Abstract
Due to their low frequency, CD4 T-cell responses to Epstein-Barr virus (EBV) lytic antigens are, so far, poorly characterized. Human peptide major histocompatibility complex (MHC) class II multimers provide a means to detect and characterize such rare T cells. Along a screening of T-cell responses to lytic or latent EBV antigens within peripheral blood leukocyte (PBL)– or synovial-derived CD4 T-cell lines, we identified an human leukocyte antigen–DR*0401 (HLA-DR*0401)–restricted epitope derived from BHRF1 (BamHI fragment H rightward open reading frame 1), a viral protein produced during the early stages of the lytic cycle. We show here that T-cell responses to this particular BHRF1 epitope are shared by most EBV-infected DR*0401+ individuals, as BHRF1-specific CD4 T cells could be sorted out from all the DRB*0401 T-cell lines analyzed, using magnetic beads coated with recombinant BHRF1/DR*0401 complexes. Sorting with these peptide MHC class II multimers was very efficient, as the yield of recovery of BHRF1-specific T cells was nearly 100%. Functional analysis of a large number of clones responding to BHRF1/DR*0401 demonstrated their cytolytic action against autologous and allogeneic DR*0401+ EBV-transformed B-lymphoblastoid cell lines (B-LCLs), with 40% to 80% killing efficiency and potent interferon γ production, thus suggesting that this CD4 T-cell population contributes to the control of EBV replication. B-LCL lysis by these T-cell clones was DR*0401 dependent, EBV dependent, and was not merely due to bystander killing. Taken together, these data provide the first demonstration that a lytic antigen can induce a direct cytolytic response against EBV-infected cells.
Introduction
Epstein-Barr virus (EBV) is a human gamma herpesvirus that establishes in 95% of the adult population a lifelong latent infection in resting memory B cells. The EBV carrier state (latency) is characterized by expression of a limited set of EBV genes and sporadic reactivation of the lytic cycle in latently infected cells, leading to viral replication. Upon reactivation in tonsillar B lymphocytes, EBV can productively infect oropharyngeal epithelium, resulting in infectious virus production and transmission.1-3 During childhood, patients'primary infection by EBV is usually asymptomatic and leads to a lifelong persistence of the virus. Host immune responses are thought to be of central importance both in limiting the primary infection and in controlling the lifelong virus carrier state. The high frequency of EBV-associated lymphoproliferative diseases or lymphomas in immunocompromised individuals strongly supports a role for anti-EBV T cells in containing EBV infection.4
The CD8 T-cell response has been the matter of intensive investigations. Large CD8+ clonal expansions have been described in infectious mononucleosis patients and there is some evidence that clones detected in the memory cytotoxic T-lymphocyte (CTL) response are selected during the acute phase and may persist at high circulating frequencies,5 contributing to the CD8+ expansion seen in healthy adults.6 Recently, the use of human leukocyte antigen (HLA) class I peptide tetramers has highlighted the large proportion of CD8 T cells directed to EBV antigens.7,8 These CD8+ CTL responses are preferentially directed toward the early lytic proteins, BZLF1 (BamH fragment Z left frame 1) and BMLF1 (BamHI-M leftward reading frame 1)9-12 and to a lesser extent, toward the latent nuclear antigens Epstein-Barr virus nuclear antigen 3A (EBNA3A), EBNA3B, and EBNA3C.12-14
Much less is known about the CD4+ T-cell responses to EBV, although there is an increasing awareness of their key role in (1) supporting high-affinity antibody (Ab) production; (2) initiating and, particularly, maintaining CTL numbers and function; and (3) performing direct effector activity. Analysis of CD4 T-cell responses has been greatly hampered by the small size of the CD4 compartment. No CD4 T-cell expansions have been detected using highly sensitive heteroduplex techniques, even during the early stages of EBV acute infection.15 The EBV specificity of some CD4+ T-cell clones has been previously reported.16-19 In more recent systematic analysis, the nuclear antigen EBNA1 was found to be a main EBV latency antigen for CD4+ T cells.20-22
Here we describe the identification and characterization of a dominant CD4 T-cell response to BHRF1 (BamHI fragment H rightward open reading frame 1) in DR*0401 individuals. We screened for anti-EBV CD4 T cells in peripheral blood leukocyte (PBL)–derived T-cell lines from healthy donors or kidney recipients and in synovial fluid (SF)–derived T-cell lines from arthritis patients, previously shown to be greatly enriched for EBV-specific CD8 T cells.11 Along this screening, an HLA-DR*0401–restricted BHRF1 epitope, an early antigen of the lytic cycle, was identified. Taking advantage of the recent availability of major histocompatibility complex (MHC) class II tetramers,23 we set up an efficient sorting strategy to isolate BHRF1-specific cells, using multimers of HLA-DR*0401/BHRF1 peptide complexes coated on immunomagnetic beads. BHRF1-specific T cells were sorted out from all DR*0401 T-cell lines studied with nearly 100% efficiency. Moreover, the direct contribution of BHRF1-specific T cells to the control of EBV replication was suggested by their ability to kill autologous B-lymphoblastoid cells (B-LCLs) and to produce interferon γ (IFN-γ).
Patients, materials, and methods
Donors and HLA-DR typing
All T-cell samples were from HLA-DR4+ and EBV+ donors11,12 (Table 1). PBLs were obtained from 2 healthy virus carriers (D3 and D5) and 2 kidney graft recipients (KRs). SF-derived lymphocytes were obtained for clinical indications from patients suffering from various forms of acute or chronic arthritis: 9 patients with rheumatoid arthritis (RA), 1 patient with ankylosing spondylitis (AS), and 1 patient with Reiter syndrome (RS). Mononuclear cells from peripheral blood (PBMCs) and from synovial fluid were isolated by Ficoll/Hypaque density gradient centrifugation. HLA-DRB1 typing was performed by using standard molecular typing techniques at the Etablissement de Transfusion Sanguine (Nantes, France). This study was approved by the Centre Hospitalier Universitaire de Nantes institutional review board.
T-cell lines and T-cell clones
PBL- or SF-derived lymphocytes were maintained in RPMI 1640 supplemented with 10% pooled human serum, 1 mM l-glutamine, and 150 IU/mL recombinant interleukin 2 (IL-2). Positive selection for CD4+ PBMCs was performed by immunomagnetic sorting using an anti-CD4–specific monoclonal Ab (mAb) as previously described.11 CD4 purity (>95%) was confirmed by staining with anti-CD8 and anti-CD4 antibodies, followed by flow cytometric analysis. CD4+ polyclonal T cells were then expanded in vitro in IL-2 culture medium supplemented with purified leukoagglutinin (0.5 μg/mL) and irradiated (30 Gy) allogeneic feeder cells (PBLs and B-LCLs at a 10:1 ratio). Clones were obtained by seeding synovial T cells at 0.3 cells/well in IL-2/CM, leukoagglutinin (0.5 μg/mL), and irradiated allogeneic feeder cells (5 × 104 PBLs and 5 × 103 B lymphoblastoid cells/well).
