Abstract
We used synthetic peptides to the extracellular loops (ECLs) of CCR5 to examine inhibitory effects on HIV infection/fusion with primary leukocytes and cells expressing recombinant CCR5. We show for the first time that peptides derived from the first, second, or third ECL caused dose-dependent inhibition of fusion and infection, although with varying potencies and specificities for envelope glycoproteins (Envs) from different strains. The first and third ECL peptides inhibited Envs from the R5 Ba-L strain and the R5X4 89.6 strain, whereas the second ECL peptide inhibited Ba-L but not 89.6 Env. None of the peptides affected fusion mediated by Env from the X4 LAV strain. Fusion mediated by Envs from several primary HIV-1 isolates was also inhibited by the peptides. These findings suggest that various HIV-1 strains use CCR5 domains in different ways. Experiments involving peptide pretreatment and washing, modulation of the expression levels of Env and CCR5, analysis of CCR5 peptide effects against different coreceptors, and inhibition of radiolabeled glycoprotein (gp) 120 binding to CCR5 suggested that the peptide-blocking activities reflect their interactions with gp120. The CCR5-derived ECL peptides thus provide a useful approach to analyze structure–function relationships involved in HIV-1 Env-coreceptor interactions and may have implications for the design of drugs that inhibit HIV infection.
Introduction
Infection by HIV-1 requires surface expression of 2 specific receptors on the target cell: CD4 (the primary receptor) and a specific chemokine receptor (the coreceptor). In the generally accepted model for HIV-1 entry (for reviews, see Wyatt and Sodroski1 and Eckert and Kim2 ), the external glycoprotein (gp) 120 subunit of the HIV-1 envelope glycoprotein (Env) initiates infection through specific binding to the first domain of CD4. The CD4 interaction induces a conformational change that exposes a highly conserved coreceptor binding site on gp120, which, along with variable loops on gp120, enables gp120 to bind to the coreceptor. The gp120-coreceptor interaction then triggers conformational changes in the gp41 transmembrane subunit of Env, presumably leading to exposure of the N-terminal fusion peptide and its insertion into the plasma membrane of the target cell to initiate the membrane fusion process.
Individual HIV-1 isolates can display markedly distinct phenotypes for infection of different CD4-expressing cell types such as primary macrophages, transformed T-cell lines, or primary T cells. Target cell tropism is determined primarily by the ability of the corresponding Env to interact with one or both of the major coreceptors (CCR5, CXCR4), coupled with the expression patterns of these receptor molecules on the different target cell types (for a review, see Berger et al3 ). HIV-1 isolates capable of using CCR5 but not CXCR4 (designated R5)4-8 are typically macrophage tropic and greatly predominate after acute infection and throughout the asymptomatic phase. Isolates able to use CXCR49 are first detected during the transition to the symptomatic phase; those that function with CXCR4 but not CCR5 (designated X4) are typically T-cell line tropic, whereas those able to function with both coreceptors (designated R5X4) are dual tropic. All these HIV-1 phenotypes readily infected activated primary CD4+ T cells. The gp120/corecptor interaction has become a major focus for the development of new anti-HIV agents for treating or preventing HIV-1 infection (for reviews, see Agrawal and Alkhatib10 and Biscone et al11 ).
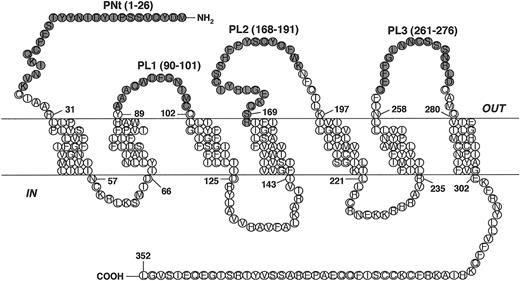
The coreceptors are members of the superfamily of G-protein–coupled receptors (GPCRs). These proteins are characterized by 7 transmembrane segments that form a helical bundle, an extracellular region that includes the N-terminal domain and 3 extracellular loops (ECLs), and an intracellular region consisting of 3 intracellular loops and the C-terminus (Figure 1 and Table 1). Identifying the coreceptor regions involved in the gp120 interaction is critical for elucidating the molecular details of the HIV-1 entry mechanism. Approaches to define such regions for CCR5 and CXCR4 have included analyzing coreceptor variants (site-directed mutants and hybrid constructs containing exchanged segments of related chemokine receptors), identifying the sites of interaction of specific coreceptor-binding agents such as chemokine ligands, monoclonal antibodies, and low-molecular-weight inhibitors, and analyzing the effects of synthetic peptides representing specific coreceptor domains (for a review, see Dragic12 ). Although there are some discrepancies between studies from different laboratories, the results indicate that multiple CCR5 regions are involved in coreceptor function and that these may vary for different HIV-1 strains. In particular, critical roles have been noted for the CCR5 N-terminus and the second ECL in HIV-1 coreceptor function.
CCR5-derived peptides. Schematic diagram illustrating the positions of the CCR5-derived peptides. The peptides, designated PNt, PL1, PL2, and PL3, correspond to portions of the predicted 4 extracellular domains of CCR5.
