Abstract
Somatic hypermutation (SHM) targets primarily the immunoglobulin variable region (IgV) genes in germinal center (GC) B cells, thereby allowing antibody affinity maturation. A malfunction of SHM, termed aberrant somatic hypermutation (ASHM), was found in about 50% of diffuse large B-cell lymphomas (DLBCLs), leading to mutations in the 5′ sequences of multiple genes, including oncogenes. Although the SHM mechanism is largely unknown, it was shown to require the activation-induced cytidine deaminase (AID) gene. AID mRNA is expressed in GC B cells and GC-derived lymphomas, but the pattern of expression of the AID protein is not known. Using 2 specific antibodies, here we show that the AID protein can be detected in GC centroblasts and their transformed counterpart (Burkitt lymphoma) but not in pre-GC B cells and post-GC neoplasms, including B-cell chronic lymphocytic leukemia and multiple myeloma. DLBCLs displayed variable levels of AID expression, which did not correlate with IgV ongoing hypermutation, ASHM, or disease subtype. Finally, both in normal and malignant B cells the AID protein appeared predominantly localized in the cytoplasm. These results indicate that the AID protein is specifically expressed in normal and transformed GC B cells; nonetheless, its predominantly cytoplasmic localization suggests that additional mechanisms may regulate its function and may be altered during lymphomagenesis. (Blood. 2004;104:3318-3325)
Introduction
Somatic hypermutation (SHM) is a specialized process that takes place in germinal center (GC) B cells in response to T cell-dependent antigen stimulation.1,2 This process introduces single nucleotide substitutions, with occasional deletions and duplications, primarily into the variable region of the immunoglobulin (IgV) heavy and light chain genes, resulting in the production of high-affinity antibodies and allowing affinity maturation of the humoral immune response.3 SHM requires active transcription of the target locus but is not IgV sequence specific and does not depend on the V region promoter.4-7 In fact, at least 3 non-Ig genes, including BCL6, the FAS/CD95 gene, and the genes encoding the 2 components of the BCR (B29 and mb1) have been shown to acquire somatic mutations during the normal GC reaction, indicating that this mechanism may target more genes than originally suspected.8-12
The molecular basis of SHM remains largely unknown. However, studies from the past 4 years have identified the AID gene, encoding for activation-induced cytidine deaminase, as an absolute requirement for both SHM and class switch recombination (CSR) in humans and mice.13,14 Activation-induced cytidine deaminase (AID) expression is also sufficient to initiate both events in fibroblasts expressing transcribed artificial constructs.15,16 Because of the high homology with the RNA-editing enzyme apolipoprotein B editing catalytic subunit 1 (APOBEC-1), it has been proposed that AID may function as a cytidine deaminase to modify a preexisting mRNA into a new one, possibly encoding an endonuclease.14,17 However, experimental evidence in Escherichia coli indicated that AID may act directly on DNA and convert deoxycytidines to uracils, which are then processed by uracil-DNA glycosylase (UNG) and endonucleases.18 AID would thereby lead to the creation of abasic sites, which may be repaired by base excision repair and putative error-prone mechanisms. Indeed, in vitro AID exhibits cytidine deamination activity on single-stranded DNA, with a base specificity similar to that reported for SHM.19-22
Recently, the SHM process has been shown to malfunction in about 50% of diffuse large B-cell lymphomas (DLBCLs) as well as in about 20% of AIDS-related non-Hodgkin lymphomas (NHLs) and in a significant fraction of primary central nervous system lymphomas derived from non-HIV patients.23-25 In these tumors, multiple somatic mutations are introduced into the 5′ region, including coding sequences, of several genes that do not represent physiologic SHM targets. These comprise the well-known proto-oncogenes PIM1, PAX5, RhoH/TTF, and cMYC, all of which have been already implicated in the pathogenesis of lymphoid malignancies. Analogous to the physiologic targets (IgV and BCL6), mutations at these loci are scattered through the first approximately 2 kb from the promoter with characteristic hotspot (RGYW/WRCY) motifs and exhibit a bias for transitions over transversions, with higher frequency at G:C pairs.23,24 Such distinctive features strongly support the hypothesis that this phenomenon, termed “aberrant somatic hypermutation” (ASHM), results from an aberrant activity, possibly a loss of target specificity, of the physiologic SHM process and therefore of AID itself. Because the mutations affect both regulatory and coding sequences of the affected genes, with several amino acid substitutions predicting a change in the protein structure, ASHM may represent a major contributor to DLBCL pathogenesis. Indeed, for some of the targeted genes, such as cMYC, the oncogenic effect of some mutations has been demonstrated.26-28
Based on RNA expression data, AID appears selectively expressed in GC B cells and GC-derived malignancies14,29-31 ; however, the pattern of expression of the AID protein has never been investigated. Using 2 specific antibodies, we examined the distribution and levels of the AID protein in purified normal B-cell subpopulations (naive and centroblasts) as well as in B-lymphoid tumors derived from GC and post-GC B cells. These results have implications for the mechanism regulating AID function and for its possible role in lymphomagenesis.