Generation of EBV B-LCLs
B-LCLs were generated for each donor by culturing 2 × 106 PBMCs in 100 μL of RPMI and 10% fetal calf serum (FCS) with 500 μL of EBV-containing supernatant from the virus-producing B95.8 marmoset cell line. Cultures were performed in the presence of 1 μg/mL cyclosporin A. After 24 hours, 2 mL of RPMI 1640 containing 10% FCS, 1mM l-glutamine, and 50 μg/mL gentamicin was added to each well.
A DR*0401+ B-LCL transformed with an EBV mutant, in which BHRF1 was deleted (BHRF1–knockout [KO]), was generated by transforming 105 CD3-depleted PBMCs from donor D3 (DR*0401+) with supernatant (104 colony-forming units) from B95/B531a cells, containing in roughly equal amounts B95.8 virus and a B95.8-derived mutant, B531a, in which cytomegalovirus–immediate early (CMVIE) neo construct replaces most of BHRF1 gene (B95/B531a).24 Two days later, G418 (GIBCO BRL, Carlsbad, CA) was added to the medium at 700 μg/mL (active drug concentration). G418-resistant LCL clones were obtained and the absence of BHRF1 was confirmed by polymerase chain reaction (PCR).
Recombinant vaccinia-EBV viruses
Recombinant vaccinia virus (rVV) vectors coding for each of the EBV latent proteins (Epstein-Barr nuclear antigens [EBNAs] 1, 2, 3A, 3B, 3C, and LP, latent membrane protein 1 [LMP1], and LMP2) or some EBV lytic proteins (BZLF1, BRLF1, BHRF1, BMLF1, BamHI-M rightward open reading frame 1 [BMRF1], BamHI left frame 1 [BHLF1], BALF2, BALF5, BCRF1, and BLLF1) have been previously described.13,14 B-LCLs were incubated with individual rVV (10 plaque-forming units [PFUs] per cell) overnight, and then effector T cells were added to infected B cells at a 10:1 effector-target (E/T) ratio. Activation of T cells was evaluated either in a tumor necrosis factor α (TNF-α) or in a 51chromium release assay.
Peptides
A set of 23 residue peptides, overlapping by 12 amino acids and spanning the whole BHRF1 protein (Chiron Mimotopes, Victoria, Australia), was used for the characterization of the BHRF1/DR*0401 epitope. The 12–amino acid peptide BHRF1122-133 (PYYVVDLSVRGM, designated PYY) was obtained from Genosys (The Woodlands, TX). Peptide stock solutions (20 mg/mL in dimethyl sulfoxide [DMSO]) were diluted first to 2 mg/mL in acetic acid (1%) and second to the final concentration in RPMI 1640 culture medium.
TNF assay
Production of TNF-α by activated lymphocytes was estimated in an autopresentation assay. To trigger TNF-α release by responding T cells, 5 × 103 clonal T cells or 5 × 104 polyclonal T cells were incubated with peptides at various concentrations. After 6 hours, the supernatant was collected and its TNF content was determined by testing its cytotoxic effect on Walter and Eliza Hall Institute (WEHI) 164 clone 13 cells in an MTT (3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) colorimetric assay.25
Generation of HLA-DR*0401– peptide tetramers
Peptide-loaded HLA-DR0401 tetramers were produced as described previously.23 Briefly, recombinant DRA1*0101 and DRB1*0401, in which the transmembrane domain was replaced by a leucine zipper, were produced in Drosophila S2 cells. At the end of the β-chain cDNA is a biotinylation sequence that allows site-specific biotinylation using the BirA enzyme. The resulting biotynilated heterodimers were loaded with peptides for 3 days at 37°C. Phycoerythrin (PE)–labeled streptavidin was used to produce fluorescent peptide–loaded DR*0401 tetramers. Peptide used corresponded to the sequence of the BHRF1 protein residues 122-133 (peptide PYY). DRA1*0101/DRB1*0401 tetramers loaded with the HA307-319 peptide (hemagglutinin) were used as a control.23
Immunomagnetic cell sorting
Immunomagnetic sorting of DRB1*0401/BHRF1-specific T cells was performed as previously described,26 using beads coated with HLA-DR*0401/BHRF1 peptide monomers. For coating immunomagnetic beads, 1 μg HLA-DR4 peptide monomers diluted in 30 μL of phosphate-buffered saline–bovine serum albumin (PBS-BSA) 0.1% were incubated for 1 hour at room temperature with 5 × 106 streptavidin-coated beads (Dynabeads M-280 streptavidin; DYNAL, Compiègne, France), followed by several washes. PBL- or SF-derived lymphocytes (5 × 106) were rotated for 4 hours at room temperature with monomer-coated beads. After 9 washes, bead-coated cells were expanded by polyclonal activation as described in “T-cell lines and T-cell clones” and then cultured for 2 to 3 weeks before testing.
Sequence analysis of T-cell receptor (TCR) transcripts
RNA from 5 × 106 T-cell clones was extracted with TRIZol reagent (Invitrogen, Cergy Pontoise, France) according to the supplier's instructions and dissolved in 15 μL of water. Reverse transcriptions, PCR amplifications, and sequencing were performed as described.11
Flow cytometric determination of cell surface and intracellular markers
Staining with DR4/PYY tetramers was performed by incubating CTL clones or T-cell lines (2.5 × 105) for 1 hour at 37°C with 1 μg of each PE-labeled tetramer in 50 μL of culture medium. Cells were then washed in PBS containing 1% FCS and 0.1% NaN3 and stained with fluorescein isothiocyanate (FITC)–conjugated monoclonal antibody against CD4 (PharMingen, San Diego, CA). After a 30-minute incubation, cells were washed again and analyzed using a Becton Dickinson FACSCan flow cytometer (San Jose, CA). T-cell lines and T-cell clones were phenotyped by indirect immunofluorescence with mAb against TCR Vβ regions.
T-cell helper 1 (Th1)/Th2 profiling was performed through intracellular detection of IFN-γ and IL-4. T cells were resuspended in RPMI 10% FCS supplemented with phorbol myristate acetate (PMA; 10–7 M; Sigma, St Louis, MO) and ionomycin (0.5 μg/mL; Sigma) and incubated at 37°C. After 2 hours, monensin (2 nM; Sigma) was added and the cells were incubated further for 3 hours. The cells were then fixed for 20 minutes in 2% paraformaldehyde, permeabilized using a 0.5% saponin buffer, and stained with the following antibodies: FITC-conjugated anti–IFN-γ (Pharmingen) and PE-conjugated anti–IL-4 (Pharmingen). The stained cells were then analyzed on the flow cytometer. A human T-cell line expressing both IFN-γ and IL-4 was included as a control to assess for the specificity of the intracellular cytokine detection assay.
