Abstract
The Runt domain transcription factor, PEBP2/CBF, is a heterodimer composed of 2 subunits. The DNA-binding α subunit, or RUNX protein, interacts with a partner PEBP2β/CBFβ through the evolutionarily conserved Runt domain. Each of the genes encoding RUNX1 and PEBP2β/CBFβ is frequently involved in acute myeloid leukemia. The chimeric protein, CBFβ(PEBP2β)/SMMHC, is generated as a result of inversion of chromosome 16 in such a way to retain the heterodimerization domain of PEBP2β at the amino-terminal side fused to the C-terminal coiled-coil region of smooth muscle myosin heavy chain (SMMHC). Here we show that, in the chimeric protein, the second heterodimerization domain is created by the fusion junction, enabling the chimeric protein to interact with RUNX1 at far greater affinity than PEBP2β and inactivate the RUNX1/AML1 function. To explain why and how heterozygous CBFB/MYH11 can inactivate homozygous RUNX1 near to completion, we propose a new model for this chimeric protein that consists of a Y-shaped dimer with unpaired N-terminal halves followed by a coiled-coil for the C-terminal region. (Blood. 2004;103:3200-3207)
Introduction
RUNX1/AML1 was originally identified as the gene on chromosome 21 involved in the chromosome translocation, t(8;21), which is the most frequent chromosome rearrangement associated with acute myeloid leukemia (AML) of French-American-British (FAB) M2 subtype.1,2 RUNX1 encodes the DNA-binding α subunit of the Runt domain transcription factor, PEBP2/CBF, and interacts with the partner protein, PEBP2β/CBFβ, through its evolutionarily conserved Runt domain.3,4 The gene encoding PEBP2β is also frequently involved in AML. Inversion of chromosome 16 is observed in almost all the cases of AML subtype FAB M4Eo,5,6 which results in the formation of the chimeric gene, CBFB(PEBP2β)/MYH11, encoding the chimeric protein, CBFβ(PEBP2β)/SMMHC (hereafter, referred to as β/MYH11 and β/SMMHC, respectively).7 These observations stress that the 2 subunits of PEBP2/CBF function together in one unit as a transcription factor. The same notion has been reaffirmed by the observations that disruption of Runx1 results in a lack of definitive hematopoiesis with massive hemorrhage and that knockout of Pebpb2/Cbfb shows nearly the identical phenotypes. 8-11
The chromosome translocation t(8;21) results in generation of chimeric protein AML1/ETO(MTG8).12,13 Studies by many laboratories revealed that, in most cases, AML1/ETO dominantly represses the function of RUNX1 and that its heterozygous knock-in mice results in embryonic lethality 13.5 days after coitum concomitant with a phenotype virtually identical to that of Runx1-/- and Pebpb2/Cbfb-/- embryos.14 Furthermore, sporadic point mutations of RUNX1, mostly incurring loss-of-function, were found among patients with AML.15,16 Almost simultaneously, hereditary cases of loss-of-function point mutations in RUNX1 were identified in patients with familial platelet disorder with propensity to acute myeloid leukemia (FPD/AML) in which the family members frequently develop AML late in their lives.17,18 These observations have culminated in the proposition that loss-of-function of RUNX1 is leukemogenic.
The chimeric protein, β/SMMHC, contains the first 165 amino acids of PEBP2β(CBFβ) on the N-terminal side fused to the C-terminal coiled-coil region of smooth muscle myosin heavy chain (SMMHC). Mice heterozygous for β/SMMHC show almost the same phenotype as do those heterozygous for AML1/ETO.19 However, chimera mice generated with β+/β/MYH11 embryonic stem (ES) cells developed leukemias at higher frequency after N-ethyl-N-nitrosourea (ENU) treatment than mice with a conditional AML1/ETO knock-in allele.20,21 These findings suggested that β/MYH11 strongly inhibits the RUNX1 function, even more potently than AML1/ETO, by dominantly competing with the residual normal CBFB.
PEBP2β, a smaller and non–DNA-binding subunit of PEBP2/CBF, is known to act as an allosteric activator of DNA binding by Runx proteins.4 It also plays additional critical roles in regulating the function of PEBP2/CBF. PEBP2β markedly augments the metabolic stability of RUNX1, which by itself is highly susceptible to proteasome-mediated degradation.22 PEBP2β per se is a cytoplasmic protein and can enter the nucleus only through association with Runx proteins. Even on coexpression with Runx proteins, however, PEBP2β does not wholly localize to the nucleus, suggesting that there operates a mechanism by which to restrict their heterodimerization.23 In contrast, coexpression of β/SMMHC and RUNX1 results in virtually complete colocalization of the 2 proteins, mostly in the cytoplasm with some part in the nucleus.23 Thus β/SMMHC appears to have an ability to override the putative regulatory mechanism of heterodimerization between RUNX1 and PEBP2β. Furthermore, β/SMMHC can stabilize RUNX1 much more strongly than PEBP2β.22
In this study, we found that β/SMMHC harbors 2 distinct functional domains that respectively confer increased abilities on the chimeric proteins to heterodimerize with RUNX1, on the one hand, and to repress the transactivation potential of RUNX1, on the other hand. The two-layered actions of these domains would constitute the molecular basis for leukemogenesis mediated by β/SMMHC.
Materials and methods
Plasmids and recombinant protein constructs
pEF-RUNX1 and pEF-PEBP2β series have been described.24,25 As a full-length β/SMMHC, we used a 611–amino acid isoform in which exon 40 is joined to exon 41 and the coding sequence stops within exon 41. To create deletions of β/SMMHC, full length of β/MYH11 (1-611) was amplified by polymerase chain reaction (PCR) by various primers. The PCR products were digested and inserted into the EcoRI-NotI site of the mammalian expression vector pEF-BOS-T7-Neo. All point mutants were generated by PCR-based mutagenesis. The integrity of the resulting constructs was verified by sequencing.
Cell culture
Mouse embryonal carcinoma cell line P19 was cultured in a 1:1 mixture of Dulbecco modified Eagle medium (DMEM; Gibco/BRL, Grand Island, NY) and Ham F12 medium (Gibco/BRL) supplemented with 10% fetal bovine serum (FBS). COS7 cells were cultured in DMEM supplemented with 10% FBS. The human myelomonoblastic leukemia cell line U937 was cultured in RPMI 1640 supplemented with 10% FBS.