CCR5-derived peptides. Schematic diagram illustrating the positions of the CCR5-derived peptides. The peptides, designated PNt, PL1, PL2, and PL3, correspond to portions of the predicted 4 extracellular domains of CCR5.
Studies with synthetic peptides corresponding to the predicted extracellular N-terminal domain have yielded particularly valuable structure–function insights into gp120 interaction with CCR5. Consistent with findings that the sulfation of tyrosine residues in this region contributes to HIV coreceptor activity and gp120 binding,13 tyrosine-sulfated peptides derived from the N-terminal domain (but not their nonsulfated counterparts) have been shown to interact specifically with gp120 and to inhibit HIV-1 entry.14,15 Mapping studies have delineated CCR5 residues 10 to 18, containing 2 sulfotyrosines, as a minimal domain required for gp120 interaction.16
In the present study, we have extended the synthetic peptide mapping approach to the 3 ECLs of CCR5. Peptides corresponding to portions of these domains were analyzed for their effects on CCR5 coreceptor function in peripheral blood mononuclear cells (PBMCs), macrophages, and cell lines expressing recombinant CCR5. Our results provide independent evidence that multiple ECLs of CCR5 are involved in coreceptor function and that these domains vary for different Envs. Moreover, our evidence suggests that peptide-blocking effects are mediated by direct interaction with Env.
Materials and methods
Cells and culture conditions
All cell lines were obtained from the American Type Culture Collection (Rockville, MD). The NIH 3T3, NIH 3T3.T4, HeLa, and HeLa.T4 cell lines were maintained in Dulbecco modified Eagle medium (DMEM; Quality Biologicals, Gaithersburg, MD) containing 10% fetal bovine serum (FBS) (Hyclone Laboratories, Logan, UT), 2 mM l-glutamine, and antibiotics.
Human PBMCs were prepared by Ficoll-Hypaque fractionation of cell concentrates obtained from healthy donors seronegative for antibodies against hepatitis B virus and HIV. PBMCs were activated with 10 μg/mL phytohemagglutinin and interleukin-2 (IL-2) for 3 days in RPMI containing 10% FBS. Primary macrophages were prepared using countercurrent centrifugation elutriation of PBMCs and differentiation of the monocyte fraction17 ; CD4 levels were augmented by infection with a recombinant vaccinia virus (see below).
Synthetic peptides
Cell fusion assay and recombinant protein expression
HIV Env-mediated cell fusion was quantitated by a previously described vaccinia-based reporter gene assay fusion.20 Effector cells expressed recombinant Env, and target cells expressed recombinant or endogenous CD4 or coreceptors; in addition, one cell population was infected with vaccinia recombinant vTF7-3 encoding bacteriophage T7 RNA polymerase,21 and the other population was infected with vCB-21R containing the Escherichia coli LacZ reporter gene linked to the T7 promoter.22 Effector and target cells were mixed in 96-well plates and incubated at 37°C for 2 hours. β-Galactosidase was produced selectively in fused cells and was quantitated in nonionic detergent cell lysates using a 96-well spectrophotometer.
Recombinant HIV-1 Envs and their receptors were produced using vaccinia virus expression technology. Vaccinia virus infections were performed at a multiplicity of infection (MOI) of 10 for each virus, and plasmid transfections were performed using DOTAP (Roche Applied Science, Indianapolis, IN). Infected cells were incubated overnight at 31°C to allow the expression of recombinant proteins and were washed before mixing for fusion. Specific details for each experiment are given in the figure legends. Envs from laboratory-adapted strains were expressed using vaccinia recombinants containing a strong vaccinia promoter—vCB-43, Ba-L; vCB-41, LAV; vSC-60, IIIB; vCB-16, Unc.23 Alternatively, where indicated, Envs were expressed by transfection with plasmids containing the Env genes linked to the same vaccinia promoter.24 Envs from primary strains were expressed by transfection with plasmids containing the Env genes linked to the T7 promoter.25 CD4 was expressed by infection with vaccinia recombinant vCB-3.26 Recombinant coreceptors were expressed by transfection with plasmids containing the genes linked to the strong vaccinia promoter24,27 ; alternatively, CCR5 was expressed using vaccinia recombinant vHC-1.28 In some experiments, recombinant CD4 was endogenously expressed on the transfected cell line HeLa.T429 or NIH 3T3.T4.30
Productive HIV-1 infection assays
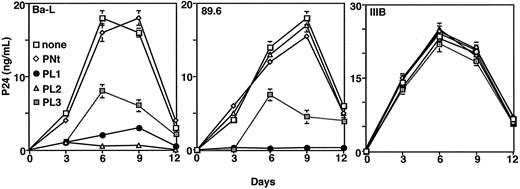
PBMCs were stimulated with phytohemagglutinin (PHA) plus recombinant IL-2 for 3 days. Infections were performed in 96-well plates (105 cells/well in 200 μL). Peptides (10 μg/mL) were preincubated with the viral inoculum (89.6, 105 infectious units (IU)/mL; Ba-L, 105.5 IU/mL; IIIB, 104.5 IU/mL) for 1 hour at 37°C. Each virus/peptide mix was added to 105 activated PBMCs, and virus adsorption was allowed to proceed for 2 hours. The cells were washed 3 times with RPMI containing 10% FBS and was resuspended in 200 μL complete medium containing the desired peptide or RANTES (regulated on activation, normal T expressed and secreted). Cell-containing aliquots were removed every 3 days and replaced with fresh medium containing fresh peptides. Levels of p24 in the culture supernatants were measured over the course of 12 days using a p24 enzyme-linked immunosorbent assay (ELISA) kit purchased from National Cancer Institute–Frederick Cancer Research and Development Center (Frederick, MD). Results are presented as the mean of duplicate samples, with error bars indicating standard deviations.