Cytolytic assays
Cytolytic assays were performed in a 4-hour chromium release assay. The targets used were unloaded B-LCLs or B-LCLs loaded with peptide at various concentrations for 30 minutes. In the mAb blocking experiments, targets were incubated with mAbs for 30 minutes at room temperature before adding effector cells. In bystander killing, HLA-DR*0401+ B-LCLs were 51Cr labeled while HLA-DR*0401– B-LCLs were left unlabeled. Assays were performed in triplicate and results were expressed as percent specific 51Cr release.
Results
Identification of a DR*0401/BHRF1 epitope.
In an attempt to characterize CD4 T-cell responses to EBV, we started from T-cell lines derived from the synovial fluid of arthritic patients, within which we previously described an enrichment for CD8 T cells responding to EBV antigens.11 CD4+ T-cell clones were obtained from SF-derived T-cell lines. Four CD4+ T-cell clones from patient RA1, which proliferated when cocultured with the autologous B-LCL, were selected for further analysis of antigen specificity. The autologous B-LCL was infected with rVV encoding 10 lytic EBV proteins and all latent proteins. One of these clones (clone 4.19 from patient RA1) was activated by the autologous B-LCL infected with rVV encoding the BHRF1 gene product, as documented by TNF-α production (Figure 1A).
Identification of a DR*0401/BHRF1 epitope recognized by the CD4 T-cell clone 4.19. (A) Recognition of the lytic EBV protein BHRF1 by clone 4.19. CTL cells were tested in a 4-hour TNF-α release assay on autologous B-LCLs expressing individual latent or lytic EBV proteins from vaccinial viral vectors. Results are shown as μg/mL. TNF release was observed at an effector-target ratio of 10:1. (B) Identification of the target epitope of BHRF1-specific clone 4.19 using a panel of peptides (23-mers, overlapping by 12 amino acids [aa's]) spanning the BHRF1 protein. These peptides were used as targets in cytotoxic assays by incubating them with 51Cr-labeled autologous EBV-B cells for 1 hour at 37°C. Clone 4.19 was added at an E/T ratio of 10:1 and Cr release was measured after 4 hours. Note the significant lysis background observed with unloaded autologous B-LCL (No pep). (C) Stimulation of clone 4.19 by the BHRF1 122-133 peptide (PYY). Cytotoxicity of clone 4.19 to Cr-labeled autologous EBV-B cells loaded with various concentrations of peptides. Clone 4.19 was added at an E/T ratio of 10:1 and Cr release was measured after 4 hours. Data obtained with the 23-mer BHRF1111-133 peptide are shown as a positive control. Data obtained with an irrelevant peptide (irr peptide) are shown as a negative control.
Identification of a DR*0401/BHRF1 epitope recognized by the CD4 T-cell clone 4.19. (A) Recognition of the lytic EBV protein BHRF1 by clone 4.19. CTL cells were tested in a 4-hour TNF-α release assay on autologous B-LCLs expressing individual latent or lytic EBV proteins from vaccinial viral vectors. Results are shown as μg/mL. TNF release was observed at an effector-target ratio of 10:1. (B) Identification of the target epitope of BHRF1-specific clone 4.19 using a panel of peptides (23-mers, overlapping by 12 amino acids [aa's]) spanning the BHRF1 protein. These peptides were used as targets in cytotoxic assays by incubating them with 51Cr-labeled autologous EBV-B cells for 1 hour at 37°C. Clone 4.19 was added at an E/T ratio of 10:1 and Cr release was measured after 4 hours. Note the significant lysis background observed with unloaded autologous B-LCL (No pep). (C) Stimulation of clone 4.19 by the BHRF1 122-133 peptide (PYY). Cytotoxicity of clone 4.19 to Cr-labeled autologous EBV-B cells loaded with various concentrations of peptides. Clone 4.19 was added at an E/T ratio of 10:1 and Cr release was measured after 4 hours. Data obtained with the 23-mer BHRF1111-133 peptide are shown as a positive control. Data obtained with an irrelevant peptide (irr peptide) are shown as a negative control.
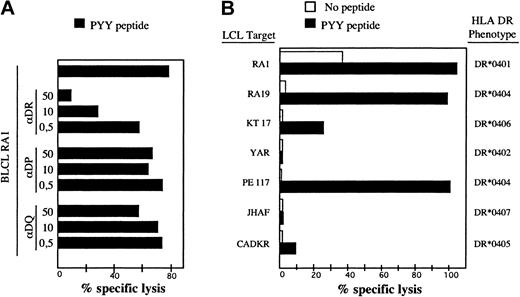
To identify the BHRF1 epitope recognized by clone 4.19, a set of synthetic peptides of 23 amino acids overlapping by 12 amino acids, and covering the entire BHRF1 protein sequence, was synthesized. Autologous EBV-B cells pulsed with each of these peptides were tested in a cytotoxic assay for recognition by clone 4.19. Two overlapping peptides scored positive, namely BHRF1111-133 and BHRF1122-144 (Figure 1B). The 12–amino acid peptide BHRF1122-133 (PYYVVDLSVRGM, designated PYY), corresponding to the overlap, was recognized by clone 4.19 (Figure 1C). The sequence suggested that tyrosine at position 123 or 124 was the peptide 1 (P1) anchor residue for MHC binding, conforming to the HLA-DR4 binding motif.27 Recognition by clone 4.19 of autologous EBV-B cells loaded with peptide PYY was inhibited by an anti–HLA-DR mAb (Figure 2A). Donor RA1 was typed DR1 and DR4 (subtype DRB1*0401). Peptide PYY was loaded onto a panel of allogeneic LCLs, sharing DR1 or DR4 with donor RA1. Clone 4.19 recognized efficiently DR*0401 (5 cell lines tested) and DR*0404 B-LCLs loaded with PYY. Peptide-loaded DR*0406 B-LCL was weakly recognized, whereas peptide-loaded DR*0402, DR*0405, and DR*0407 B-LCLs were not recognized (Figure 2B).
