/immortalized lymphocytes) was a gift from Dr Thomas Ellis (The Blood Center of South Eastern Wisconsin, Milwaukee, WI); and the MUTZ5 (acute lymphoblastic leukemia [ALL]) cell line24 was a gift from Dr Hans Drexler (DSMZ, Braunschweig, Germany). NIH 3T3 fibroblasts transfected with full-length human JAG2 cDNA (by L.M.) and the MRC5 cell line (provided by L.M.) were cultured in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), antibiotics, and 1 mg/mL G418 for NIH 3T3–JAG2 cells. All other cell lines were cultured in RPMI 1640 medium (Invitrogen) with addition of 10% FBS and antibiotics. Tonsils were collected through a clinical protocol approved by the Institutional Review Board (IRB) from Loyola University Medical Center, Maywood, IL. In addition, polyclonal plasmablasts were obtained through culture of peripheral blood cells with a panel of interleukins (IL-2, -4, -10, and -12) and anti-CD40L. A similar approach has been recently reported.25 The plasmablasts were then purified using positive selection with CD138 columns (Miltenyi Biotec, Auburn, CA). Fresh MGUS/MM (or unrelated disease [for controls]) patient samples were collected through an IRB-approved clinical protocol established at Loyola University Medical Center. Each patient signed an informed consent as requested by federal laws and the IRB committee. The MM-related patient samples presented an average of 18.5% plasma cells (3% to 66%). Fourteen of these patients were either new patients or patients seen in follow-up examinations who had MM, MGUS, or smoldering myeloma. Four samples were obtained from patients with nonplasma cell disorders. The bone marrow collected was processed to either isolate plasma cells using positive selection with CD138 columns (Miltenyi Biotec) or, when the percentage of plasma cells in the BM was too low, subjected directly to fluorescence-activated cell sorter (FACS) analysis (when the percentage of plasma cells is too low, the loss encountered during the purification process may leave us with a number of cells too small to analyze, therefore not allowing us to perform an accurate analysis [JAG2 plus isotype]).
RT-PCR
Total RNA was prepared with the ToTALLY RNA extraction kit (Ambion, Austin, TX) according to manufacturer's instructions. First-strand cDNA synthesis (MBI Fermentas, Hanover, MD) was performed at 42°C for 1 hour with 1 μg total RNA. Polymerase chain reaction (PCR) was performed with 100 ng cDNA in 200 μM for each deoxynucleotide triphosphate and 5 pmol of each JAG2 (forward: 5′-GACGTGCTCTACCAGTGCAAGAA-3′; reverse: 5′-AACAACCACAGGTGCGTCAACAG-3′), HES-1 (forward: 5′-GTCAACACGACACCGGATAA-3′; reverse: 5′-GTCACCTCGTTCATGCACTC-3′), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (human GAPDH reverse transcriptase [RT]–PCR primer, Stratagene, La Jolla, CA). The PCR cycle was as follows: 35 cycles at 92°C for 20 seconds, 57°C for 20 seconds, 72°C for 2 minutes with a 2-minute initial denaturation at 92°C, and a final 10-minute elongation step at 72°C. The expected PCR products are either a 912 bp JAG2 fragment, a 305 bp HES-1 fragment, or a 525 bp GAPDH fragment. PCR products (10 μL) were electrophoresed on a 1% agarose gel impregnated with ethidium bromide and then photographed. The RT-PCR amplification was repeated up to 3 times to assure reproducibility of the results.
Immunostaining and fluorescence
Cytospins were prepared from the same cell lines and controls and fixed with acetone for 10 minutes at room temperature and air-dried. For immunofluorescence, cells were incubated with a goat anti-JAG2 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) followed by incubation with a fluorescein isothiocyanate (FITC)–conjugated rabbit antigoat (Santa Cruz Biotechnology) coupled with a monoclonal phycoerythrin (PE)–conjugated anti-CD138 antibody (Pharmingen BD, Franklin Lakes, NJ). Nuclei were counterstained with diamidino-phenyl-indole (DAPI) (Vectashield, Vector Laboratories, Burlingame, CA). For immunocytochemistry, a double stain kit (Dako, Carpinteria, CA) was used where the anti-JAG2 antibody was revealed with DAB, whereas the anti-CD138 monoclonal antibody was revealed with Fast Red. No nuclei counterstain was applied. Fluorescent images (Figure 2B-1-2-5-6) were acquired with an Optiphot microscope (Nikon, Melville, NY) through an oil PLAN Apo (100×, 0.4 app.) and filters specific for FITC and PE. The microscope was equipped with a CCD camera (1300DS, Digital instruments, Woodbury, NY) and the EasyFISH software (Applied Spectral Imaging Inc, Vista, CA) was used for capture and background reduction, without additional modifications. Light images (Figure 2B-3-4-7-8) were acquired with a BX40 microscope (Olympus, Melville, NY) through an oil PLAN (100×, 1.25 app.) to visualize DAB and fast red stainings. The microscope was equipped with a DP10 Digital camera (Olympus) and images were captured and printed directly without alteration with the Camedia Printer software (Olympus).