Binding assays
Soluble gp120 Env from JR-FL, 89.6, and IIIB strains were obtained from Dr Chris Broder (Uniformed Services University, Bethesda, MD). Samples (5 μg) of gp120 were labeled with sodium iodide 125I Bolton-Hunter reagent (ICN, Costa Mesa, CA). NIH-3T3 cells were infected with vHC-1 (encoding CCR5) and used as targets for [I125]-labeled gp120 binding. Radiolabeled gp120 was preincubated with soluble CD4 (sCD4) at 37°C for 30 minutes to activate Env for CCR5 binding. The activated [I125]-labeled gp120 was further incubated with either PNt or PL2 for 45 minutes at 37°C before mixing with CCR5-expressing NIH 3T3 cells. After incubation, the cells were washed 3 times with binding buffer containing 0.5 M sodium chloride, and cell pellet-associated counts were measured in a gamma counter.
Results
CCR5 ECL peptide inhibition of HIV-1 Env-mediated cell fusion
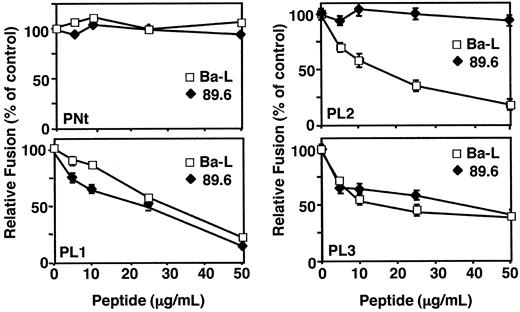
We analyzed the effects of peptides corresponding to the extracellular regions of CCR5 (Figure 1) on fusion between effector cells expressing HIV-1 Env and target cells expressing CD4 and CCR5. Figure 2 shows the dose-response analyses for each of the peptides tested against Env from the Ba-L (R5) or the 89.6 (R5X4) strain, using NIH 3T3 target cells expressing recombinant CD4 and CCR5. Peptide PNt, derived from the CCR5 amino terminus, served as a negative control because peptides from this region lacking sulfated tyrosine residues have been shown to have little effect on the Env/CCR5 functional interaction with CCR5.13,14 As expected, PNt had no effect on fusion mediated by either the Ba-L or the 89.6 Env. By contrast, the PL1, PL2, and PL3 peptides (based on the first, second, and third ECLs of CCR5, respectively) showed significant fusion-blocking activities. These activities varied between the 2 Envs: peptides PL1 and PL3 blocked fusion comparably for both, whereas peptide PL2 potently inhibited Ba-L but not 89.6. When used at suboptimal concentrations, the combination of the 3 PL peptides had an enhanced blocking effect compared with each peptide alone. For example, treatment with PL1+PL2+PL3 at 15 μg/mL for each peptide caused more than 90% inhibition of Ba-L fusion, whereas treatment with each of these peptides alone gave 30% for PL1, 50% for PL2, and 35% for PL3.
Specific Inhibition of HIV-1 Env-mediated cell fusion by the CCR5-derived peptides. NIH-3T3 cells expressing vaccinia-encoded CD4 (vCB-3), transfected CCR5, and T7 RNA polymerase (vTF7-3) were used as target cells. Peptides were added to the target cells at the indicated concentrations and incubated for 1 hour at 37°C. For effector cells, HeLa cells were infected with the vaccinia recombinant containing the T7 promoter-lacZ reporter gene (vCB-21R-lacZ) and infected with vaccinia virus encoding Ba-L (vCB-43). For 89.6 Env expression on HeLa cells, the cells were transfected with pSC59/89.6 and then infected with vCB21R-lacZ. The fusion assay was performed as described in “Materials and methods.” β-Galactosidase values (10–3 OD/min) were: Unc, 0.5; Ba-L, 8.5; 89.6 Env, 7.5. Error bars indicate SD of mean values obtained from duplicate fusion assays.
Specific Inhibition of HIV-1 Env-mediated cell fusion by the CCR5-derived peptides. NIH-3T3 cells expressing vaccinia-encoded CD4 (vCB-3), transfected CCR5, and T7 RNA polymerase (vTF7-3) were used as target cells. Peptides were added to the target cells at the indicated concentrations and incubated for 1 hour at 37°C. For effector cells, HeLa cells were infected with the vaccinia recombinant containing the T7 promoter-lacZ reporter gene (vCB-21R-lacZ) and infected with vaccinia virus encoding Ba-L (vCB-43). For 89.6 Env expression on HeLa cells, the cells were transfected with pSC59/89.6 and then infected with vCB21R-lacZ. The fusion assay was performed as described in “Materials and methods.” β-Galactosidase values (10–3 OD/min) were: Unc, 0.5; Ba-L, 8.5; 89.6 Env, 7.5. Error bars indicate SD of mean values obtained from duplicate fusion assays.