Recognition of the BHRF1 122-133 epitope by the CD4 T-cell clone 4.19 is HLA-DR*0401-restricted. (A) Clone 4.19 cells were cocultured with autologous B-LCL loaded with PYY peptide at 10 μM. Inhibition with anti-DR (L243), anti-DP (B7.21), or anti–class I (W6.32) antibodies was performed by addition of mAb at 3 different concentrations (0.5 to 50 μg/mL) during the coculture. Percentage specific lysis was determined in a standard chromium release assay at an effector-to-target ratio of 10:1. Recognition of the autologous B-LCL was blocked by an anti-DR Ab but not by anti-DP or anti–class I Ab. (B) Autologous and allogeneic DR4+ B-LCLs were precoated with peptide PYYVVDLSVRGM (10 μM final) and then exposed to clone 4.19. Clone 4.19 recognized efficiently DR*0401 and DR*0404 B-LCLs loaded with PYY. A DR*0406 B-LCL was a weakly recognized peptide but DR*0402, DR*0405, and DR*0407 B-LCLs were not.
Recognition of the BHRF1 122-133 epitope by the CD4 T-cell clone 4.19 is HLA-DR*0401-restricted. (A) Clone 4.19 cells were cocultured with autologous B-LCL loaded with PYY peptide at 10 μM. Inhibition with anti-DR (L243), anti-DP (B7.21), or anti–class I (W6.32) antibodies was performed by addition of mAb at 3 different concentrations (0.5 to 50 μg/mL) during the coculture. Percentage specific lysis was determined in a standard chromium release assay at an effector-to-target ratio of 10:1. Recognition of the autologous B-LCL was blocked by an anti-DR Ab but not by anti-DP or anti–class I Ab. (B) Autologous and allogeneic DR4+ B-LCLs were precoated with peptide PYYVVDLSVRGM (10 μM final) and then exposed to clone 4.19. Clone 4.19 recognized efficiently DR*0401 and DR*0404 B-LCLs loaded with PYY. A DR*0406 B-LCL was a weakly recognized peptide but DR*0402, DR*0405, and DR*0407 B-LCLs were not.
Staining of BHRF1122-133–specific CD4 T cells with DR*0401/PYY tetramers
DRA1*0101/DRB1*0401 tetramers loaded with BHRF1122-133 peptide were produced and their staining specificity was studied using several CD4+ T-cell clones reactive, or not, against this peptide MHC complex. The BHRF1/DR*0401-specific T-cell clone 4.19 was strongly stained by DRA1*0101/DRB1*0401 tetramers loaded with BHRF1122-133 peptide but not by tetramers loaded with irrelevant peptides (Figure 3A). In contrast, clone 4.4 (from donor RA1), which was not activated by the BHRF1 vaccine, was not stained by the BHRF1122-133 tetramer (Figure 3A). Although the PYY peptide was recognized by clone 4.19 when loaded on HLA-DR*0401+ as well as DR*0404+ or DR*0406+ B-LCLs (Figure 2B), this clone 4.19 was not stained by DRA1*0101/DRB1*0404 tetramers loaded with PYY peptide (not shown).
Staining and sorting of CD4 T cells by DR*0401/BHRF1 multimers. (A) Clone 4.19 (right) was stained by the DR*0401/BHRF1 tetramer (bottom) but not by the irrelevant DR*0401/HA tetramer (top). Another clone from donor RA1, clone 4.4 (left), that did not recognize BHRF1 in the EBV vaccinia assay, was not stained by the DR*0401/BHRF1 tetramer. Cells were double-stained with 20 μg/mL of DR*0401 tetramers and with 10 μg/mL monoclonal antibody against CD4. (B) Yield of sorting of BHRF1-specific cells by using DR*0401/BHRF1 monomers coated on magnetic beads. Synovial CD4 T cells from patient RA1 and CD4 PBLs from healthy donor D3 were stained by DR*0401/BHRF1 tetramers before (left) and after (right) sorting with DR*0401/BHRF1 monomers coated on magnetic beads. Sorted cells were expanded for 3 weeks under polyclonal stimulation before being stained with DR*0401/BHRF1 tetramers. Percentage of cells staining with tetramers is indicated. Bar indicates the range of fluorescence where the cells were scored positive.
Staining and sorting of CD4 T cells by DR*0401/BHRF1 multimers. (A) Clone 4.19 (right) was stained by the DR*0401/BHRF1 tetramer (bottom) but not by the irrelevant DR*0401/HA tetramer (top). Another clone from donor RA1, clone 4.4 (left), that did not recognize BHRF1 in the EBV vaccinia assay, was not stained by the DR*0401/BHRF1 tetramer. Cells were double-stained with 20 μg/mL of DR*0401 tetramers and with 10 μg/mL monoclonal antibody against CD4. (B) Yield of sorting of BHRF1-specific cells by using DR*0401/BHRF1 monomers coated on magnetic beads. Synovial CD4 T cells from patient RA1 and CD4 PBLs from healthy donor D3 were stained by DR*0401/BHRF1 tetramers before (left) and after (right) sorting with DR*0401/BHRF1 monomers coated on magnetic beads. Sorted cells were expanded for 3 weeks under polyclonal stimulation before being stained with DR*0401/BHRF1 tetramers. Percentage of cells staining with tetramers is indicated. Bar indicates the range of fluorescence where the cells were scored positive.
We then tested the ability of the BHRF1122-133 tetramers to detect antigen-specific cells within polyclonal CD4 T-cell lines, either PBL derived or SF derived. About 0.7% of cells in the PBL-derived CD4 T-cell line from the healthy donor D3 were stained by DR4/BHRF1 tetramers, whereas 2.2% of cells in the SF-derived CD4 T-cell line from patient RA1 scored positive (Figure 3B; Table 1). By contrast, the percentage of specific cells was below the detection threshold for other T-cell lines (Table 1). TNF assay also indicated that only some of the CD4 polyclonal T-cell lines released a detectable amount of TNF-α upon stimulation with the PYY peptide (Table 1).
Sorting of BHRF1122-133–specific CD4 T cells by using DR*0401/PYY multimers coated on magnetic beads
To investigate the presence of rare BHRF1122-133–specific T cells within polyclonal cell lines derived from EBV-seropositive HLA-DR4 donors, we performed immunomagnetic sorting using streptavidin beads coated with biotinylated DRB1*0401/BHRF1122-133 monomers (hereafter referred to as multimers), according to a multimer-based approach allowing sorting of CD8 T cells specific to defined HLA-A*0201/peptide complexes.26 After a 2-week culture under polyclonal activation, reactivity of sorted cells to the BHRF1122-133 peptide was tested both by staining with the DRB1*0401/BHRF1 tetramer and by measuring TNF production in a 6-hour self-presentation assay. As attested by TNF production upon stimulation with the PYY peptide, successful sorting of BHRF1-specific cells was achieved for all T-cell lines derived from DRB1*0401 individuals (3 PBL-derived and 6 SF-derived T-cell lines) but for none of the T-cell lines derived from DRB1*0404 or DRB1*0407 donors (5 SF-derived 1 PBL-derived T-cell lines). The frequency of BHRF1-specific cells after multimer sorting was in most cases above 95%, as indicated by staining with the BHRF1 tetramer (Table 1; Figure 3B). The BHRF1 sorting efficiency was confirmed by clonal analysis. Cloning was performed on DR4/BHRF1 sorted cells for 3 donors (RA1, RA14, D3). Thirty clones were isolated from donor RA1, 11 from donor RA14, and 37 from donor D3. All of them released a high level of TNF-α upon incubation with the PYY peptide in an autopresentation assay (not shown). For patient RA6, sorted cells showed significant TNF production upon stimulation with the PYY peptide, although they were not stained by the BHRF1 tetramer. This result probably reflects a low frequency of DR4/BHRF1-specific T cells in this line, as also suggested by the nonsaturating TNF response observed after in vitro exposure of these cells to the PYY peptide (Table 1).