Materials and methods
Cell lines and primary lymphoma cases
All B-cell lines used were cultured in Iscove modified Dulbecco medium (IMDM) or RPMI (Gibco, Grand Island, NY) supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, 100 U/mL streptomycin, and 2 mM l-glutamine. The lung carcinoma cell line H1299 was grown in Dulbecco modified Eagle medium (DMEM)/10% FCS. Ramos and H1299 were stably transduced with pBABE-puro retroviral vectors or with vectors expressing a C-terminal Flag-hemagglutinin (HA)-tagged human AID cDNA (AID-FH), followed by selection in 0.5 μg/mL puromycin. The 25 newly diagnosed DLBCL frozen tissue samples were obtained from the Department of Pathology of the Memorial Sloan-Kettering Cancer Center.32 Only cases in which the fraction of neoplastic cells corresponded to more than 70% (n = 21) were selected for the correlative analysis between AID protein expression and DLBCL subgroup or ASHM. The B-cell chronic lymphocytic leukemia (B-CLL) primary cases and their characterization for the presence of IgV mutations are described in detail elsewhere.33
Isolation of normal human B-cell subpopulations from tonsils
Naive and centroblast (CB) B-cell subpopulations were isolated by magnetic cell separation using the MidiMACS system (Miltenyi Biotec, Auburn, CA) as described.34 Briefly, naive B cells were isolated by depletion of GC B cells (CD10, CD27), memory B cells (CD27), plasma cells/blasts (CD27), and T cells (CD3), followed by a positive enrichment of IgD-positive cells. CBs were isolated in a single step by magnetically labeling CD77+ cells. The purity of the isolated fractions was determined on a FACSCalibur (Becton Dickinson, San Jose, CA) and by IgV region gene analysis.34
Antibodies
The method used for the generation of the anti-AID rabbit polyclonal antibody (Ab) is reported by Faili et al.35 The anti-AID mouse monoclonal Ab (clone 7E7), an IgG1 κ light chain, was raised against a synthetic peptide selected from the C-terminal half of the human AID protein. Additional details are provided as Supplemental Materials (at the Blood website, see the Supplemental Materials link at the top of the online article). The following Abs were also used: anti-BCL6 (N3, Santa Cruz Biotechnology, Santa Cruz, CA), anti-HA (3F10, Roche Diagnostics, Indianapolis, IN), anti-β-actin (AC15, Sigma, St Louis, MO), and anti-α-tubulin (clone B-5-1-2, Sigma).
Protein extracts, cell fractionation, and Western blot analysis
Total protein extracts were prepared from exponentially growing cell lines and snap-frozen lymphoma biopsies as described previously.36 To isolate nuclear and cytoplasmic fractions, cells were incubated for 15 minutes at 4°C in hypotonic buffer A (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] pH 7.9, 10 mM KCl, 0.1 mM EDTA [ethylenediaminetetraacetic acid], 1 mM dithiothreitol [DTT], and protease inhibitors [Sigma]); Nonidet P-40 (NP40) was added to a final concentration of 0.4%, and the samples were centrifuged for 5 minutes at 3000 rpm. Supernatants were used as cytoplasmic extracts after further centrifugation for 30 minutes at 14 000 rpm. The nuclear pellets were washed twice in buffer A, resuspended in about 10 volumes of buffer C (20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM EDTA, 0.4% NP40, 1 mM DTT, and protease inhibitors), vortexed for 30 minutes at 4°C, and centrifuged for 30 minutes at 14 000 rpm. The final salt concentration in the nuclear extracts was adjusted to 300 mM by addition of buffer D (20 mM HEPES pH 7.9, 1 mM EDTA, 1 mM DTT, and protease inhibitors). For Western blot analysis, protein extracts (30 μg whole-cell extract or 10 μg subcellular fraction) were gel electrophoresed on 4% to 20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Invitrogen Life Technologies, Carlsbad, CA), transferred to nitrocellulose membranes, and immunostained according to standard methods. Proteins were detected using the enhanced chemiluminescence (ECL) reagents (Amersham Biosciences, Freiburg, Germany) as recommended by the manufacturer.
To compare the AID protein expression levels in DLBCL samples from different experiments, equivalent amounts of GC centroblasts were loaded in each gel and used as an internal reference. Band intensities were measured using the NIH Image program 1.62, and AID expression was normalized first for β-actin, as a control for loading, and then for the CB sample loaded in the same gel.
RNA extraction, Northern blot analysis, and gene expression profiling
Total RNA was prepared using the Trizol reagent (Invitrogen Life Technologies) followed by RNeasy purification (QIAGEN, Valencia, CA), and Northern blot analysis was performed according to standard procedures, with radiolabeled probes corresponding to the full-length human AID, BCL6, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNAs. The detailed characterization of normal B-cell subpopulations by gene expression profiling has been reported previously.34 The protocol for microarray hybridization of the DLBCL samples to the Affymetrix HG95Av2 oligonucleotide array, as well as the expression data analysis, has also been reported in detail.32
PCR amplification, cloning, and sequencing analysis of PIM1, PAX5, RhoH/TTF, and cMYC
Genomic DNA from 31 DLBCL samples, including 10 cell lines and 21 primary biopsies, was extracted according to standard methods. Mutational analysis of PIM1, PAX5, RhoH/TTF, and cMYC was performed on selected genomic regions previously shown to contain more than 90% of the mutations found in DLBCL.23 Following polymerase chain reaction (PCR) amplification, purified amplicons were sequenced directly from both strands and compared with the corresponding germ line sequences as described.23 All sequence variants were confirmed on independent PCR products, and nucleotide changes due to previously reported polymorphisms or occurring more than once in separate cases were excluded from the assessment of the mutation frequencies unless their somatic origin could be proven. Cases were scored as ASHM-positive if they harbored 1 or more somatic mutations distributed in 1 or more of the 4 proto-oncogenes.
Analysis of IgV gene intraclonal variation
The rearranged IgVH genes were amplified from genomic DNA using the Pfu Turbo polymerase (Stratagene, La Jolla, CA) together with a set of 6 VH gene family-specific primers hybridizing to sequences in the framework region (FR)I and a JH primer mix in separate reactions for each VH primer.8 PCR products were gel purified and cloned into pGEM-T vector (Invitrogen Life Technologies) as described.23 For each PCR product, 25 to 58 clones were sequenced and analyzed. Mutation frequencies were calculated by dividing the total number of unique mutations (ie, changes observed in individual subclones) by the total number of base pairs sequenced.