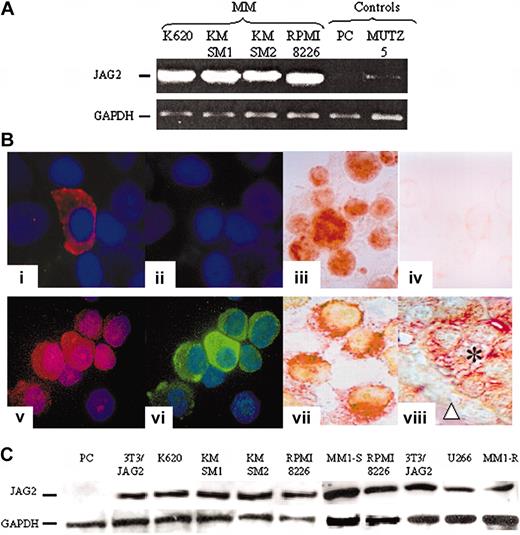
JAG2 expression. (A) Representative result of RT-PCR experiments assessing the levels of JAG2 transcripts in JAG2-negative (MUTZ5-ALL) and normal plasma cells (PC) and JAG2-positive (KMSM-1 and -2, K620, RPMI 8226) cell lines. GAPDH levels were used to normalize the amount of cDNA present in each tube. (B) Comparative immunostaining results. Normal nonpurified (1-2) and MM (5-6) plasma cells stained with an anti-CD138 (1,5) and anti-JAG2 (2,6). Panels 3 and 4 are a representative (K620) MM cell line stained with the anti-JAG2 (3) and goat isotype (4) antibodies; panels 7 and 8 show double color staining anti-CD138 (Fast Red) and -JAG2 (DAB) of an MM cell line (KMSM-1) (7) and a paraffin section from an MM patient sample (8). (C) Western blot analysis of normal PC, a NIH 3T3 cell line overexpressing JAG2, and 7 MM cell lines (K620, KMSM-1 and -2, RPMI 8226, U266, MM1-S and -R).
JAG2 expression. (A) Representative result of RT-PCR experiments assessing the levels of JAG2 transcripts in JAG2-negative (MUTZ5-ALL) and normal plasma cells (PC) and JAG2-positive (KMSM-1 and -2, K620, RPMI 8226) cell lines. GAPDH levels were used to normalize the amount of cDNA present in each tube. (B) Comparative immunostaining results. Normal nonpurified (1-2) and MM (5-6) plasma cells stained with an anti-CD138 (1,5) and anti-JAG2 (2,6). Panels 3 and 4 are a representative (K620) MM cell line stained with the anti-JAG2 (3) and goat isotype (4) antibodies; panels 7 and 8 show double color staining anti-CD138 (Fast Red) and -JAG2 (DAB) of an MM cell line (KMSM-1) (7) and a paraffin section from an MM patient sample (8). (C) Western blot analysis of normal PC, a NIH 3T3 cell line overexpressing JAG2, and 7 MM cell lines (K620, KMSM-1 and -2, RPMI 8226, U266, MM1-S and -R).
Western blots
Normal plasmablast cells as well as cells from several MM cell lines (K620, KMSM-1 and -2, RPMI 8226, U266, MM1-S, MM1-R) and our NIH 3T3–JAG2 cell line were lysed for 30 minutes on ice in lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.4; 1% Nonidet P-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 1 mM phenylmethylsulfonyl fluoride [PMSF]; 1 mg/mL aprotinin, leupeptin, pepstatin; 1 mM Na3VO4; 1 mM NaF). Cell debris was removed by centrifugation (14 000g, 15 minutes at 4°C). Samples (50 μg total protein per lane) were subjected to electrophoresis on a 3% to 7% NuPAGE Tris-Acetate gel (Invitrogen) followed by transfer to a polyvinylidene fluoride (PVDF) membrane (Amersham Pharmacia Biotech, Picataway, NJ) using NuPAGE transferring system (Invitrogen). The membranes were then incubated with a polyclonal anti-JAG2 antibody (Santa Cruz Biotechnology) for 2 hours at room temperature, followed by incubation with antigoat–horseradish peroxidase (HRP) (Santa Cruz Biotechnology). Proteins were visualized with enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia Biotech). To confirm equal loading of protein, the membranes were stripped with 2% sodium dodecyl sulfate (SDS) buffer and reprobed with anti-GAPDH antibody (Santa Cruz Biotechnology). In addition, the most established MM cell lines (RPMI 8226, U266, MM1-S and -R) were probed with anti–NOTCH1, anti-NOTCH2, anti-NOTCH3, anti-NOTCH4, and anti–Jagged-1 (Santa Cruz Biotechnology).