Transfection and Western blotting
P19 cells grown in 6-well microplates (3 × 105/well) were transfected with desired vectors using FuGENE6 (Boehringer Mannheim, Mannheim, Germany), and the cells were harvested 30 to 36 hours after transfection. Whole cell extract (WCE) was prepared by sonicating cells in a 2 × sodium dodecyl sulfate (SDS) sample buffer. After boiling for 5 minutes, WCEs were subjected to electrophoresis in 12% or 15% SDS-polyacrylamide gel. Western blotting was carried out by a standard protocol, and proteins were detected by chemiluminescence according to manufacturer's protocol (ECL; Amersham Pharmacia Biotech, Piscataway, NJ). COS7 cells (1 × 105/well) were transfected with expression plasmids for RUNX1, β, and β/SMMHC or its derivatives and WCEs were prepared 48 hours after transfection. Relative amounts of expressed proteins were evaluated by Western blotting. In transfection experiments, total input of DNA was kept constant (maximum 2 μg/culture) by supplementing appropriate amounts of backbone plasmid, thereby avoiding potential artifacts due to unbalanced DNA dosages.
In vitro binding and immunoprecipitation
Individual proteins were in vitro transcribed and translated by using TnT T7 Quick Coupled Transcription/Translation Systems (Promega, Madison, WI). In vitro binding was performed overnight at 4°C. Then immunoprecipitation (IP) was carried out under standard conditions as described.22 The reaction was terminated by adding an equal volume of 2 × SDS sample buffer. A 5-μL aliquot of the sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by exposure to x-ray film. Glutathione-S-transferase (GST) pull-down assay was performed as described previously.26 Glutathione Sepharose 4B beads and [35S]-methionine were purchased from Pharmacia Biotech (Uppsala, Sweden) and Amersham Biosciences (Tokyo, Japan), respectively.
Antibodies
For IP, monoclonal antibody that recognizes the Runt domain, anti-RD (3D9H3),22 or monoclonal anti-Cbfβ (141.4.1., a gift from Nancy Speck, Dartmouth Medical School, NH) were used. For Western blotting, polyclonal rabbit anti-αB1 (anti-Runx1) and polyclonal rabbit anti-β2 (anti-PEBP2β)23,24 were used.
Electrophoretic mobility shift assay
WCEs of COS7 cells (1 × 105) transfected with expression plasmids for RUNX1, β, and β/SMMHC or its derivatives (2 μg each) were prepared. After evaluating the relative amounts of expressed proteins by Western blotting, constant amount of RUNX1 (1 μL WCE) was mixed with equivalent amount of indicated proteins, mixed with 32P-labeled DNA probe,27 and incubated for 20 minutes on ice before electrophoresis in 6% gels.
Glutaraldehyde cross-linking assay
COS7 cells (1 × 105) transfected with expression plasmids for β1, β165, and β/SMMHC or its derivatives (2 μg each) were lysed with phosphate-buffered saline (PBS) supplemented with 0.2% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride (PMSF) for 30 minutes on ice. These lysates were diluted with the same volume of PBS and incubated at room temperature for 1 hour with or without glutaraldehyde at a final concentration of 0.025%. Cross-linking reactions were quenched by adding 2.5 M glycine to a final concentration of 250 mM followed by additional 5 minutes of incubation. Samples were resolved by 4% to 20% gradient SDS-PAGE, proteins were transferred to polyvinylidene difluoride (PVDF) membrane, and the immunoblots were performed with monoclonal anti-Cbfβ.
Transcription assay
The luciferase reporter plasmid pM-CSF-R-luc and the effecter plasmids, pEF-RUNX1, pEF encoding various β isoforms or β/SMMHC mutants were transfected at a fixed ratio into U937 cells by a nonliposomal transfection reagent, FuGENE6. WCEs were prepared 48 hours after transfection and assayed as previously described.15
Results
Mapping of the interaction domains of RUNX1 and β/SMMHC
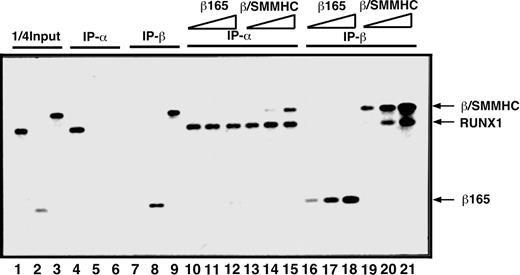
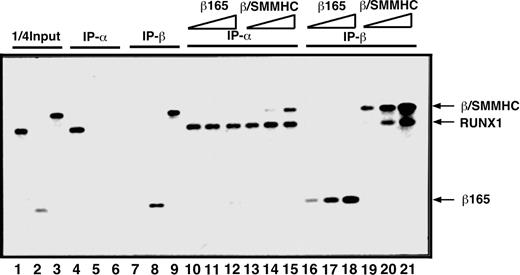
To compare β/SMMHC and β165 for the ability to heterodimerize with RUNX1, we first carried out an in vitro IP assay (Figure 1). On incubation with an anti-RUNX1 antibody, β/SMMHC was effectively coprecipitated with RUNX1 in a manner dependent on its dosage (lanes 13-15), whereas β165 was not detectably coprecipitated under any corresponding conditions (lanes 10-12). Conversely, RUNX1 was readily coprecipitated with β/SMMHC (lanes 19-21), but not with β165 (lanes 16-18), by the use of an anti-β subunit antibody. These results confirmed that β/SMMHC heterodimerizes with RUNX1 more efficiently than PEBP2β. We refer to this phenomenon as hyper-heterodimerization in this report.
Dominant binding of β/SMMHC to RUNX1 over PEBP2β COS7 cells were transfected with expression plasmids for either RUNX1, β165, or β/SMMHC, and WCE was prepared from each transfected culture as a protein source. One quarter of the amounts of RUNX1, β165, and β/SMMHC used in lanes 4 to 21 is shown in lanes 1, 2, and 3, respectively. Specificity of the antibodies is shown in lanes 4 to 9. A fixed amount of WCE containing RUNX1 (lanes 10-21) was mixed with increasing amounts (2.5 ×, 5 ×, and 25 ×) of β165 (lanes 10-12 and 16-18) or β/SMMHC (lanes 13-15 and 19-21) containing WCE and was subjected to IP/Western blotting. IP-α indicates immunoprecipitation with anti-RD; IP-β, immunoprecipitation with anti-Cbfβ (β141.4.1).