Results
Identification of the AID protein by specific antibodies
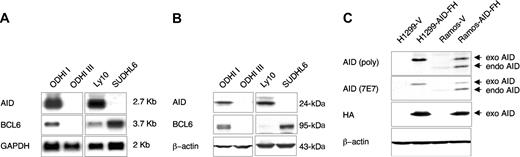
Two antibodies were used throughout the study: one is a rabbit polyclonal Ab raised in the laboratory of one of the authors (S.A.)35 ; the second is a monoclonal Ab obtained against a peptide sequence selected from the C-terminal half of the human AID polypeptide (see “Materials and methods”). To test the ability of these Abs to recognize the human AID protein, we first used 4 B-cell lines that were documented by Northern blot (NB) analysis to express (ODHI I, Ly10) or lack (ODHI III, SUDHL6) endogenous AID mRNA (Figure 1A). Western blot analysis on total protein extracts detected a specific band of the expected 24-kDa molecular weight in ODHI I and Ly10 but not in the 2 RNA-negative cell lines ODHI III and SUDHL6 (Figure 1B; the polyclonal Ab is shown). We then engineered a human epithelial cell line (H1299), lacking AID mRNA expression, and the B-cell line Ramos, which expresses low levels of AID mRNA, to stably express an exogenous, Flag-HA-tagged human AID protein (AIDFH) under the control of pBABE retroviral sequences. Figure 1C shows the presence of an approximately 27-kDa band (exo)-consistent with the predicted size of the double-tagged fusion protein—only in AID-FH-transfected cells (Figure 1C, top 2 panels, lanes 2 and 4) but not in control vector-transfected cells (Figure 1C, lanes 1 and 3). In addition, a band of smaller molecular weight was recognized in both AID-FH- and control vector-transfected Ramos cells (Figure 1C, lanes 3 and 4); this smaller band corresponded in size to the endogenous AID protein (endo) and was not present in H1299 (Figure 1C, lanes 1 and 2), as expected based on RNA data (not shown). Western blot analysis with an anti-HA Ab, which recognizes only the tagged protein, revealed the same 27-kDa band (Figure 1C, third panel), documenting its exogenous nature. Similar results were obtained using the monoclonal and the polyclonal Abs, although the latter one gave significantly higher background. The reactivity in cells expressing AID mRNA or a transfected AID cDNA indicates that both the monoclonal and polyclonal Abs can specifically recognize the AID protein.
Identification of the AID protein by specific antibodies. (A) Northern blot analysis of endogenous AID expression in 2 BL (ODHI I, ODHI III) and 2 DLBCL (Ly10, SUDHL6) cell lines. An approximately 2.7-kb message, corresponding to the AID transcript, can be detected in ODHI I and Ly10 but not in ODHI III and SUDHL6. Filters were stripped and sequentially hybridized with probes for BCL6, as marker of differentiation stage, and GAPDH as control for loading. (B) Western blot analysis of the same cell lines with anti-AID rabbit polyclonal Abs shows a specific band of the predicted 24-kDa molecular weight in ODHI I and Ly10, but not in ODHI III and SUDHL6, consistent with the RNA data in panel A. Immunoblotting with anti-BCL6 (N3) and anti-β-actin Abs is shown in the bottom panels. (C) Western blot analysis of AID expression in H1299 and Ramos cells, stably transduced with pBABE retroviral vectors (H1299-V, Ramos-V) or with vectors expressing an AID-Flag-HA protein (H1299-AID-FH and Ramos-AID-FH). Both the rabbit polyclonal antiserum (poly) and the mouse monoclonal (7E7) Abs were used (top 2 panels). An approximately 27-kDa band corresponding to the size of the exogenous protein (exo) is detected in lysates from AID-FH-transduced cells (lanes 2 and 4) but not in cells transduced with the control vector (lanes 1 and 3). In the B-cell line Ramos, the endogenous AID protein (endo) can also be seen (lanes 3 and 4). Membranes were stripped and sequentially probed with anti-HA, which recognizes the exogenous protein, and with anti-β-actin as control for loading (bottom 2 panels).
Identification of the AID protein by specific antibodies. (A) Northern blot analysis of endogenous AID expression in 2 BL (ODHI I, ODHI III) and 2 DLBCL (Ly10, SUDHL6) cell lines. An approximately 2.7-kb message, corresponding to the AID transcript, can be detected in ODHI I and Ly10 but not in ODHI III and SUDHL6. Filters were stripped and sequentially hybridized with probes for BCL6, as marker of differentiation stage, and GAPDH as control for loading. (B) Western blot analysis of the same cell lines with anti-AID rabbit polyclonal Abs shows a specific band of the predicted 24-kDa molecular weight in ODHI I and Ly10, but not in ODHI III and SUDHL6, consistent with the RNA data in panel A. Immunoblotting with anti-BCL6 (N3) and anti-β-actin Abs is shown in the bottom panels. (C) Western blot analysis of AID expression in H1299 and Ramos cells, stably transduced with pBABE retroviral vectors (H1299-V, Ramos-V) or with vectors expressing an AID-Flag-HA protein (H1299-AID-FH and Ramos-AID-FH). Both the rabbit polyclonal antiserum (poly) and the mouse monoclonal (7E7) Abs were used (top 2 panels). An approximately 27-kDa band corresponding to the size of the exogenous protein (exo) is detected in lysates from AID-FH-transduced cells (lanes 2 and 4) but not in cells transduced with the control vector (lanes 1 and 3). In the B-cell line Ramos, the endogenous AID protein (endo) can also be seen (lanes 3 and 4). Membranes were stripped and sequentially probed with anti-HA, which recognizes the exogenous protein, and with anti-β-actin as control for loading (bottom 2 panels).