BHRF1122-133–specific CD4 T cells have a restricted repertoire
We assessed the clonal diversity of BHRF1/DR4-specific T-cell lines through analysis of their TCR repertoire. TCR Vβ usage by BHRF1-specific CD4 T cells derived from 6 donors was first studied by flow cytometry using a panel of 22 TCR Vβ–specific mAbs. Each T-cell line used a predominant Vβ region, which differed from one T-cell line to another (Table 2). TCRα and β chain sequencing of several CD4 T-cell clones indicated a highly restricted T-cell repertoire, as most clones derived from a given cell line expressing identical TCRα and β chains (Table 2). Despite the absence of “public” T-cell clones (ie, expressing TCR sequences shared by distinct individuals), several recurrent TCR features were noticed within the 6 distinct clones with fully characterized TCR, such as Jβ1s1 usage (by 5/6 clones), Vα6s1 usage (by 3/6 clones), and Vβ13s1 usage (by 3/6 clones). Some of these clones were selected for further functional studies.
BHRF1122-133–specific CD4 T cells are Th1-like
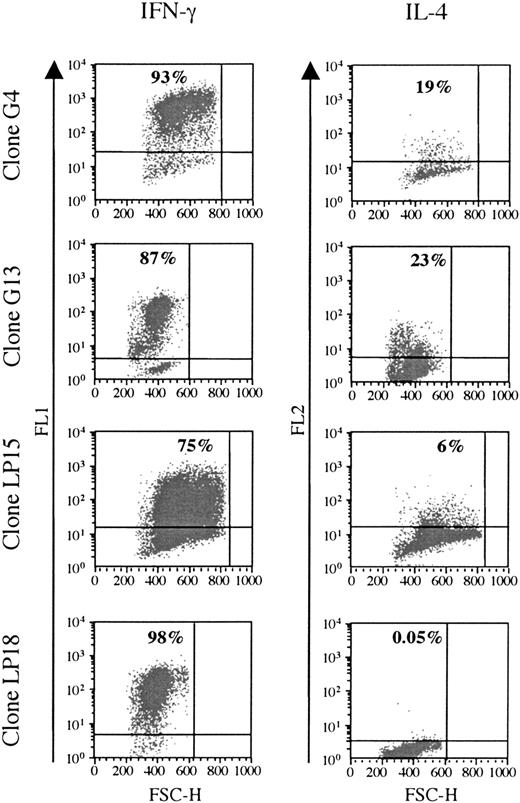
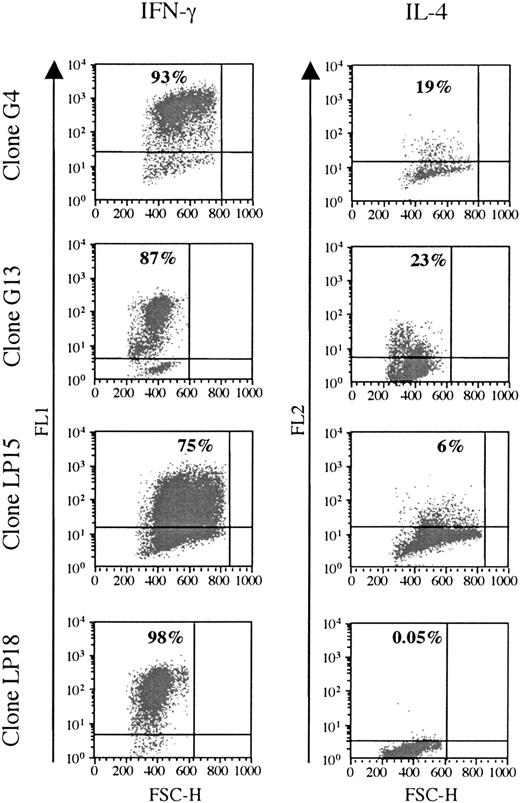
Cytokine production was analyzed in 6 clones responding to BHRF1, under unspecific stimulation with PMA/ionomycin (3 clones from the healthy donor D3: LP15, LP18, LP45; and 3 clones from the patient RA14: G4, G13, G22). All of them were Th1, as they secreted IFN-γ, but only low amounts of IL-4 (Figure 4).
BHRF1-specific CD4 T-cell clones show a Th1-like cytokines profile. Two SF-derived clones (G4 and G14) from patient RA14 and 2 PBL-derived clones (LP15 and LP18) from the healthy donor D3 (LP15 and LP18) were tested for cytokine production. IFN-γ and IL-4 production was detected after stimulation with PMA and ionomycin. The percentage of cytokine-producing cells is indicated in the quadrant. One representative experiment of 3 is shown. FSC-H indicates forward scatter height.
BHRF1-specific CD4 T-cell clones show a Th1-like cytokines profile. Two SF-derived clones (G4 and G14) from patient RA14 and 2 PBL-derived clones (LP15 and LP18) from the healthy donor D3 (LP15 and LP18) were tested for cytokine production. IFN-γ and IL-4 production was detected after stimulation with PMA and ionomycin. The percentage of cytokine-producing cells is indicated in the quadrant. One representative experiment of 3 is shown. FSC-H indicates forward scatter height.