Dominant binding of β/SMMHC to RUNX1 over PEBP2β COS7 cells were transfected with expression plasmids for either RUNX1, β165, or β/SMMHC, and WCE was prepared from each transfected culture as a protein source. One quarter of the amounts of RUNX1, β165, and β/SMMHC used in lanes 4 to 21 is shown in lanes 1, 2, and 3, respectively. Specificity of the antibodies is shown in lanes 4 to 9. A fixed amount of WCE containing RUNX1 (lanes 10-21) was mixed with increasing amounts (2.5 ×, 5 ×, and 25 ×) of β165 (lanes 10-12 and 16-18) or β/SMMHC (lanes 13-15 and 19-21) containing WCE and was subjected to IP/Western blotting. IP-α indicates immunoprecipitation with anti-RD; IP-β, immunoprecipitation with anti-Cbfβ (β141.4.1).
We next used the β/SMMHC-dependent hyperprotection of RUNX1 to define which regions on RUNX1 and β/SMMHC are required for their augmented interactions. With deletions of RUNX1 from either the N-terminus or C-terminus, or both, the hyperprotection was invariably observed so far as the Runt domain was kept intact (Figure 2A lanes 1-9 and 13-24). The results suggest that the region required for protection by β/SMMHC is the Runt domain. We then tested deletion constructs of β/SMMHC (Figure 2B). To mimic the situation found with natural variants of inv(16),28 we serially deleted β/SMMHC exon by exon, rather than cutting at arbitrarily chosen points. Diagrammatic representation of all the chimeric proteins constructed is shown in Figure 2C. C-terminal deletions up to exon 36 did not appreciably change the ability of the chimeric protein to hyperprotect RUNX1 (lanes 8-13). However, further deletions past exon 35 almost completely abolished the protection (lanes 14-17).
The minimum essential domain of β/SMMHC for protection of RUNX1. (A) Various deletion constructs of RUNX1 were coexpressed in P19 cells with either β165 or β/SMMHC, and WCEs were subjected to Western blotting. Numbers at the top represent what amino acids remain in the mutant protein. (B) RUNX1 expression plasmid (1 μg) was cotransfected with expression plasmids for various PEBP2β informs (1 μg) as well as those for deletion constructs of β/SMMHC (1 μg) and WCEs were subjected to Western blotting. (C) Schematic diagram of β/SMMHC derived constructs.
The minimum essential domain of β/SMMHC for protection of RUNX1. (A) Various deletion constructs of RUNX1 were coexpressed in P19 cells with either β165 or β/SMMHC, and WCEs were subjected to Western blotting. Numbers at the top represent what amino acids remain in the mutant protein. (B) RUNX1 expression plasmid (1 μg) was cotransfected with expression plasmids for various PEBP2β informs (1 μg) as well as those for deletion constructs of β/SMMHC (1 μg) and WCEs were subjected to Western blotting. (C) Schematic diagram of β/SMMHC derived constructs.
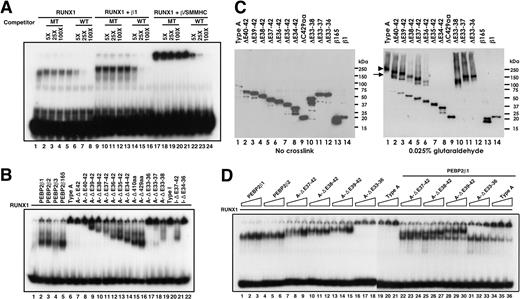
These results suggested that the myosin tail portion of β/SMMHC, spanning exons 33 to 35 at minimum, might provide an extra molecular interface for interaction with the Runt domain. This possibility was more directly confirmed by an in vitro binding study using a GST-tagged Runt domain in combination with β/SMMHC having various mutations or deletions (Figure 3A). The results are diagrammatically summarized in Figure 3B. Whereas β165 and β/SMMHC detectably bound to the Runt domain as expected (lanes 25 and 26), the SMMHC tail portion alone (exons 33-42) showed no significant binding (lane 27). However, appreciable binding was observed when the same SMMHC tail or its deletion retaining exons 33 to 36 was fused to heterodimerization-defective variants of PEBP2β, β165 carrying either double mutations amino acids (64,104; lane 28 versus 29),22 or deletion of N-terminal 11 residues (lanes 30-32).29,30 Further N-terminal deletions up to positions 80 and 103 in a fusion with SMMHC(exons 33-36) were still active in heterodimerization (lanes 33 and 34). The binding ability was eventually abolished when deletion was extended either N-terminally to position 133 of PEBP2β or C-terminally past exon 35 of SMMHC (lanes 35 and 36). Taken together, these results suggest that a new independent domain (termed hyper-heterodimerization domain) for interaction with the Runt domain is created at or around the junction between PEBP2β and SMMHC.
Mutation analysis of β/SMMHC for the interaction with the Runt domain. (A) Identification of the second binding domain in β/SMMHC. GST-tagged Runt domain (GST-RD[25-190]) was mixed with various deletion constructs of β/SMMHC, and GST pull-down experiment was performed. Lanes 1 to 12 indicate 10% of the input of in vitro– translated β/SMMHC or its derivatives; lanes 13 to 24, control pull-down experiment with GST alone; and lanes 25 to 36, GST pull-down experiment using GST-RD(25-190). (B) The schematic diagram of β/SMMHC-derived constructs and the result of the GST pull-down experiments. The binding activities of each construct are listed to the right. X marks in panel B indicate mutations.
Mutation analysis of β/SMMHC for the interaction with the Runt domain. (A) Identification of the second binding domain in β/SMMHC. GST-tagged Runt domain (GST-RD[25-190]) was mixed with various deletion constructs of β/SMMHC, and GST pull-down experiment was performed. Lanes 1 to 12 indicate 10% of the input of in vitro– translated β/SMMHC or its derivatives; lanes 13 to 24, control pull-down experiment with GST alone; and lanes 25 to 36, GST pull-down experiment using GST-RD(25-190). (B) The schematic diagram of β/SMMHC-derived constructs and the result of the GST pull-down experiments. The binding activities of each construct are listed to the right. X marks in panel B indicate mutations.
DNA-binding ability and oligomeric assembly of the RUNX1-β/SMMHC complex
It has been shown that β/SMMHC multimerizes by stacking on its coiled-coil SMMHC portion6 and that this multimerization depends on the presence of a C-terminal proximal sequence element termed ACD (assembly competence domain) located within exon 40.31-33
We next asked whether and how such hierarchical states of assembly of β/SMMHC would be correlated with its hyper-heterodimerization potential. To this end, we carried out an electrophoretic mobility shift assay (EMSA) analysis using RUNX1 and C-terminal deletions of β/SMMHC overexpressed in COS7 cells. In agreement with earlier reports,29,32,34,35 we first confirmed that the RUNX1-β/SMMHC complex bound to DNA is readily detectable as a low mobility band hardly entering the gel, presumably reflecting its multimeric state, and is specifically eliminated by an unlabeled competitor DNA (Figure 4A lane 17). Furthermore, the low-mobility band due to the RUNX1-β/SMMHC complex was much more intense than the band generated by RUNX1 alone (compare lanes 1 and 17). This implies that β/SMMHC also retains the ability to enhance DNA binding by RUNX1 just as does the normal PEBP2β.