FACS analysis
JAG2 expression was assessed by FACS using either a triple-color assay (anti-CD138–allophycocyanin [APC] coupled with anti-κ/λ–PE and with goat anti-JAG2 followed by antigoat–FITC), a double-color assay (anti-CD138–PE coupled with goat anti-JAG2 followed by antigoat–FITC), or a single-color approach (anti-JAG2 primary antibody alone) after positive purification of the plasma cells using CD138-bound magnetic beads (Miltenyi Biotec), according to manufacturer's specifications. All FACS experiments included controls reacted with either goat Ig isotype or only anti-JAG2 primary antibodies.
Genomic PCR on NotI-digested (or not) DNA from JAG2-positive and -negative cell lines
Five JAG2-positive (MM) cell lines (RPMI 8224, KMSM-1 and -2, U266, K620) and 3 JAG2-negative (MUTZ5 [ALL], DoHH2 [NHL], EBV-LIN [immortalized lymphocytes]) cell lines were used in these experiments. One microgram of DNA from each cell line was digested with 10 units of NotI restriction enzyme (Invitrogen) in the appropriate buffer. PCR amplification was subsequently performed using JAG2-specific primers (forward: 5′-CCAGCCTCAGCTTCACTCTA-3′; reverse: 5′-TCAGCCAGACCTCACGATGT-3′) as well as GAPDH-specific primers.18 The JAG2 primer pair amplifies a 1.6 kb fragment around the NotI site and encompassing the JAG2 promoter site. The PCR cycle was 94°C for 5 minutes followed by 35 cycles at 94°C for 20 seconds, 57°C for 20 seconds, and 72°C for 2 minutes, with a 10-minute extension period at 72°C. Due to the high GC content of the region to amplify, a GC melt mix was used (Invitrogen) supplemented with 200 μM for each deoxynucleotide triphosphate and 5 pmol of each primer. PCR products were electrophoresed as already described.18
Methylation blocking experiments
The JAG2-negative cell lines were cultured in RPMI medium supplemented with 10% FBS and 5% CO2 in a humidified incubator. 5-Azacytidine (5-AZA) (Sigma-Aldrich, St Louis, MO) was added to the culture medium and further cultured for 72 hours. The 5-AZA requires at least 2 cell cycles to be effective in most cells and to block methylation in the daughter cells. Subsequent FACS analyses to assay for JAG2 expression were performed as already described.
In vitro coculture assay
The MRC5 fibroblast cell line was used as a feeder layer in these experiments. Culture wells were seeded with MRC5 cells. Upon 70% confluence, the cells were irradiated (25 cGy) to stop their growth. IL-6, vascular endothelial growth factor (VEGF), and insulin-like growth factor-1 (IGF-1) levels were assayed by enzyme-linked immunosorbent assay (ELISA) (Pierce Biotechnology, Rockford, IL, for IL-6 and VEGF; and R&D Systems for IGF-1) on the culture medium when MRC5 cells were either cultured alone or cocultured with the K620, U266, or RPMI 8226 MM cell lines with or without an insert in the well, avoiding cell contacts between the 2 cell types when the insert is present. When used, the anti–NOTCH-1 monoclonal antibody was added directly to the culture without preincubation with the MM cells. When HES-1 expression was assessed in the MRC5 cells, MM cells were removed by shaking the culture plates and aspiration and the MRC5 cells were collected for RNA extraction followed by RT-PCR. In some experiments, soluble peptides corresponding to the binding sequence of Jagged-1 and JAG2, which mimic function of the ligands (L.M., personal communication, 2003), were used to activate NOTCH proteins in the MM and stromal/MRC5 cells. Cells were incubated with increasing amounts of binding peptides, and IL-6, VEGF, and IGF-1 secretion was measured by ELISA as already described.
Results
JAG2 overexpression in malignant but not in normal plasma cells
Using reverse-transcriptase polymerase chain reaction (RT-PCR), we evaluated 100 ng cDNA from 4 MM cell lines and 1 non-MM cell line as well as in normal plasmablast cells for the transcript expression level of JAG2. High JAG2 expression was detected in 4 of 4 MM cell lines, whereas this expression was barely detectable in the non-MM cells (Figure 2A). To confirm this putative overexpression at the protein level, we studied the same cell lines by immunofluorescence/immunocytochemistry. Similarly, JAG2 overexpression was detected in all the MM cell lines, whereas in non-MM cells JAG2 levels were below the detection limit of our method (signal showed no difference compared with goat isotype control) (Figure 2B). Western blot analysis of the same 4 MM cell lines in addition to U266, MM1-S, and MM1-R (Figure 2C) confirmed JAG2 overexpression. Controls included normal plasmablast cells (negative) and our NIH 3T3 cell line that constitutively expresses a retrovirally transduced JAG2 (positive). Our results indicate that the overexpression observed in our MM cell lines is comparable to the ectopic expression directed by a retroviral long terminal repeat (LTR) in the NIH 3T3 cell line. To assess JAG2 protein levels in fresh samples, we developed a FACS approach. We were able to quantify JAG2 expression levels in malignant (cell lines) and plasmablast cells isolated from normal BM as well as from tonsils (plasmablasts). The normal plasmablast cells showed a comparable level of JAG2 expression with the goat isotype control antibody (Figure 3A, left panel), whereas all the MM cell lines showed a marked JAG2 overexpression (Figure 3A, right panel). Therefore, high-level expression of the JAG2 protein appears frequently in MM cell lines and thus distinguishes MM from nonmalignant plasma cells.