AID protein expression in normal B-cell subpopulations
To examine the expression pattern of the AID protein in normal B-cell subpopulations, pre-GC naive B cells (IgD+ CD27-) and GC CBs (CD77+) were purified from the reactive tonsil of normal individuals, and Western blot analysis was performed on total protein extracts as well as on nuclear and cytoplasmic cell fractions. Elevated AID protein levels were detected in the CB population (ie, the cells deputed to SHM) but not in the pre-GC naive B cells, confirming RNA data generated by gene expression profiling (Figure 2A-B). Unexpectedly for a protein that is presumed to function as a DNA mutator, Western blot analysis of GC B cells after cell fractionation revealed a predominantly cytoplasmic localization of AID, with only traces being detected in the nuclear fraction (Figure 2C). The purity of the extracts was monitored using anti-α-tubulin and anti-BCL6 Abs, respectively. Thus, the AID protein is expressed in GC B cells, where it resides predominantly in the cytoplasm.
AID protein expression and subcellular localization in normal B lymphocytes. (A) AID mRNA expression in purified normal B-cell subpopulations (5 samples each of N [naive], CB [centroblasts], CC [centrocytes], M [memory]). AID was represented by 2 probe sets in the Affymetrix U133Plus GeneChip array. BCL6, BCL7A, and the proliferation-associated gene Ki67 are shown along with BCL2 and TOSO as markers for the identity of the purified subpopulations. (B) Western blot analysis of AID and β-actin expression in whole cell extracts obtained from naive and CB populations. (C) Western blot analysis of AID expression in nuclear (nu) and cytoplasmic (cy) extracts from tonsillar GC B cells. The purity of the fractions was monitored using anti-BCL6 and anti-α-tubulin Abs, respectively.
AID protein expression and subcellular localization in normal B lymphocytes. (A) AID mRNA expression in purified normal B-cell subpopulations (5 samples each of N [naive], CB [centroblasts], CC [centrocytes], M [memory]). AID was represented by 2 probe sets in the Affymetrix U133Plus GeneChip array. BCL6, BCL7A, and the proliferation-associated gene Ki67 are shown along with BCL2 and TOSO as markers for the identity of the purified subpopulations. (B) Western blot analysis of AID and β-actin expression in whole cell extracts obtained from naive and CB populations. (C) Western blot analysis of AID expression in nuclear (nu) and cytoplasmic (cy) extracts from tonsillar GC B cells. The purity of the fractions was monitored using anti-BCL6 and anti-α-tubulin Abs, respectively.
AID protein expression in B-cell malignancies
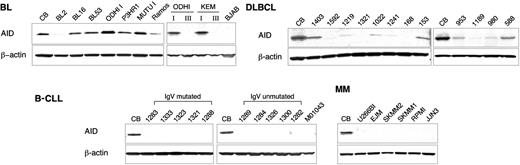
In B-cell neoplasms, the AID protein was found restricted to cells displaying a mature GC phenotype, including 10 of 10 group I Burkitt lymphoma (BL) cell lines, where the levels were mostly comparable to CBs (Figure 3, top left panel). AID expression was also examined in 3 pairs of BL lines (KEM, MUTU, ODHI), each one derived from the same tumor case but selected in vitro to represent either a group I GC phenotype or a group III post-GC immunoblastic phenotype.37 Elevated AID protein levels were detected in the group I but not in the group III BL lines (Figure 3, top left panel), suggesting that AID expression may be down-regulated in this later stage of differentiation. Among DLBCL cases, which included 25 primary biopsies and 10 cell lines, AID expression was very heterogeneous, ranging from complete absence (3 of 10 cell lines and 10 of 25 biopsies; overall, 13 of 35 samples, corresponding to 37% of the cases) to levels comparable to the ones observed in CBs (Figure 3, top 2 right panels, shows representative results). No evidence of AID protein expression was found in post-GC tumors such as MM (0 of 7 cell lines) and B-CLL (0 of 12 samples, including 10 primary cases and 2 cell lines) regardless of their IgV mutational status (Figure 3, bottom panels). In all samples studied, NB analysis documented the complete concordance between RNA and protein levels in terms of relative distribution (data not shown). Collectively, these data indicate that expression of the AID protein is restricted to B-cell malignancies derived from GC cells.
The AID protein is expressed in GC but not in post-GC-derived B-cell malignancies. Western blot analysis of AID expression in BL (top left panels), DLBCL (top right panels), and post-GC-derived B-CLL and MM samples (bottom panels). Both IgV mutated and unmutated cases were included in the B-CLL filter. The top right panel shows 2 pairs of isogenic BL cell lines representative of the group I/latency I and group III/latency III phenotype (see text for a detailed explanation); the B-cell lymphoma line BJAB serves as a negative control, because it does not express AID mRNA (L.P., unpublished results, 2003). In each membrane, equivalent amounts of total protein extracts from purified CBs were included; β-actin controls for loading.
The AID protein is expressed in GC but not in post-GC-derived B-cell malignancies. Western blot analysis of AID expression in BL (top left panels), DLBCL (top right panels), and post-GC-derived B-CLL and MM samples (bottom panels). Both IgV mutated and unmutated cases were included in the B-CLL filter. The top right panel shows 2 pairs of isogenic BL cell lines representative of the group I/latency I and group III/latency III phenotype (see text for a detailed explanation); the B-cell lymphoma line BJAB serves as a negative control, because it does not express AID mRNA (L.P., unpublished results, 2003). In each membrane, equivalent amounts of total protein extracts from purified CBs were included; β-actin controls for loading.