DNA binding and oligomeric assembly of the RUNX1-β/SMMHC complex. COS7 cells were transfected with each of the indicated expression plasmid (2 μg), and WCE was prepared. (A) A fixed amount of RUNX1 and WCE containing equivalent amount of either PEBP2β1 or β/SMMHC together with wild-type DNA probe were mixed as indicated and incubated, and EMSA was performed. Competitor oligonucleotides with wild-type (WT) or mutated RUNX-binding sites (MT) were incubated in the reaction mixture at 5-, 25-, and 100-fold molar excess. Migration of RUNX1 or RUNX1/β heterodimer is shown in lanes 1 and 9, respectively. Note that the complex formed by β/SMMHC, RUNX1, and DNA did not migrate into the gels but was eliminated specifically by WT (lanes 16-24). (B) A fixed amount of RUNX1 was mixed with an equivalent amount of the indicated protein and subjected to EMSA. (C) Assessment of the oligomeric state of β/SMMHC by glutaraldehyde cross-linking. Proteins overexpressed in COS7 cells were cross-linked in vitro with glutaraldehyde and analyzed by 4% to 20% gradient SDS-PAGE followed by immunoblotting with monoclonal anti-Cbfβ, as described in “Materials and methods.” Symbols on the right panel indicate the oligomeric state of cross-linked products: arrowhead indicates tetramer; and arrow, dimer. (D) Dominant binding of β/SMMHC over PEBP2β depends on the hyperdimerization domain. A fixed amount of RUNX1 (1 unit) and increasing relative amounts (0.25, 0.5, and 1 unit) of PEBP2β or β/SMMHC or its derivatives were subjected to EMSA (lanes 1-21). Similar experiments were carried out with increasing amounts of β/SMMHC constructs and a fixed amount of PEBP2β1 (1 unit; lanes 22-36). Note that A-ΔE33-36 has largely lost the ability to act dominantly over PEBP2β1.
DNA binding and oligomeric assembly of the RUNX1-β/SMMHC complex. COS7 cells were transfected with each of the indicated expression plasmid (2 μg), and WCE was prepared. (A) A fixed amount of RUNX1 and WCE containing equivalent amount of either PEBP2β1 or β/SMMHC together with wild-type DNA probe were mixed as indicated and incubated, and EMSA was performed. Competitor oligonucleotides with wild-type (WT) or mutated RUNX-binding sites (MT) were incubated in the reaction mixture at 5-, 25-, and 100-fold molar excess. Migration of RUNX1 or RUNX1/β heterodimer is shown in lanes 1 and 9, respectively. Note that the complex formed by β/SMMHC, RUNX1, and DNA did not migrate into the gels but was eliminated specifically by WT (lanes 16-24). (B) A fixed amount of RUNX1 was mixed with an equivalent amount of the indicated protein and subjected to EMSA. (C) Assessment of the oligomeric state of β/SMMHC by glutaraldehyde cross-linking. Proteins overexpressed in COS7 cells were cross-linked in vitro with glutaraldehyde and analyzed by 4% to 20% gradient SDS-PAGE followed by immunoblotting with monoclonal anti-Cbfβ, as described in “Materials and methods.” Symbols on the right panel indicate the oligomeric state of cross-linked products: arrowhead indicates tetramer; and arrow, dimer. (D) Dominant binding of β/SMMHC over PEBP2β depends on the hyperdimerization domain. A fixed amount of RUNX1 (1 unit) and increasing relative amounts (0.25, 0.5, and 1 unit) of PEBP2β or β/SMMHC or its derivatives were subjected to EMSA (lanes 1-21). Similar experiments were carried out with increasing amounts of β/SMMHC constructs and a fixed amount of PEBP2β1 (1 unit; lanes 22-36). Note that A-ΔE33-36 has largely lost the ability to act dominantly over PEBP2β1.
When C-terminal deletion constructs were tested, A-ΔE42 and A-ΔE40-42 still produced low-mobility bands as did the intact type A chimera (Figure 4B lanes 6-8). With further extensions of the C-terminal deletion beyond exon 39, DNA-bound complexes began to enter the gel in increasing fractions at accelerated mobilities (lanes 9-16). Similarly, an internal deletion lacking exons 33 to 36 still showed a low-mobility band, whereas further internal deletions up to exons 38 resulted in increasing generation of faster migrating bands (lanes 17-19). Parallel trends were also observed with a different deletion series starting from another variant of β/SMMHC, designated type I, in which exon 4 of PEBP2β was fused to exon 34 of β/SMMHC. Taken altogether, these results indicate that the C-terminal proximal region spanning exons 37 through 39 is minimally sufficient for the formation of a low-mobility complex, albeit the ACD is absent in this region.
The EMSA data in Figure 4B further provided an important insight into the state of molecular assembly of β/SMMHC in high-mobility complexes. As the SMMHC tail was shortened, the mobility of the RUNX1-β/SMMHC-DNA complex tended to increase in a manner extrapolating to that of the corresponding complexes formed with PEBP2β (compare lanes 9-16 and 2-5). This implies that these high-mobility complexes share the same oligomeric state, namely, heterodimer. In other words, the β/SMMHC molecules within the high-mobility complexes would exist as monomers, rather than coiled-coil homodimers.
To assess the oligomeric state of β/SMMHC more directly, we conducted glutaraldehyde cross-linking analysis as follows. By incubation with glutaraldehyde at an appropriate concentration, full-length β/SMMHC (Figure 4C lane 1) was completely converted into covalently linked tetramers and higher order multimers. Three internal deletions retaining exons 39 to 42 at minimum also yielded similar results (lanes 10-12). On the other hand, increasing C-terminal deletions past exon 40 up to exon 36 resulted in progressive decreases in the extent and overall efficiency of multimerization as accompanied by the reciprocal emergence of dimer and monomer bands (lanes 2-6). Further C-terminal deletions past exon 35 totally abolished the ability to dimerize and multimerize (lanes 7-9). These results are consistent with and lend support to our preceding interpretations that the ACD is dispensable, though definitely beneficial, for the multimerization of β/SMMHC and that the high-mobility complex represents a heterodimer involving a monomeric β/SMMHC protein.