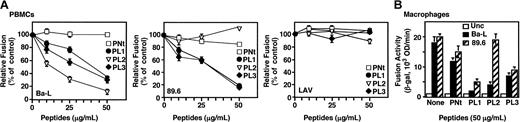
We extended the analysis of the peptide effects on fusion with primary target cell types, including PHA-activated PBMCs and monocyte-derived macrophages (Figure 3). Because activated PBMCs express both CCR5 and CXCR4, these cells enabled us to test the specificities of the peptides against R5, R5X4, and X4 Envs (Figure 3A). The dose-response effects for the Ba-L (R5) and 89.6 (R5X4) Envs paralleled the findings above with target cells expressing recombinant CCR5. Thus for Ba-L, the control PNt peptide had no effect, whereas PL1, PL2, and PL3 inhibited (with PL2 showing the most potent activity); for 89.6, inhibition was observed with PL1 and PL3, but not with PNt or PL2. Different results were obtained with the LAV (X4) Env; none of the peptides inhibited fusion. Thus the CCR5-derived peptides inhibited only those Envs capable of using that coreceptor. The peptides (at 50 μg/mL) were also examined for their effects on fusion with primary macrophage target cells, mediated by the Envs capable of using CCR5 (Figure 3B). Results were similar to those obtained with CCR5-transfected and PMBC target cells. Thus the PNt peptide had minimal effect on the Ba-L and 89.6 Envs; the PL1 and PL3 peptides showed comparable inhibition of both Envs, and the PL2 peptide inhibited Ba-L but not 89.6.
Specific peptide inhibition of HIV-1 Env-mediated fusion with primary target cell types. Designated primary target cells infected with vCB-21R-LacZ were preincubated with peptides at the indicated concentrations for 1 hour at 37°C. Target cells were then mixed with HeLa effector cells coinfected with vTF7-3 and vCB43 (Ba-L) or vCB-41 (LAV). Effector 89.6 Env-expressing HeLa cells were prepared by transfection of pSC59/89.6 and infection with vTF7-3. Fusion assay was performed as described in “Materials and methods.” (A) PBMC targets. (B) Primary macrophage targets. Error bars indicate SD of mean values obtained from duplicate fusion assays.
Specific peptide inhibition of HIV-1 Env-mediated fusion with primary target cell types. Designated primary target cells infected with vCB-21R-LacZ were preincubated with peptides at the indicated concentrations for 1 hour at 37°C. Target cells were then mixed with HeLa effector cells coinfected with vTF7-3 and vCB43 (Ba-L) or vCB-41 (LAV). Effector 89.6 Env-expressing HeLa cells were prepared by transfection of pSC59/89.6 and infection with vTF7-3. Fusion assay was performed as described in “Materials and methods.” (A) PBMC targets. (B) Primary macrophage targets. Error bars indicate SD of mean values obtained from duplicate fusion assays.
Potent activity of CCR5 ECL peptide against Envs from primary HIV-1 isolates
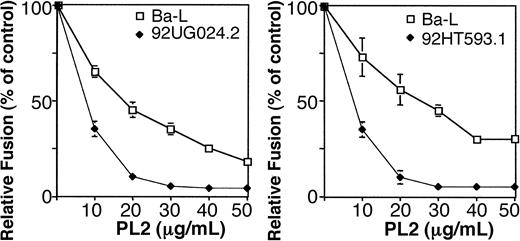
The strains analyzed have all been adapted to replication in the laboratory (PBMCs for Ba-L and 89.6; continuous T-cell lines for LAV). We wanted to extend the analyses to Envs derived from primary isolates. Effector cells expressing the Env from Ba-L or the primary isolates 92HT593.1 and 92UG024.2 (both previously shown to use CCR5 in the cell fusion assay27 ) were tested for fusion with target cells expressing recombinant CD4 and CCR5, in the absence or presence of the PL2 peptide. As shown in Figure 4, the primary Envs were highly sensitive to inhibition. We concluded that peptide inhibition was not restricted to laboratory-adapted isolates and was not confined to a particular genetic subtype because Ba-L, 89.6, and 92HT593.1 are clade B, and 92UG024.2 is clade D.
Activity of CCR5 ECL peptides on Envs from primary HIV-1 isolates. Effector cells (HeLa cells transfected with plasmids containing the designated primary Envs linked to the T7 promoter and infected with vTF7-3) were preincubated for 1 hour at 37°C with the P3 peptide at the indicated concentrations. Target cells were prepared by coinfecting NIH-3T3.T4 cells with the vaccinia recombinants encoding CCR5 (vHC-1) plus vCB21R-LacZ. Fusion assay was performed as described in “Materials and methods.” β-Galactosidase values (10–3 OD/min) were: Ba-L, 9.5; 92UG024.2, 7.5; 92HT93.1, 9.0. Error bars indicate SD of mean values obtained from duplicate fusion assays.