Expression of the AID protein in DLBCL subgroups
Based on gene expression profiling studies, DLBCLs have been classified into 3 biologically and clinically distinct subgroups: the GC B-cell-like (GCB), the activated B-cell-like (ABC), and the type 3 DLBCL.38,39 To investigate whether the AID protein was differentially expressed among these phenotypic groups, we used the Affymetrix U95Av2 oligonucleotide microarray to generate gene expression profiles from 30 DLBCL samples, including 9 cell lines and the 21 primary biopsies that contained more than 70% tumor cells, and then measured the AID protein levels in each group identified. The analysis classified 15 samples (50%) into the GCB subgroup, 9 samples (30%) into the ABC subgroup, and 6 samples (20%) into the type 3 group. In all of them, expression of the AID protein was very heterogeneous, with 9 of 15 (60%) positive cases in the GCB-like, 7 of 9 (77.8%) positive cases in the ABC-like, and 4 of 6 (66.6%) positive cases in the type 3 DLBCL (Figure 4, top panel); however, when compared with the GCB type, the ABC DLBCLs had on average higher expression levels (about 3-fold) both in primary biopsies and in cell lines (Figure 4, bottom panel). Moreover, the overall higher levels of AID in ABC DLBCLs tend to inversely correlate with the expression of BCL6, a well-established marker of GC (Figure 1A-B and data not shown), differently from what is observed in GC B cells or in BL, where the 2 proteins are coexpressed (see “Discussion”).
The AID protein is expressed in both GCB- and ABC-like DLBCLs. (Top) AID protein levels, measured by Western blotting and normalized for β-actin as described in “Materials and methods,” are shown in individual DLBCL samples classified by gene expression profiling as GCB-like (○), ABC-like (•), and type 3 ( ). Two different scales are used for the primary biopsies (left) and the cell lines (right), because lower AID levels were consistently observed in the former ones, likely due to the presence of contaminating nontumor cells. (Bottom) Histograms show the average AID protein expression levels in the 3 groups. Error bars indicate standard deviations.
). Two different scales are used for the primary biopsies (left) and the cell lines (right), because lower AID levels were consistently observed in the former ones, likely due to the presence of contaminating nontumor cells. (Bottom) Histograms show the average AID protein expression levels in the 3 groups. Error bars indicate standard deviations.
The AID protein is expressed in both GCB- and ABC-like DLBCLs. (Top) AID protein levels, measured by Western blotting and normalized for β-actin as described in “Materials and methods,” are shown in individual DLBCL samples classified by gene expression profiling as GCB-like (○), ABC-like (•), and type 3 ( ). Two different scales are used for the primary biopsies (left) and the cell lines (right), because lower AID levels were consistently observed in the former ones, likely due to the presence of contaminating nontumor cells. (Bottom) Histograms show the average AID protein expression levels in the 3 groups. Error bars indicate standard deviations.
). Two different scales are used for the primary biopsies (left) and the cell lines (right), because lower AID levels were consistently observed in the former ones, likely due to the presence of contaminating nontumor cells. (Bottom) Histograms show the average AID protein expression levels in the 3 groups. Error bars indicate standard deviations.
AID expression in DLBCL does not correlate with ongoing SHM
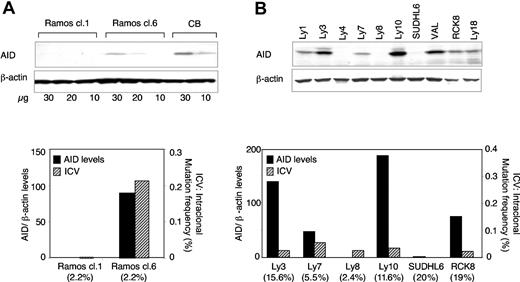
To examine whether the differential expression of AID in DLBCL cases reflects the ongoing activity of the SHM machinery, we investigated a number of samples for the presence of intraclonal variation (ICV) in their IgV sequences, an indicator that SHM has been active during clonal expansion. The rearranged IgV genes of 6 cell lines, representative of AID-positive (Ly3, Ly7, Ly10, RCK8) and -negative (Ly8, SUDHL6) cases, were amplified from genomic DNA using the proofreading Pfu Turbo polymerase and then cloned and sequenced (n = 25 to 58 clones per sample). The BL Ramos clone 6, which is known to undergo SHM in culture constitutively, was used as positive control, while Ramos clone 1, which has lost the ability to hypermutate, served as a negative control.40 Indeed, detectable AID protein levels were observed in Ramos clone 6 and were associated with significant intraclonal variation in its IgV sequences (22 mutational events distributed in 14 of 27 clones, corresponding to a frequency of 0.239 × 10-2/bp; P < .0001); conversely, only 1 nucleotide difference was found in 31 DNA clones sequenced from the AID-negative Ramos clone 1 (frequency, 0.009 × 10-2/bp, which is not significantly different from the expected error rate of the Pfu polymerase) (Figure 5A; see also Supplemental Table S1). Of the DLBCL cell lines analyzed, only 1 (Ly7) displayed a significant number of IgV intraclonal variants (11 of 19 426 bp, corresponding to a mutation frequency of 0.057 × 10-2/bp), while the frequency of mutations in the remaining 5 lines was not significantly higher than the polymerase error rate (range, 2 of 5957 bp to 5 of 23 142 bp sequenced) (Figure 5B and Supplemental Table S1). These extremely low frequencies were detected regardless of the AID protein levels and even in cell lines, such as Ly3 and Ly10, which express protein amounts comparable to GC B cells and higher than Ramos clone 6 (Figure 5B). Thus, differently from GC CB and BL cases, the heterogeneous expression of AID in DLBCLs does not reflect the level of activity of the SHM machinery.