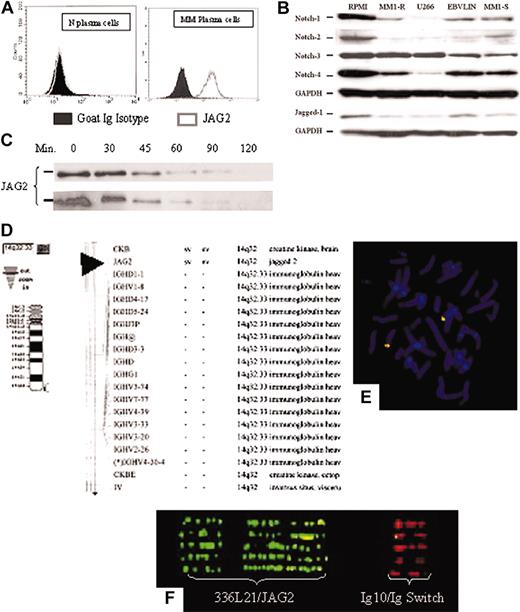
JAG2 expression. (A) FACS analysis of normal (left panel) and MM (right panel) plasma cells for JAG2 expression. Shown are the JAG2 expression profiles of the CD138+ plasma cells using either a goat Ig isotype or anti-JAG2 primary antibodies. (B) Western blot analysis of 4 MM (RPMI 8226, U266, MM1-S and -R) and 1 lymphoblastoid (EBV-LIN) cell lines for the Notch-1, -2, -3, -4, and Jagged-1 proteins with GAPDH as loading control. (C) Time-course Western blot on the RPMI 8224 cell line (MM, JAG2-positive) (top panel) and the NIH 3T3 cell line constitutively expressing a retrovirally transduced JAG2 (bottom panel) after incubation with puromycin. No difference in JAG2 protein stability is detected between the two cell lines. If a stabilized protein were to be present in the MM cell lines, the JAG2 protein would be still present after the production of new protein has been blocked by puromycin. (D-F) Physical mapping of the JAG2 gene. (D) National Center for Biotechnology Information (NCBI)–related mapping showing the proximity of JAG2 with IgH and (E) confirmation of the physical mapping using the RP11-336L21 BAC clone in combination with the Ig10 cosmid on normal chromosome metaphase. (F) Fiber-FISH profile obtained with extended DNA from both normal controls and MM cell lines using the RP11-336L21 and the Ig10 probes.
JAG2 expression. (A) FACS analysis of normal (left panel) and MM (right panel) plasma cells for JAG2 expression. Shown are the JAG2 expression profiles of the CD138+ plasma cells using either a goat Ig isotype or anti-JAG2 primary antibodies. (B) Western blot analysis of 4 MM (RPMI 8226, U266, MM1-S and -R) and 1 lymphoblastoid (EBV-LIN) cell lines for the Notch-1, -2, -3, -4, and Jagged-1 proteins with GAPDH as loading control. (C) Time-course Western blot on the RPMI 8224 cell line (MM, JAG2-positive) (top panel) and the NIH 3T3 cell line constitutively expressing a retrovirally transduced JAG2 (bottom panel) after incubation with puromycin. No difference in JAG2 protein stability is detected between the two cell lines. If a stabilized protein were to be present in the MM cell lines, the JAG2 protein would be still present after the production of new protein has been blocked by puromycin. (D-F) Physical mapping of the JAG2 gene. (D) National Center for Biotechnology Information (NCBI)–related mapping showing the proximity of JAG2 with IgH and (E) confirmation of the physical mapping using the RP11-336L21 BAC clone in combination with the Ig10 cosmid on normal chromosome metaphase. (F) Fiber-FISH profile obtained with extended DNA from both normal controls and MM cell lines using the RP11-336L21 and the Ig10 probes.
Finally, we assessed the expression levels of other members of the NOTCH family (receptors: NOTCH1, NOTCH2, NOTCH3, NOTCH4; ligands: Jagged-1) (Figure 3B) in RPMI 8226, U266, MM1-S, and MM1-R MM cell lines and a lymphoblastoid cell line (EBV-LIN) by Western blot. Our results show that NOTCH-2 was detectable only in RPMI 8226; NOTCH-1, -3, and -4 is detected in all cell lines (MM and lymphoblastoid lines); and Jagged-1 is detected in RPMI 8226 and EBV-LIN (lymphoblastoid line) whereas only a faint band is detected in the other 3 MM cell lines.