DR*0401-dependent killing of B-LCLs by CD4 T-cell clones specific to BHRF1/DR*0401
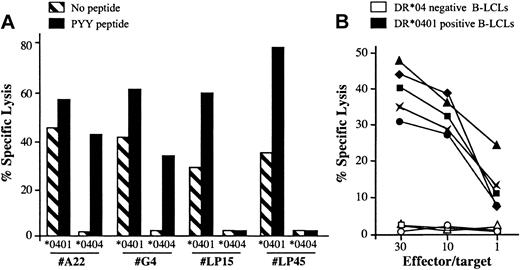
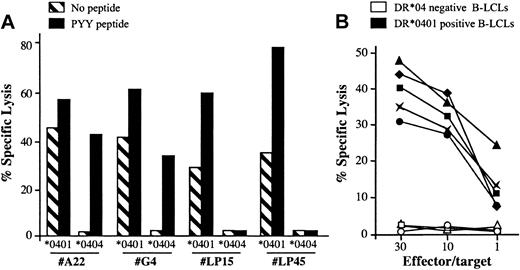
Along the characterization of the BHRF1/DR*0401 epitope, we noticed that the CD4 clone 4.19 lysed unloaded autologous B-LCLs with a 20% to 40% lysis efficiency but failed to recognize DR*0404 or DR*0406 B-LCLs (Figures 1, 2). To determine whether the spontaneous killing of DR*0401+ B-LCLs was a general feature of DR*0401-restricted BHRF1-specific CD4 T-cell clones, 5 clones isolated from 3 DR*0401+ donors were tested against DR*0401 or DR*0404 B-LCLs. Among these clones, 2 were derived from PBLs of the healthy virus carrier D3 (LP15 and LP45, with different Vβ sequences; Table 2) and 3 were SF-derived (clone A22 from patient RA1 and clones G4 and G22 from patient RA14). All clones significantly killed unloaded DR*0401+ B-LCLs. Percentage of specific lysis was slightly increased when the DR*0401 B-LCL was loaded with the PYY peptide at 10 μM (Figure 5A and data not shown). In contrast, these clones did not kill an unloaded DR*0404 B-LCL. The SF-derived, but not the PBL-derived clones, killed DR*0404 B-LCLs loaded with PYY peptide at 10 μM (Figure 5A). This suggested lower affinity of the DR*0401-restricted clones for PYY/DR*0404 than for PYY/DR*0401 complexes. Accordingly while SF-derived clones killed DR*0401 B-LCLs loaded with as low as 1 nM of the PYY peptide, significant killing of DR*0404 B-LCLs was observed after loading with at least 10 μM of this peptide (not shown). Spontaneous killing of DR*0401 B-LCLs by BHRF1-specific clones was confirmed by testing the lytic activity of these 5 clones against a panel of 5 DR*0401+, 2 DR*0404+, and 3 DR4– B-LCLs. All clones killed all the DR*0401+ but not the DR*0404+ nor the DR4– B-LCLs (see representative data obtained with clone A22 in Figure 5B). Altogether these results indicated that all BHRF1/DR4-specific T-cell clones killed autologous B-LCLs in a DR*0401-dependent fashion. Accordingly, lysis of peptide-unloaded autologous B-LCLs was blocked by anti–HLA-DR–specific mAb (data not shown).
DR*0401-dependent killing of B-LCLs by BHRF1-specific CD4 T-cell clones. (A) Two PBL-derived clones from the healthy donor D3 (LP15 and LP45) and 2 SF-derived clones (A22 and G4) killed a DR*0401 B-LCL, either unloaded or loaded with PYY peptide, while they did not kill an unloaded DR*0404 B-LCL. The 2 SF-derived clones, but not the PBL-derived clones, killed the DR*0404 B-LCL after loading with PYY peptide. Results are expressed as percent specific lysis at E/T ratio 10:1. (B) The CD4 T-cell clone A22 was tested against a panel of 5 DR*0401+ (black symbols) and 3 DR4– (open symbols) unloaded B-LCLs. All DR*0401+ B-LCLs were killed, while DR4– B-LCLs were not.
DR*0401-dependent killing of B-LCLs by BHRF1-specific CD4 T-cell clones. (A) Two PBL-derived clones from the healthy donor D3 (LP15 and LP45) and 2 SF-derived clones (A22 and G4) killed a DR*0401 B-LCL, either unloaded or loaded with PYY peptide, while they did not kill an unloaded DR*0404 B-LCL. The 2 SF-derived clones, but not the PBL-derived clones, killed the DR*0404 B-LCL after loading with PYY peptide. Results are expressed as percent specific lysis at E/T ratio 10:1. (B) The CD4 T-cell clone A22 was tested against a panel of 5 DR*0401+ (black symbols) and 3 DR4– (open symbols) unloaded B-LCLs. All DR*0401+ B-LCLs were killed, while DR4– B-LCLs were not.
Killing of B-LCLs by CD4 T-cell clones specific to BHRF1/DR*0401 is EBV dependent
Owing to the low frequency of cells in lytic cycle within a B-LCL (classically below 5%), efficient lysis of peptide-unloaded B-LCL by T-cell clones directed against an EBV lytic epitope was unexpected and led us to further investigate this phenomenon. To investigate whether lysis of DR*0401+ B-LCLs was due to bystander killing rather than through direct recognition of BHRF1 in the DR4 context, we studied the cytotoxicity of BHRF1-specific CTLs toward labeled DR4– B-LCLs mixed with cold DR4+ B-LCLs. None of the mismatched LCLs were killed in the presence of DR4-matched target cells, thus ruling out a mere “bystander” killing effect (not shown).
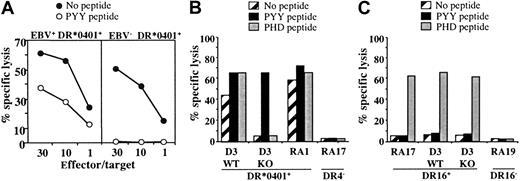
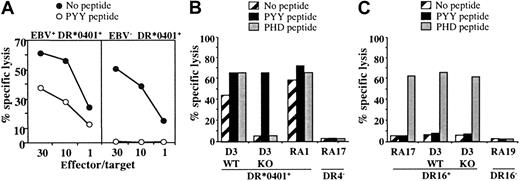
We then tested the capacity of DR*0401-restricted, BHRF1-specific CD4 T-cell clones to recognize, or not, EBV– DR*0401+-presenting cells. As shown in Figure 6A, an EBV– DR*0401+-activated CD8 T-cell clone (which was brightly stained by DR-specific mAb) was not killed by DR4/BHRF1-specific T-cell clones, unless loaded with the relevant peptide (Figure 6A).
Cytolytic activity of the DR*0401-restricted BHRF1-specific CD4 T-cell clone A22 toward DR*0401+ B-LCL is EBV dependent. (A) Cytolytic activity of clone A22 toward the autologous B-LCL (EBV+, DR*0401+) or toward the CD8 T-cell clone A2.10 directed against BMLF1/HLA-A*0201 (EBV–, DR*0401+). Unlike the autologous B-LCL (left), clone A2.10 (right) was not killed by the BHRF1-specific CD4 T-cell clone A.22, unless it was loaded with PYY peptide. (B) The CTL clone A22 recognized the DR*0401+ B-LCL from donor D3 transformed with the wild-type EBV (WT) but not the same B-LCL transformed with a viral mutant with a KO for BHRF1 gene (KO). The 2 B-LCLs were recognized after loading with PYY peptide. The DR*0401+ B-LCL from donor RA1 and the DR4– B-LCL from donor RA17 were included as positive or negative control, respectively. (C) Clone P4.2 (donor RA17), responding to EBNA3C in the HLA-DR16 context, was used as a control to verify that the BHRF1-KO B-LCL from donor D3 was able to present an epitope from the latent protein EBNA3C. Clone P4.2 did not exhibit spontaneous lysis toward DR16+ B-LCL, but it killed its autologous B-LCL, as well as the WT and the BHRF1-KO B-LCLs from donor D3 (DR4+, DR16+), once loaded with the EBNA3C100-111 peptide (PHD). Results are expressed as percent specific lysis.