Activity of CCR5 ECL peptides on Envs from primary HIV-1 isolates. Effector cells (HeLa cells transfected with plasmids containing the designated primary Envs linked to the T7 promoter and infected with vTF7-3) were preincubated for 1 hour at 37°C with the P3 peptide at the indicated concentrations. Target cells were prepared by coinfecting NIH-3T3.T4 cells with the vaccinia recombinants encoding CCR5 (vHC-1) plus vCB21R-LacZ. Fusion assay was performed as described in “Materials and methods.” β-Galactosidase values (10–3 OD/min) were: Ba-L, 9.5; 92UG024.2, 7.5; 92HT93.1, 9.0. Error bars indicate SD of mean values obtained from duplicate fusion assays.
Inhibition of HIV-1 productive infection by CCR5 ECL peptides
To extend the peptide inhibition studies beyond Env-mediated cell fusion, we examined their effects on HIV-1 productive infection. The peptides (10 μg/mL) were preincubated with the virus preparation before they were added to activated PBMCs, and measuring p24 production followed the time course of infection. As shown in Figure 5, the Ba-L isolate was unaffected by PNt but was inhibited by all ECL peptides, with PL2 showing the greatest potency; the 89.6 isolate was inhibited by PL1 and PL3, but not by PNt or PL2; the IIIB strain (X4, closely related to LAV) was not inhibited by any of the peptides. We concluded that the general patterns of infectivity inhibition paralleled the fusion inhibition results described.
Effects of CCR5 ECL peptides on HIV-1 productive infection. Indicated HIV-1 isolates were pretreated with the designated peptides (10 μg/mL) and then adsorbed to activated PBMCs. Cells were washed, plated, and analyzed for p24 production over time, as described in “Materials and methods.” Error bars indicate SD of mean values obtained from duplicate infection assays.
Effects of CCR5 ECL peptides on HIV-1 productive infection. Indicated HIV-1 isolates were pretreated with the designated peptides (10 μg/mL) and then adsorbed to activated PBMCs. Cells were washed, plated, and analyzed for p24 production over time, as described in “Materials and methods.” Error bars indicate SD of mean values obtained from duplicate infection assays.
Comparison of the effects of peptide pretreatment of Env-expressing versus CCR5-expressing cells
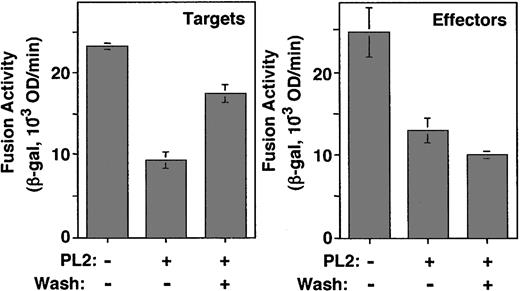
We wanted to determine whether the peptide inhibitory effects reflected their interactions with the Env-expressing or the CCR5-expressing cells. In one approach, we compared the peptide fusion-blocking activities in a protocol involving pretreating the effector versus the target cells, washing away excess peptide, then measuring fusion. Because of the potent inhibitory activities described for PL2 against Ba-L, we focused on the effects of this peptide/Env combination. In the experiment shown in Figure 6, the PL2 peptide was added to either cell type at 25 μg/mL, a concentration deliberately chosen to be suboptimal for fusion inhibition. The treated cells were then mixed with the untreated partner cells, either in the continued presence of the peptide or after washing the pretreated cells to remove excess peptide. Results show that washing the pretreated target cells resulted in a significant reversal of the blocking effect (Figure 6, top). By contrast, washing the pretreated Env-expressing cells did not reverse the blocking activity (Figure 6, bottom). These results suggest that the blocking activity reflected peptide interaction with Env, which persisted after washing.
Comparison of the effects of peptide pretreatment of Env-expressing versus CCR5-expressing cells. (left) Target cells (HeLa.T4 infected with vTF7-3 and transfected with the CCR5 plasmid) or (right) effector cells (HeLa cells coinfected with vCB-21R-LacZ and vCB-43) were preincubated in the absence or presence of the PL2 peptide (25 μg/mL). After incubation for 1 hour at 37°C, the cells were mixed with their fusion partners without or with washing to remove excess peptide. Fusion assay was performed as described in “Materials and methods.” Error bars indicate SD of mean values obtained from duplicate fusion assays.
Comparison of the effects of peptide pretreatment of Env-expressing versus CCR5-expressing cells. (left) Target cells (HeLa.T4 infected with vTF7-3 and transfected with the CCR5 plasmid) or (right) effector cells (HeLa cells coinfected with vCB-21R-LacZ and vCB-43) were preincubated in the absence or presence of the PL2 peptide (25 μg/mL). After incubation for 1 hour at 37°C, the cells were mixed with their fusion partners without or with washing to remove excess peptide. Fusion assay was performed as described in “Materials and methods.” Error bars indicate SD of mean values obtained from duplicate fusion assays.