JAG2 overexpression is driven by promoter hypomethylation
To assess whether the JAG2 overexpression is due to transcriptional or posttranscriptional processes, we performed a series of experiments to verify that the JAG2 protein was not stabilized by, for example, a specific mutation and that the level of expression would reflect both transcriptional deregulation as well as protein stabilization. The RPMI 8226 and U266 MM cell lines, as well as the NIH 3T3 cell line constitutively expressing JAG2, were cultured in the presence of puromycin to prevent the production of new protein. Proteins were harvested at different times after introduction of puromycin in the culture medium, and SDS–polyacrylamide gel electrophoresis (PAGE) and Western blotting were performed. We did not detect any stabilization of the protein (Figure 3C) with similar results being obtained from the MM cell lines RPMI 8226 and U266 and the NIH 3T3–JAG2 cell line. These experiments indicated that the JAG2 protein overexpression was likely due to the alteration of transcriptional regulation of the gene and not to stabilization of the protein.
To evaluate whether any potential genomic rearrangements could be responsible for the observed overexpression, we first confirmed the chromosomal assignment of the JAG2 gene at 14q32.18 A BAC clone containing the entire JAG2 gene (RPCI11-336L21) was identified and used in fluorescence in situ hybridization (FISH) experiments26,27 in combination with the Ig10 cosmid that encompasses the IgH C/switch region (Figure 3D-F). These results confirmed the localization of JAG2 to 14q32, just upstream of the IgH switch region. The JAG2-specific BAC clone has been used in FISH experiments to identify potential genomic rearrangements in the MM cell lines. We were unable to detect any gross rearrangement in either chromosome preparations or in interphase cells (data not shown). In addition, extended DNA was prepared from normal peripheral blood lymphocytes (PBLs) as well as from MM cell lines and used for FISH experiments.28,29 No abnormal fiber-FISH profile could be detected in the cell lines studied (RPMI 8226, U266, KMSM-1 and -2) (Figure 3F).
Because no gross genomic rearrangement could be detected, we analyzed the JAG2 promoter region for altered sequences. The hypothesis was that a submicroscopic or point mutation in a functionally critical area (a repressor binding site, for example) could be responsible for the lack of repression of JAG2, thus leading to overexpression. The sequence of the 4 kb region upstream of the first JAG2 exon did not reveal any mutations that could conceivably affect the transcription regulation of JAG2. Therefore, these experiments were interpreted to mean that JAG2 overexpression is not due to a genomic rearrangement or sequence alteration.
We identified by sequence analysis that the 1.2 kb region upstream of the ATG start codon contains about 160 CpG dinucleotides. In addition, we identified a NotI site situated 252 bp upstream of the ATG start codon, halfway between the core promoter region7 and the ATG. We therefore studied the methylation status of this region because (hyper-) methylation of a promoter region usually results in the blocking of its transcription.
DNA extracted from both JAG2-positive (MM) and -negative (ALL) cell lines were digested with NotI, a methylation-sensitive restriction enzyme. Following digestion, PCR was performed using primers located on either side of the NotI site. If the targeted region is hypomethylated, the DNA should be digested and no PCR product would be obtained. If the targeted region is hypermethylated, the DNA should remain intact, and therefore a PCR product could be obtained. The results on our positive and negative cell lines (Figure 4A-B) showed that the JAG2-positive (MM) cell lines did not produce any PCR amplicon with the digested DNA, indicating a hypomethylated state, whereas the JAG2-negative lines produced a PCR product. Therefore, these experiments showed that the differential of JAG2 expression correlates with a differential in the methylation status of the JAG2 promoter region.