We then examined how the hyper-heterodimerization potential of β/SMMHC would be affected by its C-terminal or internal deletions. We first conducted a simple titration assay in which a fixed amount of RUNX1 was mixed with increasing amounts of β/SMMHC proteins. Most of the tested constructs showed dose-dependent increases in DNA binding in a manner comparable to those observed with PEBP2β1 and PEBP2β2 (Figure 4D, compare lanes 7-15 and 19-21 with lanes 1-6). To evaluate their relative heterodimerization abilities more precisely, we next performed a competitive titration assay using a mixture containing fixed amounts of RUNX1 and PEBP2β1 with increasing amounts of β/SMMHC constructs. In this protocol, the chimeric proteins retaining the hyperdimerization domain invariably exhibited stronger competitive abilities than did the normal PEBP2β (lanes 22-30 and 34-36). Particularly, the RUNX1-PEBPβ1 band became barely visible by the addition of A-ΔE39-42 or the intact type A chimera at the highest, equivalent dose (lanes 30 and 36), whereas these chimeras themselves together with RUNX1produced very intense bands (lanes 15 and 21). Thus, the affinities of these chimeric proteins for RUNX1 are estimated to be higher than that of PEBPβ1 by one order of magnitude or more. On the other hand, A-ΔE33-36, which lacked the hyperdimerization domain, only showed a modest competitive ability against PEBP2β1 (lanes 31-33). These results indicated that the dominant binding of β/SMMHC over PEBP2β critically depends on the hyperdimerization domain. Worth commenting on, however, was a consistent tendency that the competitive ability of a chimeric protein increases as more exons are retained beyond exon 36. This implies that the hyperdimerization domain is not a completely independent functional unit but is subject to contextual influence from surrounding regions.
Transcription repression by β/SMMHC
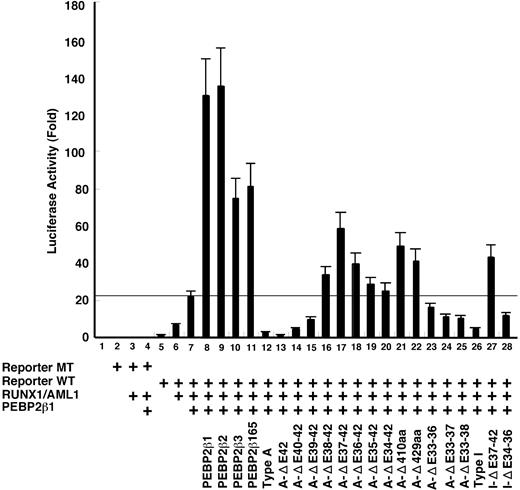
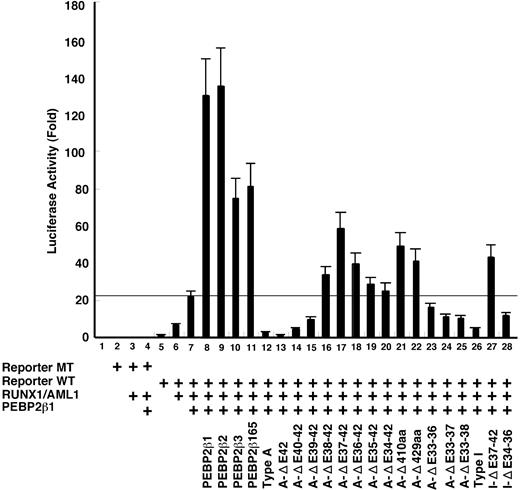
To map the regions on β/SMMHC required for its dominant repressive action against PEBP2β,19,25,36 we conducted a transcription assay using a macrophage colony-stimulating factor receptor (M-CSFR) promoter-based reporter. The transcription from this M-CSFR promoter is marginally stimulated by RUNX1 alone (Figure 5 bar 6) and requires coexpression of PEBP2β for its maximal activation (bar 7). Under this experimental setting, PEBP2β1 and each test construct were cotransfected at the same respective doses in weight (approximately equimolar to each other). The exact dose of PEBP2β was cautiously chosen to be well below a saturating level, as envisaged by a large extra transcriptional stimulation on doubling its dose (compare bars 7 and 8 in Figure 5). Among PEBP2β variants, PEBP2β1 and PEBP2β2 showed about the same stimulatory activities (bars 8-9), whereas PEBP2β3 and PEBP2β165 were approximately half as active as PEBP2β1 (bars 10-11). As expected, type A β/SMMHC caused a strong inhibition of transactivation to a level several-fold below that attained with a half dose of PEBP2β1 alone (compare bars 7 and 12). With incremental C-terminal deletions, the repression was noticeably strengthened at first (ΔE42, bar 13; approximately 2-fold), and then gradually dropped but still remained significant for the next 2 steps (ΔE40-42 and ΔE42-39, bars 14-15). Lutterbach et al36 also observed a similar rise in repression on a partial deletion of exon 42 in their transcriptional analysis using a different reporter system. Further C-terminal deletions abolished such dominant negative effects, leading instead to moderate enhancements of transactivation, which peaked in A-ΔE37-42, turned to decline progressively for the next 3 deletions, and went up again in A-ΔC410 and A-ΔC429 (bars 16-22). Coincidentally, it has been reported that similar C-terminal deletion mutants are partially functional in enhancing transactivation (ΔC283 in Cao et al,29 comparable to A-ΔE36-42) as well as restoring definitive hematopoiesis by Pebpb2-/- ES cells (ΔC317 in Miller et al,37 comparable to A-ΔE35-42). However, internal deletions spanning exons 33 to 36 or more resulted in decreased, rather than increased repression (bars 23-25). Type I chimera also showed a strong inhibition, and its C-terminal and internal deletions effected a moderate stimulation or inhibition of transcription in a manner very similar to the corresponding deletions of type A chimera (compare bars 26-29).