Comparison of the effects of modulating Env versus coreceptor expression levels on the efficiency of peptide blocking
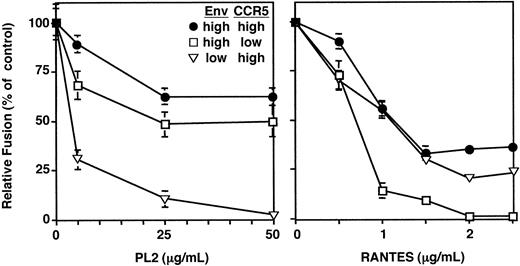
To further test the hypothesis that the blocking activity reflects peptide interaction with Env, experiments were performed comparing the effects of modulating the expression levels of Env versus CCR5 on inhibitory efficacy. In the vaccinia expression system, this can be achieved by introducing the gene of interest by recombinant vaccinia virus infection (high expression) or by plasmid transfection (low expression). Each method results in uniform expression levels within the cell population: with infection, a high MOI ensures infection of the entire cell population; with transfection, the plasmid need only be introduced into the cytoplasm, thereby giving rise to high transfection efficiencies. Cell surface staining analyses (data not shown) indicated a 10-fold higher expression level for infection compared with transfection for Ba-L Env (expressed in HeLa cells) and CCR5 (expressed in HeLa-T4 cells constitutively expressing recombinant CD4). In the experiment shown in Figure 7, the effects of inhibitors on fusion reactions with high levels of Ba-L Env and CCR5 were compared with the effects on reactions with reduced levels of either Env or CCR5. The activities of the PL2 peptide (left panel) were compared with those of RANTES (right panel), a chemokine ligand that directly interacted with CCR5. The data indicated that the efficacy of PL2 was enhanced more effectively by reducing Env expression than by reducing CCR5 expression, as reflected in terms of lower IC50 and greater inhibition. The opposite results were obtained with RANTES, for which greater enhancement of efficacy was achieved by reducing CCR5 expression than by reducing Env expression; again, these differences were observed with respect to IC50 and extent of inhibition. These data provide additional support for the conclusion that the peptide inhibitory activity reflects its interaction with Env.
Comparison of the effects of modulating Env versus coreceptor expression levels on the efficiency of peptide blocking. Effector cells were prepared by infecting HeLa cells with vCB21R-LacZ and expressing the Ba-L Env either by plasmid transfection (Low Ba-L) or recombinant vaccinia virus infection (Hi Ba-L). Target cells were prepared by infecting HeLa.T4 cells with vTF7-3 and expressing CCR5 either by plasmid transfection (Low CCR5) or recombinant vaccinia virus infection (Hi CCR5). The P3 peptide was preincubated with the effector cells, and RANTES was preincubated with the target cells for 45 minutes at 37°C. Effector and target cells were mixed in the indicated combinations, and cell fusion was measured as described in “Materials and methods.” Error bars indicate SD of mean values obtained from duplicate fusion assays.
Comparison of the effects of modulating Env versus coreceptor expression levels on the efficiency of peptide blocking. Effector cells were prepared by infecting HeLa cells with vCB21R-LacZ and expressing the Ba-L Env either by plasmid transfection (Low Ba-L) or recombinant vaccinia virus infection (Hi Ba-L). Target cells were prepared by infecting HeLa.T4 cells with vTF7-3 and expressing CCR5 either by plasmid transfection (Low CCR5) or recombinant vaccinia virus infection (Hi CCR5). The P3 peptide was preincubated with the effector cells, and RANTES was preincubated with the target cells for 45 minutes at 37°C. Effector and target cells were mixed in the indicated combinations, and cell fusion was measured as described in “Materials and methods.” Error bars indicate SD of mean values obtained from duplicate fusion assays.
Independence of peptide inhibition from the coreceptor used
Although CCR5 and CXCR4 are the relevant coreceptors for HIV-1 infection of natural CD4+ target cells (T lymphocytes and macrophages), many Envs are capable of functioning with other chemokine receptors such as CCR3.27,31-33 If the CCR5 ECL peptides block fusion by interacting with Env, it might be predicted that their inhibitory activities would be independent of the coreceptor used. This was indeed the case, as shown in Table 2 for different Envs mediating fusion with target cells expressing recombinant CD4 plus either CCR5, CXCR4, or CCR3. The PL1 peptide was chosen for this experiment because it was active against all the CCR5-using Envs examined above. For each Env tested, inhibition by the PL1 peptide was similar when different coreceptors were used. Thus Ba-L was efficiently inhibited with target cells expressing CCR5 or CCR3, 89.6 was blocked when using CCR5, CXCR4, or CCR3, and IIIB (closely related to LAV) was unaffected when using CXCR4 or CCR3. Consistent observations were made with Envs from the primary isolates 92UG024.2 and 92HT593 (both R5X4), which is to say the PL1 peptide inhibited fusion with target cells expressing CCR5, CXCR4, or CCR3. The dependence of the inhibitory effect on the Env rather than the coreceptor provides added evidence that the peptides exert their inhibitory activities by acting on Env.