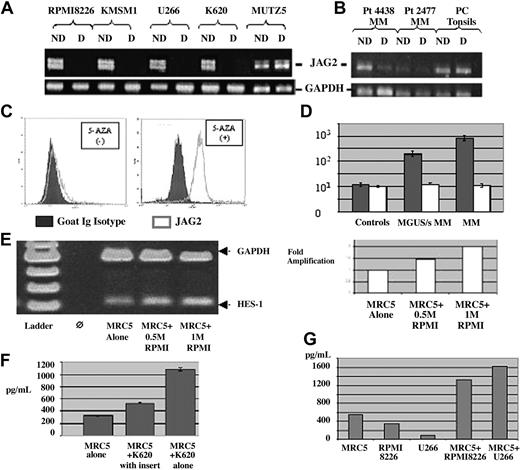
Methylation status. PCR on NotI-digested and undigested DNA from (A) MM JAG2-positive and -negative (MUTZ5) cell lines with primers located on either side of the NotI site. The JAG2-positive line shows the absence of a band when digested (hypomethylated promoter region), whereas the negative line shows no difference (hypermethylated promoter region). (B) A similar PCR pattern was obtained on NotI-digested and undigested DNA from 2 MM JAG2-positive samples (4438, 2477) and (JAG2-negative) tonsil plasma cells using the same primers. The JAG2-positive samples show a reduction of band when digested (hypomethylated promoter region), whereas the JAG2-negative cells show no difference (hypermethylated promoter region). The profile with MM patient DNA presents a reduction of band intensity, certainly due to the nonpurity of the plasma cells after purification. ND indicates not digested; D, digested. (C) JAG2 expression profile in the MUTZ5 cell line before (left panel) or after incubation with 5-azacytidine (right panel). Blocking of methylation with 5-AZA induced JAG2 expression in the negative cell line, comparable to the levels observed in MM. (D) Representation of the JAG2 expression levels differential between controls, MGUS/smoldering MM, and MM samples per cell lines. (E) RT-PCR with HS-1 and GAPDH-specific primers on RNA extracted from MRC5 cells after coculture with 0.5 million and 1 million MM cells. The induction of HES-1 expression is proportional to the activation signal per number of MM cells in the coculture (right panel). (F) IL-6 detection assay by ELISA for the assessment of IL-6 secretion with MRC5 cells alone, MRC5 cells cocultured with K620 cells (MM) in an insert (no contact between the two cell types), and MRC5 cells cocultured with K620 cells. (G) VEGF secretion measured by ELISA in the MM cells alone (RPMI 8226 and U266) or when cocultured with the MRC5 cells. Error bars (panels D and F) represent standard deviation.
Methylation status. PCR on NotI-digested and undigested DNA from (A) MM JAG2-positive and -negative (MUTZ5) cell lines with primers located on either side of the NotI site. The JAG2-positive line shows the absence of a band when digested (hypomethylated promoter region), whereas the negative line shows no difference (hypermethylated promoter region). (B) A similar PCR pattern was obtained on NotI-digested and undigested DNA from 2 MM JAG2-positive samples (4438, 2477) and (JAG2-negative) tonsil plasma cells using the same primers. The JAG2-positive samples show a reduction of band when digested (hypomethylated promoter region), whereas the JAG2-negative cells show no difference (hypermethylated promoter region). The profile with MM patient DNA presents a reduction of band intensity, certainly due to the nonpurity of the plasma cells after purification. ND indicates not digested; D, digested. (C) JAG2 expression profile in the MUTZ5 cell line before (left panel) or after incubation with 5-azacytidine (right panel). Blocking of methylation with 5-AZA induced JAG2 expression in the negative cell line, comparable to the levels observed in MM. (D) Representation of the JAG2 expression levels differential between controls, MGUS/smoldering MM, and MM samples per cell lines. (E) RT-PCR with HS-1 and GAPDH-specific primers on RNA extracted from MRC5 cells after coculture with 0.5 million and 1 million MM cells. The induction of HES-1 expression is proportional to the activation signal per number of MM cells in the coculture (right panel). (F) IL-6 detection assay by ELISA for the assessment of IL-6 secretion with MRC5 cells alone, MRC5 cells cocultured with K620 cells (MM) in an insert (no contact between the two cell types), and MRC5 cells cocultured with K620 cells. (G) VEGF secretion measured by ELISA in the MM cells alone (RPMI 8226 and U266) or when cocultured with the MRC5 cells. Error bars (panels D and F) represent standard deviation.
To demonstrate that this methylation status is causal for elevated JAG2 expression, we treated the JAG2-negative cell lines (Figure 4C, left panel) with 5-azacytidine (which inhibits DNA methylation). FACS analysis of these cell lines (initially JAG2-negative) after 5-AZA treatment revealed an elevated level of JAG2 expression comparable to the MM cell lines (Figure 4C, right panel). In addition, DNA analysis on the DNA from 5-AZA–treated cells showed a profile similar to that of the MM cell lines (hypomethylation resulting in absence of band when digested) (data not shown).
JAG2 overexpression in MGUS and MM patient samples
To relate our observations made on cell lines to alterations in primary tumors, we studied a series of MGUS/MM samples (Table 1) by FACS and immunofluorescence. FACS analysis on patient samples was performed using a 3-color detection system to ensure an accurate analysis of JAG2 expression in the malignant plasma cells. In all cases, immunofluorescence or immunohistochemistry was also performed in double color as already described (Figure 2B). All the MM and MGUS samples showed a marked JAG2 overexpression, whereas the non-MM samples did not. These results perfectly reproduce those obtained with our cell lines. Moreover, JAG2 expression levels seem to increase with the stage of the disease (Figure 4D). RT-PCR performed on some of the samples (for which we obtained enough cells for both experiments after CD138 selection) showed a similar transcript level increase (data not shown). Therefore, this series of experiments on fresh patient samples confirmed our data obtained on cell lines, showing that JAG2 overexpression is characteristic for MM cell lines and patient samples. We used the same approach as described with our cell lines to check the methylation status of at least some of the patient samples we obtained. DNA was extracted from the purified CD138+ plasma cells and digested (or not as control) with NotI. PCR was performed as previously described. The 4 patient samples tested showed the same pattern as that obtained with the cell lines. Plasma cells purified from tonsils presented the same hypermethylated profile as our negative cell line (Figure 4B).