Cytolytic activity of the DR*0401-restricted BHRF1-specific CD4 T-cell clone A22 toward DR*0401+ B-LCL is EBV dependent. (A) Cytolytic activity of clone A22 toward the autologous B-LCL (EBV+, DR*0401+) or toward the CD8 T-cell clone A2.10 directed against BMLF1/HLA-A*0201 (EBV–, DR*0401+). Unlike the autologous B-LCL (left), clone A2.10 (right) was not killed by the BHRF1-specific CD4 T-cell clone A.22, unless it was loaded with PYY peptide. (B) The CTL clone A22 recognized the DR*0401+ B-LCL from donor D3 transformed with the wild-type EBV (WT) but not the same B-LCL transformed with a viral mutant with a KO for BHRF1 gene (KO). The 2 B-LCLs were recognized after loading with PYY peptide. The DR*0401+ B-LCL from donor RA1 and the DR4– B-LCL from donor RA17 were included as positive or negative control, respectively. (C) Clone P4.2 (donor RA17), responding to EBNA3C in the HLA-DR16 context, was used as a control to verify that the BHRF1-KO B-LCL from donor D3 was able to present an epitope from the latent protein EBNA3C. Clone P4.2 did not exhibit spontaneous lysis toward DR16+ B-LCL, but it killed its autologous B-LCL, as well as the WT and the BHRF1-KO B-LCLs from donor D3 (DR4+, DR16+), once loaded with the EBNA3C100-111 peptide (PHD). Results are expressed as percent specific lysis.
In another series of experiments, we used a DR*0401+ B-LCL transformed with an EBV mutant, in which BHRF1 was disrupted by insertional mutagenesis (BHRF1-KO).24 While BHRF1-specific T-cell clones efficiently recognized (both in terms of cytolysis and TNF production) DR*0401+ B-LCLs transformed with wild-type EBV, they neither killed nor produced TNF (data not shown) when incubated with the BHRF1-KO B-LCL from the same donor. The experiment was done twice with 3 BHRF1-specific clones originating from 3 different donors. A representative experiment is shown in Figure 6B. In contrast, the BHRF1-KO B-LCL (DR4+, DR16+) could activate a CD4 T-cell clone from donor RA17 (clone P4.2) that recognizes the EBV latent protein EBNA3C in the HLA-DR16 context, thus indicating efficient EBV infection of the BHRF1-KO B-LCL. As shown in Figure 6C, this clone did not exhibit spontaneous lysis toward DR16+ B-LCLs, but it killed DR16+ B-LCLs once loaded with the EBNA3C100-111 peptide. The BHRF1-KO B-LCL was killed, as well as the autologous B-LCL from donor RA17. Together with the HLA-DR4–dependent killing of autologous B-LCLs (Figure 5), these results strongly suggest that the spontaneous killing of autologous B-LCLs by DR4/BHRF1-specific CTLs is linked to recognition of an HLA-DR–restricted BHRF1 epitope.
Discussion
Because of the importance of CD4+ T cells in antiviral immunity, much effort is now directed toward identifying MHC class II–restricted viral antigens. The data presented in this paper uncover a CD4 T-cell response to the early lytic EBV protein BHRF1 in the HLA-DR*0401 context (epitope BHRF1122-133). We show that the response to BHRF1/DR*0401 is recurrent as it was found in all T-cell lines tested (9 at the total). This response was found in peripheral blood of healthy EBV carriers and elevated in synovial fluid of arthritis patients. DRB1*0401/BHRF1122-133 tetramers stained a small number of cells in only 4 DR*0401+ T-cell lines out of the 9 that were tested. It should be underlined that in contrast to most, if not all, previous studies aiming at isolating antiviral or antitumoral CD4 T cells, we did not perform any in vitro antigen (Ag) stimulation prior to testing. BHRF1-specific cells were efficiently sorted out through immunomagnetic sorting, using DRB1*0401/BHRF1122-133 monomers coated on magnetic beads, and these cells could be rapidly expanded under nonspecific stimulation. The expanded CD4 T cells were nearly 100% specific to BHRF1, in 4 of 5 cases, thus underlining the efficiency of this sorting approach with DR4 multimers.
The DR*0401-restricted BHRF1-specific clones, described in this study, exhibit a strict specificity for DR*0401. BHRF1/DR*0401-specific clones were not stained by BHRF1/DR*0404 tetramers and they did not kill DR*0404 B-LCLs. This is in accordance with previous data indicating that although DRA1*0101/DRB1*0401 and DRA1*0101/DRB1*0404 are structurally very similar, the limited polymorphism between these alleles at codons 86 and 71 of the DRB1 chain may dictate unique binding patterns.28
The response to BHRF1 appears to be not specifically linked to arthritis, as this CTL response was also found in peripheral blood of healthy EBV carriers. A number of synovial-derived CD4 T-cell lines were included in our screening on the basis of our previous observation that synovial fluid–derived T-cell lines were enriched for CD8 T cells responding to EBV antigens in the synovial fluid of RA patients.11 As a greater number of Ag-specific CD4+ T cells are expected to be found in the tissues affected by autoimmune diseases or in an infection, we used these synovial fluid–derived T-cell lines to screen for CD4 T-cell responses to EBV. In fact, one of the difficulties in analyzing CD4 T-cell responses is inherent to the rare occurrence of memory CD4 T cells. CD4+ T cells of a given antigen specificity seem to be less abundant than their CD8+ counterparts. This may be because peptides from some antigens are generated at low efficiency, which results in low density on antigen-presenting cells, or it might reflect localized tissue-specific expression. In this regard, differential regulation of antiviral T-cell immunity results in stable CD8+, but declining CD4+ T-cell memory.29 Thus, the frequency of virus-specific CD4 T cells is at least 35-fold lower than CD8 cells during acute and memory phases of lymphocytic choriomeningitis virus infection.30 This explains why the few published studies on human class II–restricted responses using HLA-peptide tetramers required prior in vitro antigenic selection to detect CD4 T cells specific for an influenza or herpes simplex virus 2 (HSV-2) epitope in chronic virus carriers.23,31 This also explains why until recently, the CD4 T-cell response to EBV has been poorly characterized.