Binding of radiolabeled gp120 to CCR5
HIV-1 gp120 from R5 isolates complexed with sCD4 binds to CCR5, as shown by competition for the binding of radiolabeled β-chemokines and by direct binding studies with radiolabeled gp120.34,35 We therefore tested whether binding of sCD4-activated [I125]-labeled gp120 to CCR5 was subject to CCR5 peptide inhibition (Figure 8). We selected PL2 for these experiments because it showed the most potency in our inhibition assays and it displayed differential effects on different Envs. Figure 8A shows that PL2, but not PNt, dramatically reduced binding to CCR5 of sCD4-activated [I125]-labeled gp120 from the JR-FL isolate (R5); by contrast, binding of the gp120 from the 89.6 (R5X4) strain was unaffected. As expected, the β-chemokine RANTES caused potent binding inhibition of gp120 from both strains. The sCD4-activated gp120 from the IIIB (X4) strain showed only background binding, which was unaffected by either of the peptides or by RANTES. Figure 8B shows the dose-response effects of the PL2 peptide on CCR5 binding of sCD4-activated [I125]-labeled gp120 from the different strains. Again, the JR-FL gp120 was inhibited while the 89.6 gp120 was not; IIIB gp120 showed only negligible binding that was unaffected by the PL2 peptide. Thus the strain specificities of PL2 inhibition of sCD4-activated gp120 binding to CCR5 paralleled those described above for cell fusion and viral infectivity.
Inhibition of direct binding of [I125]-labeled gp120 to CCR5. NIH-3T3 cells expressing recombinant CCR5 were prepared by infection with vHC-1. [I125]-labeled gp120 molecules derived from the indicated HIV-1 strains were preincubated with 100 nM sCD4 then treated with (A) 50 μg/mL of PNt or PL2 or (B) increasing concentrations of PL2 before mixing with CCR5-expressing cells. As a background control for specificity of gp120 binding to CCR5, samples of CCR5-expressing cells were preincubated with 500 nM cold RANTES before mixing with the sCD4-activated [I125]-labeled gp120 (A). Error bars indicate SD of mean values obtained from duplicate experiments.
Inhibition of direct binding of [I125]-labeled gp120 to CCR5. NIH-3T3 cells expressing recombinant CCR5 were prepared by infection with vHC-1. [I125]-labeled gp120 molecules derived from the indicated HIV-1 strains were preincubated with 100 nM sCD4 then treated with (A) 50 μg/mL of PNt or PL2 or (B) increasing concentrations of PL2 before mixing with CCR5-expressing cells. As a background control for specificity of gp120 binding to CCR5, samples of CCR5-expressing cells were preincubated with 500 nM cold RANTES before mixing with the sCD4-activated [I125]-labeled gp120 (A). Error bars indicate SD of mean values obtained from duplicate experiments.
Discussion
In the studies presented here, the synthetic peptide approach previously used to analyze the N-terminal14-16,36,37 and transmembrane38 regions of CCR5 has been extended to the ECLs. We demonstrate that synthetic peptides derived from each of the CCR5 ECLs can block the fusogenic activity of HIV-1 Env and that the inhibition patterns vary for different Envs. Our findings that fusion inhibition by the ECL peptides is restricted to those Envs capable of using CCR5 as a coreceptor (R5 or R5X4) attest to the specificity of these activities; parallel selectivity has been reported previously for tyrosine-sulfated N-terminal CCR5 peptides.14,15 Our data support the notion that multiple extracellular regions of the coreceptors contribute not only to their activities but also to their specificities for different Envs.
Our results with the ECL peptides are consistent with data in the literature indicating the importance of the CCR5 ECLs.12 Particularly noteworthy are the data emphasizing the activity and selectivity of ECL2. For example, a mutant virus adapted to use CCR5 lacking the N-terminus was found to have increased affinity for ECL2,39 perhaps reflecting a direct interaction of gp120 with this region of CCR5. Our finding of the particular potency of PL2 (for the susceptible R5 Envs) compared with PL1 and PL3 proved interesting. Important correlations were also observed regarding the strain specificities of the ECL2 region. For example, grafting of the CCR5 ECL2 onto the coreceptor-inactive murine CCR5 conferred coreceptor activity for the R5 strains ADA and YU2, but not for 89.640 ; those reports are consistent with our demonstration of the activities of the PL2 peptide against the R5 strains Ba-L and JR-FL, but not the 89.6 strain, in assays of cell fusion, infectivity, and inhibition of gp120 binding to CCR5.