AID protein expression is associated with ongoing SHM in Ramos but not in DLBCL cell lines. (A) Total cell extracts were prepared from 2 Ramos clones that express (cl.6) or lack (cl.1) AID mRNA expression40 and, from each of them, 3 dilutions were analyzed by immunoblotting using anti-AID Abs and β-actin as control (top). In the bottom panel, the AID protein levels from the top panel, quantitated using the NIH Image software and normalized for β-actin (▪), are compared with the levels of ICV in the corresponding rearranged IgV genes (▨), expressed as frequency of mutations and calculated as described in “Materials and methods.” The mutation frequency in the consensus sequence, defined as the sequence identified by direct sequencing and common to all DNA clones analyzed, is indicated in parentheses below the chart (also see Supplemental Table S1). (B) AID protein expression in 10 DLBCL cell lines (top). The correlation between AID protein levels (▪) and the level of IgV genes ICV (▨), assessed as in panel A, is shown in the bottom panel for 6 representative lines. Numbers in parentheses indicate the mutation frequency in the IgV consensus sequence.
AID protein expression is associated with ongoing SHM in Ramos but not in DLBCL cell lines. (A) Total cell extracts were prepared from 2 Ramos clones that express (cl.6) or lack (cl.1) AID mRNA expression40 and, from each of them, 3 dilutions were analyzed by immunoblotting using anti-AID Abs and β-actin as control (top). In the bottom panel, the AID protein levels from the top panel, quantitated using the NIH Image software and normalized for β-actin (▪), are compared with the levels of ICV in the corresponding rearranged IgV genes (▨), expressed as frequency of mutations and calculated as described in “Materials and methods.” The mutation frequency in the consensus sequence, defined as the sequence identified by direct sequencing and common to all DNA clones analyzed, is indicated in parentheses below the chart (also see Supplemental Table S1). (B) AID protein expression in 10 DLBCL cell lines (top). The correlation between AID protein levels (▪) and the level of IgV genes ICV (▨), assessed as in panel A, is shown in the bottom panel for 6 representative lines. Numbers in parentheses indicate the mutation frequency in the IgV consensus sequence.
AID expression levels do not correlate with ASHM in DLBCL
To investigate whether AID overexpression is involved in the pathogenesis of DLBCL-associated ASHM, we comparatively analyzed AID protein levels and the presence of somatic mutations in 4 genes previously identified as aberrant targets: PIM1, PAX5, RhoH/TTF, and cMYC.23 To this end, 31 DLBCL samples were first subjected to PCR amplification and direct sequencing of selected regions representing the hypermutable area in these genes.23 Because PCR products were sequenced directly in all experiments, the contribution of Taq polymerase error to mutations in the final sequence is insignificant. The overall mutation load in each case was then plotted against the normalized AID protein levels, measured by Western blotting, and quantitated using the NIH Image software.
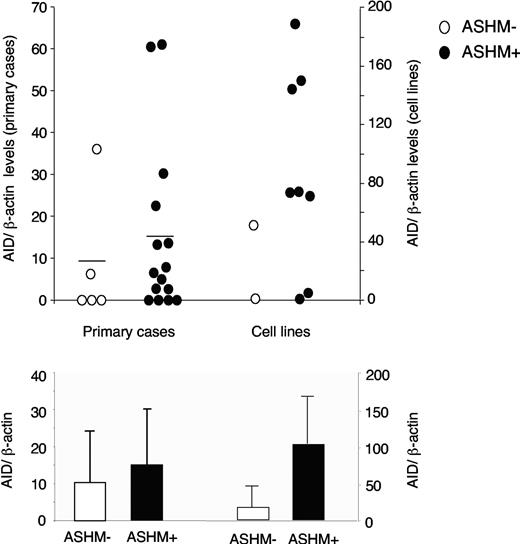
PCR products of PIM1, PAX5, RhoH/TTF, and cMYC were obtained from all DLBCL cell lines and from 20 of 21 primary cases (30 total evaluable samples). The analysis revealed the complete absence of somatic mutations in 2 of 10 cell lines (20%) and 5 of 20 primary biopsies (25%), while the remaining 23 samples, including 8 cell lines and 15 biopsies, carried 1 or more somatic mutations distributed in 1 or more of the 4 target genes (range, 1 to 28 mutational events per case in the cell lines; 1 to 18 mutational events per case in the primary biopsies) (Supplemental Table S2). All the mutations, which included mostly single nucleotide substitutions but also 4 deletions and 1 duplication, were confirmed on independent PCR products. These results are analogous to those originally reported for an independent cohort of DLBCL samples.23 When we compared the mutation status with the AID protein levels, we found that, both in cell lines and in primary biopsies, AID expression did not correlate with the presence of ASHM (Figure 6). In particular, AID was expressed in 3 of 7 (43%) unmutated and 17 of 23 (74%) aberrantly mutated samples (Figure 6, top). Moreover, no direct correlation was observed in the mutated samples between AID protein amount and the total number of mutations (not shown), indicating that a simple increase in AID expression levels is not sufficient to explain ASHM.
Lack of correlation between AID protein levels and ASHM in DLBCL. (Top) AID protein expression levels in DLBCL samples displaying (•) or lacking (○) ASHM, as defined by the presence of mutations in PIM1, PAX5, cMYC, and RhoH/TTF. AID protein levels were measured by Western blot analysis and normalized for β-actin as described in “Materials and methods.” (Bottom) Histograms show the average expression levels in the 2 groups. Error bars indicate standard deviations.
Lack of correlation between AID protein levels and ASHM in DLBCL. (Top) AID protein expression levels in DLBCL samples displaying (•) or lacking (○) ASHM, as defined by the presence of mutations in PIM1, PAX5, cMYC, and RhoH/TTF. AID protein levels were measured by Western blot analysis and normalized for β-actin as described in “Materials and methods.” (Bottom) Histograms show the average expression levels in the 2 groups. Error bars indicate standard deviations.