The specificity of a few CD4 T-cell clones reacting to lytic or latent proteins has been described previously.16-19 Moreover, a systematic analysis of CD4 T-cell responses to latent antigens was performed by 2 different groups, who reported highly recurrent responses to EBNA1 and EBNA3C epitopes in most donors, though at a very low frequency.20,21 The magnitude of responses specific to EBNA1-derived CD4 epitopes ranged from 60 to 350 IFN-γ–producing cells per 106 CD8-depleted PBMCs,21 but these responses could be efficiently reactivated with autologous dendritic cells transduced with EBNA1-vaccinia constructs.20 The response to BHRF1 described in this paper was found, at least in 2 donors, at rather high frequencies (0.2% of the CD4 T cells among the PBLs of an healthy virus carrier and 2.2% of the CD4 synovial T cells of an arthritic patient). A recent analysis of CD4+ and CD8+ T-cell responses during primary infection showed that lytic and latent EBV protein–specific CD4+ T cells were readily detected at presentation with acute infectious mononucleosis and declined rapidly thereafter.32
BHRF1-specific clones, be they PBL- or SF-derived, spontaneously killed autologous or HLA-matched B-LCLs (ie, without further addition of the BHRF1 peptide). Preliminary data indicate that these clones express perforin and granzyme A. Some,22 but not all,17 EBNA1-specific CD4 T-cell clones have also been shown to kill the autologous B-LCLs. However, while EBNA-1 is expressed during latency, BHRF1 is supposed to be expressed during the lytic cycle only. The consistent killing of B-LCLs by BHRF1-specific CD4 T-cell clones was rather unexpected. In fact, the percentage of spontaneous killing (30%-60%) far exceeds the small percentage of B cells, within a B-LCL, that enter lytic infection (eg, 5%). The underlying mechanism of the spontaneous killing is clearly beyond the scope of the present study and will need further investigation. One mechanism could be that most cells take up BHRF1 or its fragments from the medium after its release from a few lytic cells. Another possibility is that a small amount of BHRF1 is expressed during latent infection and presented directly. While it is clear that BHRF1 is expressed at high levels during early lytic infection, its expression during latency remains a controversial issue. Latently spliced BHRF1 cDNAs have been reported,33 but attempts to detect BHRF1 protein expression in latently infected cells have failed.1 Nevertheless, the lack of recognition of BHRF1-KO B-LCLs by BHRF1-specific CD4 T-cell clones demonstrates that the spontaneous killing of autologous B-LCLs by DR4/BHRF1-specific CTLs is linked to direct recognition of an epitope of the lytic protein BHRF1. DR2-restricted BHRF1-specific CD4 T-cell clones had previously been isolated from patients with infectious mononucleosis and it was shown that they killed the autologous B-LCLs.19
These BHRF1-specific cells showed a Th1 cytokine profile, consistent with their cytotoxic potential. Therefore they could directly take part in the control of EBV replication. Along this line, the importance of CD4 T cells in the resistance to persistent viral infections is supported by several observations.34-38 Adoptively transferred CD8 CMV-specific T-cell clones do not persist in the long-term without endogenous recovery of CD4 T cells.35 In HIV infection, a minority of patients are long-term nonprogressors and this correlates with high CD4 T-cell counts and vigorous CD4 T-cell proliferative responses to HIV proteins.36 Similar observations were made in a mouse model of lymphocytic choriomeningitis virus.37,38 CD4 T cells are believed to be instrumental in the initiation of CD8 T-cell expansion via stimulation of dendritic cells,39,40 but their role in maintaining adequate numbers and function of specific CD8 T cells is less understood. Zajac and colleagues38 observed that in CD4–/– mice, effector functions rather than frequency of lymphocytic choriomeningitis virus–specific CD8 T cells were impaired.
In summary, the data presented here show that it is possible to efficiently sort out and expand CD4 T cells specific to defined epitopes. Such peptide/MHC class II multimer sorting strategy could be of potential therapeutic interest for the immunotherapy of viral infections or cancers.
Prepublished online as Blood First Edition Paper, October 16, 2003; DOI 10.1182/blood-2003-03-0930.
Supported by grants from the Association pour la Recherche sur la Polyarthrite (ARP), Ligue Nationale Contre le Cancer (LNCC), Centre Hospitalier Universitaire de Nantes, European Community (grant QLK2-CT-2001-01205) and by institutional grants from INSERM.
E.L. and X.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs A. Rickinson and M. Kurilla for the gift of recombinant vaccinia constructs. We also thank Dr H. Vié for critically reading the manuscript.

![Figure 1. Identification of a DR*0401/BHRF1 epitope recognized by the CD4 T-cell clone 4.19. (A) Recognition of the lytic EBV protein BHRF1 by clone 4.19. CTL cells were tested in a 4-hour TNF-α release assay on autologous B-LCLs expressing individual latent or lytic EBV proteins from vaccinial viral vectors. Results are shown as μg/mL. TNF release was observed at an effector-target ratio of 10:1. (B) Identification of the target epitope of BHRF1-specific clone 4.19 using a panel of peptides (23-mers, overlapping by 12 amino acids [aa's]) spanning the BHRF1 protein. These peptides were used as targets in cytotoxic assays by incubating them with 51Cr-labeled autologous EBV-B cells for 1 hour at 37°C. Clone 4.19 was added at an E/T ratio of 10:1 and Cr release was measured after 4 hours. Note the significant lysis background observed with unloaded autologous B-LCL (No pep). (C) Stimulation of clone 4.19 by the BHRF1 122-133 peptide (PYY). Cytotoxicity of clone 4.19 to Cr-labeled autologous EBV-B cells loaded with various concentrations of peptides. Clone 4.19 was added at an E/T ratio of 10:1 and Cr release was measured after 4 hours. Data obtained with the 23-mer BHRF1111-133 peptide are shown as a positive control. Data obtained with an irrelevant peptide (irr peptide) are shown as a negative control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-03-0930/6/m_zh80040456650001.jpeg?Expires=1769432236&Signature=R-Qgy0F2N~tI8z27tsJxEd3BqUJgWY3Od40GE1p59zyrFVNO15o07YX32mNq-O~YHCS1ZhVCzQYed4EKhOA6gu3ZT4uwf7NIC0XFmnqUDETq7YadTIDN8-znLLn4qG1IM3aGKPoKWjRFLDijiTZClEuIXOyCR8VdTkJDkaIDzAIdokQda90rQxY4GxRifTE48GWZXDh7xb8jSQo7B2xwg4K0Aolhe4J9BbnSKqAsmUB15jZVB2UYeG4d52T-Ja7Gr7iauDDAEXVo141SpX6GyyULPjRkK4Wx9hkvM~IFMJZoLL4qi8VW09YRQFEVd1IGtd4AJFUt5O5hPVoAN46tvA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)