Several independent lines of evidence suggest that the fusion-blocking activities of the ECL peptides reflect their direct interactions with Env: (1) the PL2 inhibitory effect persisted after pretreating and washing the Env-expressing effector cells, but not the receptor-expressing target cells; (2) the potency of PL2 fusion inhibition correlated inversely with the expression level of Env, but not of CCR5; (3) for a given Env, the PL1 sensitivity was comparable when different coreceptors were used; and (4) the PL2 peptide inhibited CCR5 binding of sCD4-activated [I125]-labeled gp120 from an R5 virus. The simplest interpretation is that the ECL peptides associated with the specific regions of gp120 that normally interact with the corresponding CCR5 domains, thereby blocking the gp120/CCR5 interactions required for fusion. Recent studies with CCR5 N-terminal peptides are illuminating in this regard. The tyrosine-sulfated N-terminal CCR5 peptides (but not their unsulfated counterparts) have been shown to associate with gp120, based on direct binding studies of gp120/CD4 complexes to the peptides and peptide inhibition of the binding of these complexes to surface CCR5 or to anti-gp120 monoclonal antibodies.14,15 Critical determinants on gp120 for the N-terminal peptide binding appear to reside in the fourth constant (C4) region and the third variable (V3) loop,14-16 each of which is known to play a central role in gp120 binding to coreceptor.1,3,12 Residues in the V3 stem appear to be important for gp120 binding to N-terminal sulfated peptides, whereas the V3 stem and crown appear to be required for gp120 binding to cell surface CCR5.16 Such findings have led to the notion that gp120 interaction with coreceptor might be a multistep process, perhaps first involving binding to the N-terminal region, followed by the ECLs. Thus it will be most important to determine the regions on gp120 to which each of the ECL peptides binds.
In considering the use of synthetic peptides as biochemical probes for the gp120/coreceptor interaction, it is important to note that the critical feature of each ECL region (and perhaps also the N-terminus) is likely to be a structural motif rather than a specific amino acid sequence. This notion is based on the fact that many HIV-1 isolates can function comparably well with CCR5 and CXCR4, which have negligible sequence homology. Moreover, potent coreceptor activity has been observed for chimeras in which the N-terminal region of CCR5 is replaced by the corresponding region of a different chemokine receptor lacking coreceptor activity for a particular Env and for chimeras in which the CCR5 N-terminus is grafted to a noncoreceptor chemokine receptor.12 An even more dramatic example is the recent demonstration of coreceptor activity (albeit relatively weak; P.E. Kennedy and E.A.B., unpublished observations, July 2003) for a chimera containing the N-terminus of CCR5 grafted onto the corresponding region of bacteriorhodopsin,41 a 7-transmembrane receptor with structural features dissimilar to those of mammalian GPCRs. The recent demonstration of ordered structures for soluble peptides representing each of the extracellular regions of bovine rhodopsin42 suggests that considerable insight might be gained from structural analysis of peptides derived from the extracellular regions of CCR5 and from comparisons with the structures of peptides from the corresponding regions of CXCR4 and other GPCRs, with and without coreceptor activity. The synthetic peptide approach thus has potential to bring order to the bewildering complexity of HIV-1 Env/coreceptor interactions.
Prepublished online as Blood First Edition Paper, October 23, 2003; DOI 10.1182/blood-2003-08-2669.
Supported by start-up funds provided by Indiana University School of Medicine and the Walther Cancer Institute and by the National Institutes of Health Intramural AIDS Targeted Antiviral Program.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Paul Kennedy for excellent technical assistance, Kevin Ridge for helpful discussion, and Christopher Broder for soluble gp120 molecules. The use of human peripheral blood mononuclear cells was approved by the Indiana University Institutional Biosafety Committee. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: plasmids encoding the primary HIV-1 envelope glycoproteins 92HT593.1 and 92UG024.2 from Drs Gao Feng and Beatrice Hahn (Department of Medicine, Duke University Medical Center, Durham, NC); HeLa.T4 cell line from Dr Richard Axel (Columbia University, New York, NY); NIH-3T3.T4 cell line from Dan Littman (New York Medical Center, New York, NY).

![Figure 8. Inhibition of direct binding of [I125]-labeled gp120 to CCR5. NIH-3T3 cells expressing recombinant CCR5 were prepared by infection with vHC-1. [I125]-labeled gp120 molecules derived from the indicated HIV-1 strains were preincubated with 100 nM sCD4 then treated with (A) 50 μg/mL of PNt or PL2 or (B) increasing concentrations of PL2 before mixing with CCR5-expressing cells. As a background control for specificity of gp120 binding to CCR5, samples of CCR5-expressing cells were preincubated with 500 nM cold RANTES before mixing with the sCD4-activated [I125]-labeled gp120 (A). Error bars indicate SD of mean values obtained from duplicate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-08-2669/6/m_zh80040456550008.jpeg?Expires=1767815998&Signature=P4bUh6nmG7RjxbDe6gVFDRJbuToJ5uE4iF3taTTLZGFOUdpEmjjnKmQrSRsDYhJwIpQBj4Og6y-STQKCK-ZhGTMrm2lw5s~9tytd4dOX8koOZQAFG8Swz00VWByCC45OZC8DCg2WLOaI2GTmnlX4Wz~RjZDTDMUFYwZ3EyHS-x8wAXgrw5IqGOR6rrCEZPtKIcecqDvJYc3lAaLncD1n4UgzBa~D0fuWZDh7YjK~vcvV07IbPMhndOHei60fwqJZtjN9P0ItgMR02oO4SaVEcRJEkghM9qU04fUDaojwlH-i2ieeztIv7XC6HKwXvixU6mqzEkg5GMDTUjEe8iUrDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