Transcription repression by β/SMMHC. U937 cells were transfected with M-CSF-R-luc plasmid (1 μg) with mutated RUNX binding site (lanes 2-4) or wild-type site (lanes 5-28), together with expression plasmids for RUNX1 (0.5 μg, a full dose) and PEBP2β1 (0.25 μg, a half dose) as indicated. On top of it, a half dose of PEBP2β isoforms or β/SMMHC mutant expression vectors were transfected in such a way as to mimic the conditions of inv(16) cells. A total of 2 μg DNA including 1 ng Renilla luciferase expression plasmid as an internal control was used. Luciferase activities are expressed as fold changes relative to the control transfected with M-CSF-R-luc and the backbone expression vector. The data are mean of 3 independent experiments and the values were normalized using luciferase activities. Standard deviations are indicated by error bars. Horizontal line indicates the level of transactivation attained by a full dose of RUNX1 and a half dose of PEBP2β1. Above this line is considered to be transcription stimulation by each construct and below the line, repression.
Transcription repression by β/SMMHC. U937 cells were transfected with M-CSF-R-luc plasmid (1 μg) with mutated RUNX binding site (lanes 2-4) or wild-type site (lanes 5-28), together with expression plasmids for RUNX1 (0.5 μg, a full dose) and PEBP2β1 (0.25 μg, a half dose) as indicated. On top of it, a half dose of PEBP2β isoforms or β/SMMHC mutant expression vectors were transfected in such a way as to mimic the conditions of inv(16) cells. A total of 2 μg DNA including 1 ng Renilla luciferase expression plasmid as an internal control was used. Luciferase activities are expressed as fold changes relative to the control transfected with M-CSF-R-luc and the backbone expression vector. The data are mean of 3 independent experiments and the values were normalized using luciferase activities. Standard deviations are indicated by error bars. Horizontal line indicates the level of transactivation attained by a full dose of RUNX1 and a half dose of PEBP2β1. Above this line is considered to be transcription stimulation by each construct and below the line, repression.
These results altogether suggest that the C-terminal region of β/SMMHC spanning exons 37 to 40 is responsible for its strong repressive function in a length- and context-dependent manner without showing any clear-cut domain structure. On the other hand, the hyperdimerization domain by itself has no intrinsic repressive function, but it nevertheless is shown to play a critical role in maximizing the β/SMMHC-mediated repression.
Discussion
In this study, we have identified 2 functional domains in β/SMMHC that cooperate to achieve a full repression of the RUNX1 function. The first one, newly discovered and termed “hyperdimerization domain” herein, spans the fusion junction and provides an extra interface for interaction with the Runt domain. The second one, which has been implicated previously,6,36 is located in the C-terminal proximal region and confers on β/SMMHC the ability to act dedicatedly for repression. While this work was in progress, a few other groups have reported related studies focusing, respectively, on each of these 2 aspects.32,38,39 Their results are consistent or complementary with ours in general aspects, but seemingly discrepant in some important details. In the following, we will make close comparisons between those reports and ours with a view to construct a unifying model for the mechanism of repression mediated by β/SMMHC (Figure 6).
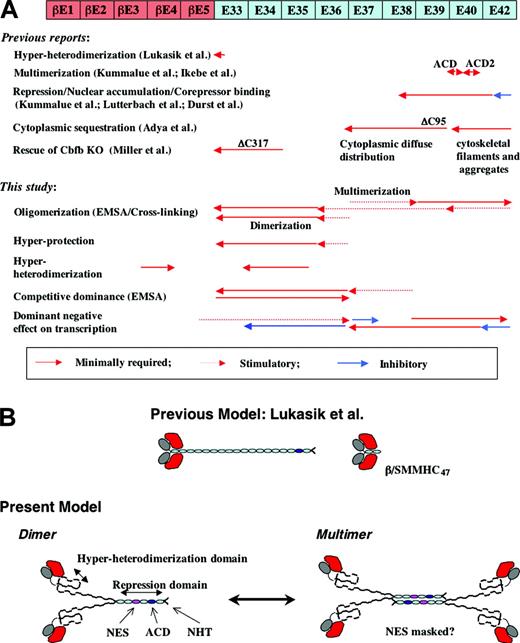
Summary of domain analysis of β/SMMHC. (A) Summary of earlier studies (top half) and this study (bottom half). Diagram shown at the top represents β/SMMHC. Red-colored segments—βE1 to βE5—represent exons encoding CBFβ/PEBP2β. ACD indicates assembly competence domain. (B) Diagrammatic models for RUNX1-β/SMMHC complex with varying SMMHC deletions. Center top shows the model proposed by Lukasik et al.38 Lower drawings represent the models proposed by this study. NES indicates presumptive nuclear export signal; ACD, assembly competence domain; NHT, nonhelical terminus.
Summary of domain analysis of β/SMMHC. (A) Summary of earlier studies (top half) and this study (bottom half). Diagram shown at the top represents β/SMMHC. Red-colored segments—βE1 to βE5—represent exons encoding CBFβ/PEBP2β. ACD indicates assembly competence domain. (B) Diagrammatic models for RUNX1-β/SMMHC complex with varying SMMHC deletions. Center top shows the model proposed by Lukasik et al.38 Lower drawings represent the models proposed by this study. NES indicates presumptive nuclear export signal; ACD, assembly competence domain; NHT, nonhelical terminus.
The hyperdimerization domain
The SMMHC tail as such does not significantly bind to the Runt domain. However, fusion of PEBP2β to SMMHC creates a new binding interface designated hyperdimerization domain, which can function independently of, and act in concert with, the original heterodimerization interface on PEBP2β. Putting the results with type A and type I chimeras, we may define the minimal domain for hyperheterodimerization to reside within exons 34 to 35 on the SMMHC side. As another important functional feature, β/SMMHC retains virtually the same allosteric activity to enhance DNA binding by RUNX1 as does the normal PEBP2β. This property could have a profound relevance to the efficiency of transcriptional repression by β/SMMHC as will be discussed.
The analysis by EMSA further led us to the unexpected finding that β/SMMHC deleted C-terminally past exon 39 largely exist as monomers, rather than coiled-coil dimers or higher order multimers. Retrospectively, a number of reports have been consistent with this view. Previous cross-linking studies suggested that the intact β/SMMHC as well as its derivative lacking C-terminal 95 amino acids (almost equivalent to A-ΔE40-42) form stable dimers or multimers, whereas shorter ones lacking C-terminal 175 amino acids (deleted to the middle of exon 38) or more no longer do so.29,30 The same trends were also confirmed herein by cross-linking analysis using a more extended series of deletion constructs. Furthermore, the coiled-coiled rod of SMMHC is known to be quite flexible and rather unstable under physiologic salt conditions, readily tending to transform into a folded hairpin structure40 or to dissociate into a monomer on truncation.41
In the light of these observations, the hyperdimerization domain would plausibly exist and function in an unpaired state. If so, how can we explain the fact that hyperdimerization was also observed with the full-length β/SMMHC and minimally truncated derivatives, which were supposed to exist as coiled-coil dimers or higher order multimers? The simplest model resolving this puzzle may be a Y-shaped dimer consisting of unpaired N-terminal halves followed by a coiled-coil for the C-terminal remainder (Figure 6B). However, because of the limitation of the method used, we could not completely rule out the possibility that the SMMHC portion of β/SMMHC forms coiled-coil structure in its entirety. With this reservation in mind, we will subsequently look into possible structural and functional implications of the Y-shaped model.