The NOTCH target gene HES-1 expression is induced in the stromal cells upon cell-to-cell contact
To assess the induction of HES-1 expression in the MRC5 cells, we performed an RT-PCR using HES-1–specific primers with RNA extracted from the MRC5 cells after coculture with the RPMI 8226 MM cell line. Our results (Figure 4E) show that the HES-1 expression is induced upon coculture with the MM cell line and that this induction is proportional to the number of MM cells used in the coculture.
IL-6 secretion blocked by specific anti-Notch monoclonal antibody
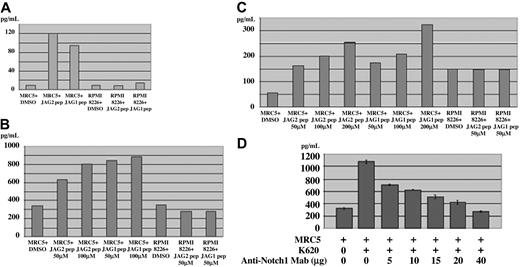
To demonstrate that IL-6 secretion is a direct consequence of JAG2 overexpression in the MM context, we developed a coculture in vitro assay using fibroblast cells and our multiple myeloma cell lines (which overexpress JAG2 as already shown). IL-6 levels were assayed on the culture medium when MRC5 cells were cultured alone, when they were cocultured with an MM cell line (K620), or when they were cocultured with the MM cell line but separated from them by an insert, avoiding cell contacts between the 2 cell types. Coculture of MRC5 and the MM cell line induced a 3.5-fold increase in IL-6 secretion (Figure 4F) whereas the same coculture without contact between the 2 cell types only marginally affected IL-6 secretion (1.6-fold). In addition, we showed that interaction between MM cells and the MRC5 cells induced the secretion of VEGF and IGF-1, 2 other cytokines important in MM (Figure 4G). To provide confirmatory evidence for the role of JAG2 in the induction of cytokine secretion, we incubated the MRC5 cells with the JAG2-binding peptide. As shown in Figure 5A-C, we were able to show that incubation with the JAG2-binding peptide induces IL-6, VEGF, and IGF-1 secretion. Interestingly, incubation of the MM cells alone with the JAG2-binding peptide did not induce cytokine secretion. The same results were obtained when a Jagged-1–binding peptide was used.
Incubation with binding peptide. (A) IL-6, (B) VEGF, and (C) IGF-1 secretion measured by ELISA when the MRC5 cells or MM cells are incubated with JAG2- and Jagged-1–binding peptides. (D) IL-6 secretion measured by ELISA with MRC5 cells cultured alone, MRC5 cells cocultured with K620 (MM) cells alone, or with incremental amounts of anti–NOTCH-1 mAb (5, 10, 15, 20, 40 μg). The IL-6 secretion is inversely proportional to the amount of mAb used. Error bars represent standard deviation.
Incubation with binding peptide. (A) IL-6, (B) VEGF, and (C) IGF-1 secretion measured by ELISA when the MRC5 cells or MM cells are incubated with JAG2- and Jagged-1–binding peptides. (D) IL-6 secretion measured by ELISA with MRC5 cells cultured alone, MRC5 cells cocultured with K620 (MM) cells alone, or with incremental amounts of anti–NOTCH-1 mAb (5, 10, 15, 20, 40 μg). The IL-6 secretion is inversely proportional to the amount of mAb used. Error bars represent standard deviation.
To demonstrate the role of the NOTCH pathway in the induction of cytokine by the MM cells, we used an anti–NOTCH-1 monoclonal antibody as a blocking agent. This specific antibody was raised against the NOTCH-1 domain responsible for the binding with its ligand and has been shown to block NOTCH signaling in thymocytes.30 This monoclonal antibody (mAb) blocking reagent was used in the coculture in vitro assay described above, with coculture of the MRC5 and K620 cell lines either alone or in the presence of an increasing amount of anti–NOTCH-1 mAb. We showed that the IL-6 secretion was inversely proportional to the amount of mAb added to the culture medium (Figure 5D).
Discussion
In this paper, we have shown that malignant plasma cells from multiple myeloma cell lines and MM and MGUS patient samples display marked JAG2 overexpression. In comparison, plasma cells from normal controls had barely detectable JAG2 expression, even by RT-PCR. The detection of JAG2 overexpression in our MGUS samples seems to indicate that JAG2 overexpression may represent an early event in MGUS/MM pathogenesis. The demethylation of the JAG2 promoter was shown to drive the observed overexpression. The physical location of the gene proximal to the IgH switch region at 14q32 7 places it in a very unstable locus. The numerous physiological rearrangements plasma cells undergo during switch recombinations might affect the methylation integrity of the surrounding genes, especially JAG2 and its promoter region.