Predominant cytoplasmic localization of the AID protein in normal and transformed B cells
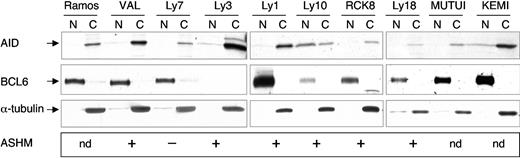
To examine whether abnormal subcellular localization, rather than overexpression of AID, could be responsible for loss of target specificity in DLBCL cases with ASHM, we performed Western blot analysis on nuclear and cytoplasmic fractions prepared from 10 AID-expressing cell lines, including 7 DLBCL and 3 BL. The DLBCL lines were representative of both the ABC (Ly3, Ly10) and the GCB (Ly7, Ly1, Ly18, RCK8) type and displayed ASHM in 6 of 7 cases. The results shown in Figure 7 indicate that—with the exception of the ABC-like, ASHM-positive Ly10 cell line, where AID appears to be evenly distributed in the nuclear and cytoplasmic fraction—AID is mostly retained in the cytoplasm (more than 90%), and only trace amounts of the protein can be detected in the nuclear fraction, analogous to what observed in normal GC B cells. This distribution was observed regardless of the histologic type of lymphoma (BL versus DLBCL) and also of the evidence for ongoing SHM (see CB and Ramos clone 6). Thus, in steady state conditions, the AID protein appears to be localized mostly in the cytoplasm, both in normal and transformed B cells.
Predominant cytoplasmic localization of the AID protein in BL and DLBCL. Nuclear (N) and cytoplasmic (C) fractions were prepared from 10 AID-positive lymphoma cell lines, including 3 BL and 7 DLBCL, and immunoblotted with anti-AID Abs, followed by anti-BCL6 (nuclear) and anti-α-tubulin (cytoplasmic) as controls for the purity of the fractions. The presence or absence of ASHM is also given; nd indicates not determined.
Predominant cytoplasmic localization of the AID protein in BL and DLBCL. Nuclear (N) and cytoplasmic (C) fractions were prepared from 10 AID-positive lymphoma cell lines, including 3 BL and 7 DLBCL, and immunoblotted with anti-AID Abs, followed by anti-BCL6 (nuclear) and anti-α-tubulin (cytoplasmic) as controls for the purity of the fractions. The presence or absence of ASHM is also given; nd indicates not determined.
Discussion
In the past few years, the AID protein has been the object of extensive studies due to its primary role in SHM and CSR. As a mutator, AID has also been implicated in the pathogenesis of B-cell lymphomas, and indeed its deregulated expression may represent a good candidate to explain the loss of target specificity that seems to derange the SHM mechanism in DLBCL.23,24 Based on RNA data, AID appears selectively expressed in GC B cells and GC-derived malignancies. However, the expression pattern of the AID protein has never been shown, mainly due to unavailability of specific Abs. This is the first study to report the distribution of the AID protein in normal and transformed human B cells, its relationship with IgV SHM, and its correlation with ASHM. Our data have implications for the understanding of the AID function in general and its specific role in lymphomagenesis.
The results of our study complement available data on AID mRNA expression by showing a close correlation between AID transcripts and protein levels in GC B cells and GC-derived malignancies29,30 (data not shown). Thus, no major control mechanism appears to be involved in the expression of AID at the level of RNA translation or protein stability. Nonetheless, neither RNA nor protein studies on bulk cell populations can address the issue of AID expression in cell subpopulations. Therefore, our results cannot determine whether AID is expressed in all GC B cells or in selected subpopulations, a question requiring the use of immunohistochemistry techniques that, unfortunately, could not be performed with the 2 Abs described.
Consistent with its distribution in normal B-cell subpopulations, the AID protein was not detected in B-CLL samples. B-CLL represents a post-GC-derived malignancy in at least 60% of the cases, which carry somatically mutated IgV genes; moreover, based on gene expression profiling analysis, all B-CLL cases appear to originate from a post-GC memory B cell.41 Our data are in apparent contradiction with recent studies reporting expression of the AID mRNA, mostly in the IgV unmutated subgroup of the disease.29,42,43 However, these studies were performed using reverse transcriptase (RT)-PCR methods, while more sensitive quantitative assays, including limiting dilution assays, revealed that only 0.01% to 0.2% of the B-CLL tumor cells are AID positive44 and that the total mRNA levels in B-CLL cases correspond to only about 5% of those observed in GC B cells,45 the population where AID typically exerts its activity. Thus, our data are consistent with previous reports in that the bulk of the B-CLL cells do not express AID. The biologic relevance of these low and very sparse AID mRNA expression levels in B-CLL remains to be proven.
Our results show that the AID protein is expressed—in heterogeneous fashion—in all phenotypic subtypes of DLBCL (GC-like, ABC-like, and type 3) and that the AID amounts are tendentially higher in the ABC type. Analogous results have been obtained by comparing AID mRNA levels in larger DLBCL databases generated by gene expression profiling at different institutions (http://llmpp.nih.gov/lymphoma). The presence of AID—in some cases at supraphysiological levels—in the ABC DLBCLs was somehow unexpected. In fact, this tumor subtype has been typically defined by the down-regulation of GC-specific expression markers (eg, BCL6) and up-regulation of post-GC markers such as IRF4 and XBP1, overall suggesting a derivation from a later, post-GC stage of differentiation.38,46 In addition, both the SHM machinery and the CSR mechanism have been presumably shut off in this disease type, because the tumor cells lack intraclonal diversity in their rearranged IgV genes (our data and Lossos et al47 ) and express high levels surface IgM.38,39,48 Thus, the presence of AID in the ABC DLBCL subgroup may suggest deregulation of AID expression; alternatively, this may reflect an activation status or the origin from a distinct B-cell subpopulation characterized by the indicated phenotypic features, possibly at the exit of the GC. Because the presently available Abs are not suitable for immunohistochemistry/immunofluorescence studies of AID, this question remains unanswered.