A partial destruction of the coiled-coil could be facilitated by its fusion to a structurally different protein, PEBP2β, in addition to its intrinsic local instability. This putative situation would also explain why the presence of the PEBP2β portion, for at least its exons 4 and 5, is required to make the chimeric protein accessible for interaction by RUNX1 (exon 5 may actually be dispensable as it is missing in some β/SMMHC variants). As depicted in Figure 6B, we further infer that the unpaired region of SMMHC would not be so stiff and thus could take a somewhat folded structure. In this model, only a part of the hyperdimerization domain would directly contact RUNX1 and the remainder would indirectly serve to keep the domain in a proper conformation. This folded domain might well make additional contacts with its C-terminally flanking, supposedly unpaired region, thereby gaining an extra stabilization. This view is consistent with the observation that the hyperheterodimerization potential visibly tends to increase progressively as the more of C-terminal exons were added to A-ΔE37-42 (Figure 4D). Thermodynamically, it then follows that the presence of RUNX1 should reciprocally promote the integration of such extra tail segments into the hyperdimerization domain with a consequential destabilization of the coiled-coil structure. Concordant with this prediction, the relative intensities of high-mobility complexes (heterodimers) compared with those of low-mobility complexes (multimerized heterodimers) observed in the EMSA, particularly in Figure 4D (lanes 7-15 and 22-30), were by far stronger than expected from the cross-linking pattern presented in Figure 4C.
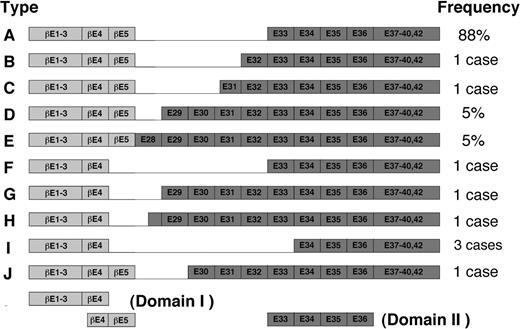
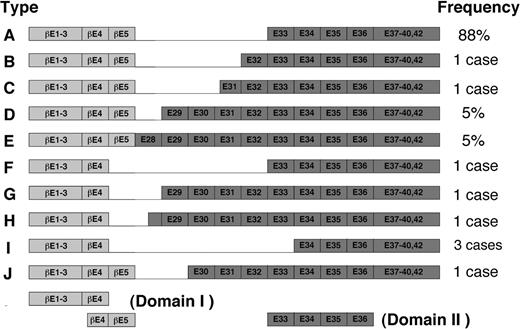
The proposed model could also be extended with slight modifications to other types of inv(16) fusion products thus far identified (Figure 7). In these variants, exon 4 or exon 5 in PEBP2β is joined to any of the exons between 28 and 34 in SMMHC in nearly all possible combinations (Figure 7). Interestingly, however, all the variants contain exons 34 to 35, the above-noted minimal domain for the hyper-heterodimerization. Thus, kinking or folding in the N-terminal proximal part of SMMHC tail, similar to that postulated in the model, could persistently allow the Runt domain to interact with exons 34 to 35, regardless of how far this region is separated from the chimeric junction on the polypeptide sequence. Nevertheless, the possibility remains that subtle variations in the attainable efficiency of hyperdimerization could make difference in terms of pathogenicity. Interesting in this regard, 2 of the 3 patients with type I chimera so far reported showed FAB M2 phenotype instead of M4Eo phenotype that is characteristic to most cases of inv(16).42,43
Schematic diagram of fusion variants of β/SMMHC described by van Dongen et al.28 The exon numbers have been changed due to recent completion of the structural analysis of MYH11 gene (GenBank accession no. AB020673). Adapted from van Dongen et al28 with permission; copyright Nature Publishing Group (http://www.nature.com/).
Schematic diagram of fusion variants of β/SMMHC described by van Dongen et al.28 The exon numbers have been changed due to recent completion of the structural analysis of MYH11 gene (GenBank accession no. AB020673). Adapted from van Dongen et al28 with permission; copyright Nature Publishing Group (http://www.nature.com/).
In an apparent contradiction with this model, Lukasik et al38 reported that a 47–amino acid portion of SMMHC ending within exon 33 was sufficient to support its enhanced heterodimerization with the Runt domain as well as its homodimerization into a coiled-coil. Moreover, they described that the enhanced heterodimerization of β/SMMHC47 was hindered in the presence of DNA, which disagrees with our results. These discrepancies would be possibly attributable to differences in the analytical methods and experimental conditions used. In our in vivo and in vitro binding assays, the concentrations of the Runt domain and β/SMMHC were roughly in nanomolar ranges, and salts were invariably present at moderate concentrations. On the other hand, Lukasik et al38 used physical-chemical methods such as microcalorimetry, dynamic light scattering, and nuclear magnetic resonance (NMR) spectroscopy, which inevitably required the use of those proteins at micromolar to submillimolar concentrations together with buffers of minimal ionic strengths. These conditions must have greatly facilitated protein-protein associations, either homologous or heterologous ones, which could be hardly detectable if otherwise.
The repression domain and underlying mechanisms
In this study, we demonstrated that a broad region spanning exons 38 to 40 is required for the dominant repression of RUNX1-dependent transcription by β/SMMHC in competition with PEBP2β. This repression domain approximately coincides with those previously identified by other groups.29,30,32,36,39 However, there have been split views as to how β/SMMHC mediates repression.