However, we cannot exclude the possibility that this demethylation is the consequence of a primary genetic event yet to be characterized. Further studies are currently ongoing to dissect the mechanism of this demethylation (it may be worth noting that histone acetylation in this region regulates VDJ and switch recombination and may also have an indirect effect in the methylation status of this promoter).
Very low-level JAG2 expression was detected in plasma cells from patients with other malignancies and from healthy individuals, whereas JAG2 up-regulation was present in all MGUS/MM patient samples studied. In addition, we demonstrated the specific role of the JAG2/NOTCH binding in the IL-6, VEGF, and IGF-1 secretion. Indeed, the use of JAG2-binding peptide induced the cytokine secretion from the stromal cells. In addition, an incremental amount of anti–NOTCH-1 monoclonal antibody accordingly decreased the secretion of IL-6 in our coculture assay. These experiments showed that secretion of IL-6 is specifically activated by JAG2/NOTCH ligation in adjacent cell layers, as demonstrated by the use of specific anti–NOTCH-1 monoclonal antibodies, and that the induction of IL-6 secretion is dependent on cell-to-cell contact. These data are in agreement with the induced paracrine model, where the malignant plasma cells and the stromal cells in the BM microenvironment act in a symbiotic fashion for the malignant plasma cells' survival and further proliferation.
Recently, JAG2 was reported to be overexpressed in most MM cell lines studied by cDNA array.31 In addition, the NOTCH pathway has been shown to play a critical role in lymphomagenesis. NOTCH1 and Jagged-1 (another NOTCH ligand) are highly expressed in Hodgkin and anaplastic large cell lymphoma,32 demonstrating a novel mechanism for the oncogenic capacity of NOTCH1. The interaction between intact NOTCH1 and Jagged-1 has been shown to dramatically induce proliferation and inhibit apoptosis in vitro. NOTCH1 signaling may therefore be activated by Jagged-1 through homotypic or heterotypic cell-to-cell interactions, likely contributing to lymphomagenesis in vivo.
Finally, 2 new studies33,34 reported on the NOTCH pathway in MM. Jundt et al33 showed that Jagged-1, NOTCH-1, and NOTCH-2 are overexpressed in MM samples. Five representative cell lines showed variable levels of expression for these 3 proteins, but a lack of Western blot loading controls does not allow an accurate interpretation. In any event, 1 of 5 cell lines is negative for Jagged-1, 2 of 5 show a faint band, 1 of 5 a stronger band, and NCI-H929 is highly positive, whereas the same cell line is almost negative in the second study. In a study by Nefedova et al,34 4 MM cell lines as well as 4 lymphoblastoid cell lines established from MM patients were studied for NOTCH-1, -2, -3, -4, and Jagged-1. The 4 MM cell lines present a positive band for NOTCH-1 and -3, except for U266 negative for NOTCH-1. Only one (RPMI 8226) MM cell line has a detectable band for Jagged-1. The authors also assessed the expression levels of the NOTCH family members in BM stromal cells from healthy donors where they show detectable expression of all proteins in all the samples. JAG2 was not detectable in these stromal cells. Unfortunately, JAG2 levels were not assessed in the MM cells.
Taken together, these data suggest that overexpression of the NOTCH ligand JAG2 may play a causative role in the development of plasmacytosis, giving rise to MGUS and/or to the propagation of fully malignant plasma cells in MM. JAG2 overexpression activates NOTCH, which drives the secretion and release of IL-6, VEGF, and IGF-1 in the microenvironment supporting myeloma cell growth. This study provides evidence that JAG2 overexpression by MM cells can potentially induce IL-6, VEGF, and IGF-1 production in a paracrine fashion and therefore can be a key early event in the pathogenesis of MGUS/MM. The study of a larger number of MGUS samples will be necessary to determine whether either all MGUS or only a subpopulation shows JAG2 overexpression. If JAG2 overexpression is detected in all MGUS, an additional event would be necessary to induce the expansion of a malignant clone. If not, the outlined subpopulation might correspond to those cases that transform to MM. Finally, our findings should allow us and others to develop new therapeutic approaches involving interference with the NOTCH pathway as a means to block malignant plasma cell proliferation and, it is hoped, induce their apoptosis, either alone or in combination with a low-dose chemotherapy regimen.
Prepublished online as Blood First Edition Paper, August 3, 2004; DOI 10.1182/blood-2003-12-4114.
Supported in part by a grant from the American Cancer Society, Illinois Division (grant 02-14), by Loyola University Medical Center/Cardinal Bernardin Cancer Center, and by Dr Ralph and Marian Falk Medical Research Trust (L.J.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank H. Baumann, A. Chanan-Khan, T. Shows, P. Reczek (Roswell Park Cancer Institute), R. Fonseca (Mayo Clinic, Rochester, MN), and S. Morris (St. Jude Research Hospital, Memphis, TN) for critical comments; C. Brigaudeau, T. Ellis, P. Simms for technical assistance; and E. Sanders-Noonan for editorial assistance.