We could not find a correlation between AID expression levels and evidence of ASHM in the DLBCL primary cases analyzed. Considering that no structural abnormalities of the AID coding sequence are present in DLBCL, based on DNA sequencing data,23,29 several possibilities may explain this finding: (1) AID may have acted at earlier stages, before the biopsy was taken; however, tumors like BL consistently express high levels of AID but do not show evidence of ASHM23 ; (2) because the extent of the ASHM target is not known, it is possible that cases presently classified as “unmutated” may in fact carry mutations in genes other than the ones identified so far and therefore represent “false negatives”; and (3) regulation of AID activity (eg, gene target recognition and accessibility; shuttling of the protein to the target sequence) rather than the absolute expression levels may be implicated in ASHM (see next paragraph). For instance, the ASHM defect may reside primarily in other molecules associated with AID.
One perhaps surprising finding of this study is represented by the predominant cytoplasmic localization of the AID protein in all samples analyzed, including CBs. This result is in agreement with the original report by Rada et al, who used confocal microscopy to detect an exogenous GFP-AID fusion protein stably transfected in the Ramos B-cell line,49 and with 2 more recent studies also performed by transduction of GFP-AID retroviral constructs in B and non-B cells.50,51 However, the localization of AID in the cytoplasm is in disagreement with its presumed activity as a mutator, whether directly on DNA or on pre-mRNA, because the RNA-editing process is also thought to take place in the nucleus.52 While we cannot exclude the possibility that the biochemical procedure used may have allowed some leakage of AID into the cytoplasm, it is also conceivable that very small amounts of AID in the nucleus at any given time may be sufficient to exert its activity. Nevertheless, one may wonder why these cells would produce such a large amount of AID when not necessary. A more plausible explanation is that nuclear/cytoplasmic shuttling of AID constitutes a major point of regulation in its function, where sequestration of AID in the cytoplasm would serve as an active mechanism to protect genetic information from uncontrolled mutational activity. Indeed, it has been recently reported that a GFP-AID fusion protein can accumulate in the nucleus of stably transfected cells following treatment with the inhibitor of chromosome region maintenance/exportin 1 (CRM-1)-dependent nuclear export leptomycin B (LMB).50,51 Notably, mutants that accumulate in the nucleus were associated with higher mutation activity on cotransfected artificial target constructs, while mutants that could not shuttle to the nucleus upon LMB treatment lost their activities in both SHM and CSR.50,51 These results suggest that AID nuclear localization quantitatively correlates with its biologic function. Further studies aimed at the identification of signals controlling AID subcellular localization are necessary in order to understand its regulation in GC B cells and its possible deregulation in DLBCL.
Prepublished online as Blood First Edition Paper, August 10, 2004; DOI 10.1182/blood-2004-04-1558.
Supported by a Leukemia and Lymphoma Society Specialized Center of Research (SCOR) grant with contributions from the Cathy and Scott Zeilinger Philanthropic Fund of the Jewish Community Federation of Cleveland, the Carl and Ruth Shapiro Family Foundation, Lesley Goldwasser and Jonathan Plutzik and Friends, and Marla Kaminski and Friends. L.P. is a Special Fellow of the Leukemia and Lymphoma Society. Also supported by the Ligue Nationale Contre le Cancer (Equipe labellisée) (S.A.).
Two of the authors (R.P. and J.M.) are employed by a company (Cell Signaling Technology, Inc, Beverly, MA) whose potential product was studied in the present work.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank U. Klein for the isolation of the normal B-cell subsets, V. Miljkovic and A. Babiac for assistance in DNA sequencing, Claude-Agnès Reynaud for kindly providing us with the anti-AID polyclonal antibody, and A. Martin for the Ramos clone 6 and clone 1.

![Figure 2. AID protein expression and subcellular localization in normal B lymphocytes. (A) AID mRNA expression in purified normal B-cell subpopulations (5 samples each of N [naive], CB [centroblasts], CC [centrocytes], M [memory]). AID was represented by 2 probe sets in the Affymetrix U133Plus GeneChip array. BCL6, BCL7A, and the proliferation-associated gene Ki67 are shown along with BCL2 and TOSO as markers for the identity of the purified subpopulations. (B) Western blot analysis of AID and β-actin expression in whole cell extracts obtained from naive and CB populations. (C) Western blot analysis of AID expression in nuclear (nu) and cytoplasmic (cy) extracts from tonsillar GC B cells. The purity of the fractions was monitored using anti-BCL6 and anti-α-tubulin Abs, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/10/10.1182_blood-2004-04-1558/6/m_zh80220469530002.jpeg?Expires=1769277125&Signature=UqVLto0vRr0qUSGlAamukCf7EHyBvgwBe4rtc~gVgGI19GVhEVuthfvM6jvj3MuXySwNeaujHKYSPDqr0vYrkJIAyyCe-~~33k1xN5Ygj6BmSDm11nCPTFaoIDQ1HObNzt4ALxe10OQbxpDlmzGWTWl-uIBf0Elp0HA2JYCecKsp0A7WhJj2GGOX6a7xjJy3iCYxn-Txw9ZqQaNdJO74vJiwhpkJo3ERJSWJ5f~CmhFke8MXWRSF50u5eT84n~NKnlS0sfiVIlWgodHThX434qzE5dyMmmfG1IQ~9n9Ic9M5fYA4GYwR-aieKWiDR2PeBK4LIUbRkaJFXOWVRLQqwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)