One view, first implicated by Lu et al23 and Liu et al6 and experimentally substantiated by Adya et al30 and Kanno et al,25 suggests that β/SMMHC sequesters RUNX1 to the cytoplasm through its intrinsic ability to interact with actin (the sequestration model). On the other hand, a second view proposed by Lutterbach et al36 suggests that β/SMMHC acts for repression in the nucleus by its newly uncovered ability to interact with a corepressor, mSin3A (the corepressor-recruitment model). In an apparent support to the corepressor-recruitment model, Kummalue et al32 reported that β/SMMHC tends to accumulate preferentially in the nucleus as multimerized complexes, which involve RUNX1 and can specifically bind to DNA containing a RUNX consensus site. They also demonstrated that the ability of β/SMMHC to form multimers depends on the presence of the ACD motif and is tightly correlated with its repression activity. To further extend the corepressor-recruitment model, Durst et al39 recently described that the C-terminal 163 amino acids of the SMMHC region harbors the abilities to repress transcription and bind histone deacetylase, HDAC8, as well as the mSin3A corepressor, all in an ACD-dependent manner.
Suggestive in resolving these controversies is our present finding that C-terminal deletion mutants, A-ΔE40-42 and A-ΔE39-42, showed considerable repression activities. These proteins lacked the ACD and hence should no longer be able to form multimers nor to accumulate in the nucleus according to the Kummalue et al.32 Indeed, Adya et al30 previously showed that β/SMMHCΔ95 (nearly equivalent to A-ΔE40-42), together with RUNX1, was localized almost entirely to the cytoplasm in a diffuse pattern, instead of attaching to cytoskeletal filaments. Thus it seems as though β/SMMHCΔ95 had a cryptic nuclear export signal that could be made active by removal of the ACD (colored pink in the model of Figure 6B). Collectively, these observations favor the sequestration model, rather than the corepressor-recruitment model, at least for those mutants that were C-terminally deleted only moderately and retained considerable abilities to form dimers, but not multimers. However, it should be pointed out here that the intact β/SMMHC, unlike β/SMMHCΔ95, actually tended to localize in part to the nucleus as well, when it was highly overexpressed beyond the available level of actin to result in progressive disorganizations of the cytoskeletal structure. Under such conditions, the corepressor-recruitment model could work as an additional or alternative mechanism for repression. This may explain why the intact β/SMMHC and A-ΔE42 showed stronger repression abilities than that seen with A-ΔE40-42.
A crucial question emerging from these consideration is which of the 2 alternative repression mechanisms would be at work in the original inv(16) leukemic cells as well as their murine model, which contain a single allele each of intact PEBPB2 gene and β/MYH11 on the background of 2 intact RUNX1 alleles. As one most straightforward approach to this question, work is under way to construct and analyze gene-targeted mice carrying β/SMMHC with various C-terminal and internal deletions used herein.
Prepublished online as Blood First Edition Paper, December 24, 2003; DOI 10.1182/blood-2003-07-2188.
Supported by a research grant from Human Frontier Science Program (Y.I., coinvestigator of RGP0375/22001-M).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Nancy Speck and Dong-Er Zhang for generous gift of anti-Cbfβ monoclonal antibodies (β141.4.1) and M-CSF-R-luc reporter, respectively. We also thank Zhi-Jian Xiao for construction of β/SMMHC/AA.


![Figure 3. Mutation analysis of β/SMMHC for the interaction with the Runt domain. (A) Identification of the second binding domain in β/SMMHC. GST-tagged Runt domain (GST-RD[25-190]) was mixed with various deletion constructs of β/SMMHC, and GST pull-down experiment was performed. Lanes 1 to 12 indicate 10% of the input of in vitro– translated β/SMMHC or its derivatives; lanes 13 to 24, control pull-down experiment with GST alone; and lanes 25 to 36, GST pull-down experiment using GST-RD(25-190). (B) The schematic diagram of β/SMMHC-derived constructs and the result of the GST pull-down experiments. The binding activities of each construct are listed to the right. X marks in panel B indicate mutations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/8/10.1182_blood-2003-07-2188/6/m_zh80080459760003.jpeg?Expires=1763505692&Signature=yfmcwrN4Wd~NDI~gjdEy061r3kPWD3xMrCL1s41vHzUy-OdRiRQV8x-F6hq~RDBabeXi0bCtoluVpNN8gFvixB815fsxMtwrE6xqUcyB4CkwK48-z8fGchHGTecxg7ODTHx5huyyk8a-SfH7LZ-zTsNiTaoFscZ~q8bt0fwScgPj5-NhjoEKpMlYevVeIKc7ww7ld-zQlyflVJAEmZYUK-HTqPGHablReAlI6A7~1HcGB~hb~Ata6ASmFi6vyZ3xDy4sJfL6IzYRf4VNDqYrXO2VHweAt1f44dWv4dLfZlOKVCvAQAtPQz2in7b92DhWXgrUuPtNI~vFzRW67RrZyw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

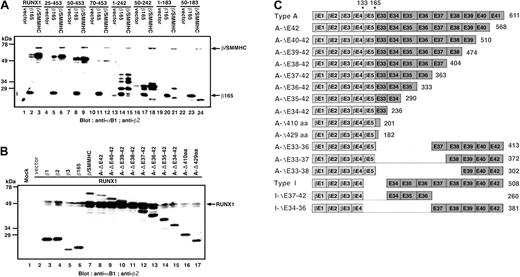
![Figure 3. Mutation analysis of β/SMMHC for the interaction with the Runt domain. (A) Identification of the second binding domain in β/SMMHC. GST-tagged Runt domain (GST-RD[25-190]) was mixed with various deletion constructs of β/SMMHC, and GST pull-down experiment was performed. Lanes 1 to 12 indicate 10% of the input of in vitro– translated β/SMMHC or its derivatives; lanes 13 to 24, control pull-down experiment with GST alone; and lanes 25 to 36, GST pull-down experiment using GST-RD(25-190). (B) The schematic diagram of β/SMMHC-derived constructs and the result of the GST pull-down experiments. The binding activities of each construct are listed to the right. X marks in panel B indicate mutations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/8/10.1182_blood-2003-07-2188/6/m_zh80080459760003.jpeg?Expires=1763505693&Signature=qEQJDGjA5TMwFC1pbJqjtjOluir9KLb6YLPYzi5whPTBa4cdzDIcCOV4IqNgL5oA8F3idm-pG1H-22mqKYP8taMDfzzGr6HqF2O9xijZd204gA2ueBRtQOdRjy4c7DyGbnhcUIyQgWM999bBhe71b8D-dw~lUzajd2jVKMyVjOrt1KuZFkGbvxm38Yqs2NRQQTwdvRcm3UkizBVV~UYuuQsQvdNecNiRY0kfHAcdj28n~32X6RUFQSL2iZiQ0eujCg2IR4RASJGZDrKj-En8nyLDM0X~xBpwr4x5J2NRuizIyu-Az5C081vR0zp2hgfU9QYR1HTkULNnd06Xg29GSQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)