Abstract
Transcription of natural killer (NK) cell antigen receptors (NKRs), such as CD94, NKG2, and killer immunoglobulin-like receptors (KIRs), is developmentally regulated and clonally distributed. We have shown a restricted KIR repertoire (rKIR-R) without monoclonal T-cell receptor rearrangement (mTCR-R) supports a NK lineage in nasal-type extranodal NK/T-cell lymphoma (NTENL) but does not correlate with clinical outcomes. Developing NK cells express first CD94, then NKG2A, NKG2E, and finally NKG2C. This sequence suggests an immature CD94- and a mature CD94+ subtype of NTENL. Using a rKIR-R without a mTCR-R as a criterion in 25 cases of NTENL, we confirmed a true NK lineage in 19 cases, including 10 CD94+ and 9 CD94- patients by reverse transcriptase-polymerase chain reaction (RT-PCR). Eight of the 10 CD94+ patients but only 2 of the 9 CD94- patients survived beyond 1 year (median survival, 60 months versus 10 months by Meier-Kaplan survival analysis, P = .026 by Cox F test). The remaining 6 patients had a rKIR-R plus a mTCR-R, suggesting mixed NK/T differentiation. They were CD94- by RT-PCR, found predominantly in young women, and had a median survival of 35 months. Thus, on the basis of the transcripts of NKRs, a division of NTENLs into CD94+, CD94-, and mixed NK/T types reflects a true biologic divergence with different clinical behaviors. (Blood. 2003;102:2623-2631)
Introduction
Nasal-type extranodal NK/T-cell lymphoma (NTENL) is a rare disease that presents both diagnostic and therapeutic difficulties. The lymphoma may cause extensive ulceration and necrosis of the nasal cavity, often leaving little viable tissue for diagnosis. The lymphoma can pursue an aggressive course, sometimes terminating unexpectedly in fatal hemophagocytosis.1,2 At present, the only known prognostic factor for NTENLs is the stage at diagnosis. However, patients with the same initial stage have greatly variable clinical courses.3 It is highly desirable to have molecular tests to confirm the diagnosis and predict the outcome.
The cellular origin of NTENLs has been controversial because of the difficulty in separating the T lineage from the natural killer (NK) lineage, both being derived from the same bipotential T/NK precursor.4,5 NTENL is cytoplasmic CD3+, suggesting a T lineage, but, unlike other T-cell lymphomas, it is surface CD3- and does not have a monoclonal T-cell receptor (TCR) rearrangement.6 Because cytoplasmic CD3 can also be found in fetal NK cells,7,8 and CD56, an NK cell marker, is frequently expressed by NTENLs, most researchers now believe NTENLs have an NK cell origin.
Like TCRs or immunoglobulins, which are the antigen receptors for T cells or B cells, respectively, NK cells use CD94, NKG2, and killer immunoglobulin-like receptors (KIRs) as NK-cell antigen receptors (NKRs).9 Furthermore, the expression of NKRs, with CD94/NKG2 preceding KIRs, is developmentally regulated and clonally distributed.10 Because a monoclonal TCR rearrangement is the reference standard for T-cell lymphoma and a monoclonal immunoglobulin gene rearrangement is characteristic of B-cell lymphoma, the patterns of NKRs might be used for establishing monoclonality for an NTENL of the NK lineage and further identifies developmental stages within the NK lineage.
The KIRs have a consensus sequence composed of 3 immunoglobulin-like domains, D0, D1, and D2.11-14 Alternative splicing of the consensus sequence results in 3 groups, KIR2DL4 (D0-D2), KIR2D (D1-D2), or KIR3D (D0-D1-D2). A polyclonal NK cell population is expected to express all 3 groups, but a monoclonal NK cell population would have a KIR repertoire restricted to 1 or 2 groups. As expected, we have shown that a restricted KIR repertoire is typically found in NTENLs of NK cell origin.15 The demonstration of a restricted KIR repertoire, however, could not be used further to specify distinct developmental stages that might predict outcomes.16
Unlike KIRs, CD94 and NKG2 are C-type lectins.17 The NKG2 family includes 2A, 2B, 2C, 2D, 2E, and 2F. During NK cell development, CD94 is expressed first, followed by 2A, 2E, and finally 2C.18-21 The expressed CD94 forms heterodimers with NKG2, usually 2A, but sometimes with 2E or 2C.22 NKG2B is a splicing variant of 2A. NKG2F is highly homologous to 2C and 2E, and is a 3′-truncated variant. NKG2D is divergent from the other members and is expressed as homodimer. This developmental regulation implies an immature CD94- precursor cell lymphoma and a mature CD94+ NK cell lymphoma, and, possibly, a better clinical outcome associated with the mature phenotype.23
To support the hypothesis that expression of CD94/NKG2 receptors is of prognostic value, we developed a novel reverse transcriptase-polymerase chain reaction (RT-PCR) approach to characterize the transcripts of CD94 and the NKG2 family in 25 cases of NTENL. In conjunction with the patterns of KIRs for lineage assignment, we hope to identify mature and immature subsets and to correlate the phenotype and genotype with the clinical course of NTENLs.
Materials and methods
Tissue samples
Tissue sections and medical records of all NTENLs at the Department of Pathology, National Taiwan University Hospital, in the period from 1995 to 2002, were reviewed by 2 independent hematopathologists (C.W.L. and S.M.H.). The diagnosis was made by a combination of morphology, immunohistochemistry, and clinical features according to the World Health Organization (WHO) classification. Twenty-five consecutive cases with sufficient tissue for studies were collected. Nine consecutive lymphoblastic lymphomas (LBLs) involving the mediastinum or the lymph nodes were used in a search for possible NK differentiation at the immature stage. Ten peripheral T-cell lymphomas (PTCLs) with a monoclonal TCR rearrangement were randomly chosen as negative controls.
All the NTENLs were cytoplasmic CD3+ by immunohistochemistry on paraffin sections and EBER (EB virus-encoded RNA) positive by in situ hybridization. These NTENLs were characterized molecularly by a restricted KIR repertoire (rKIR-R) with or without a monoclonal TCR rearrangement (mTCR-R). Parts of the KIR and TCR data have been published in 2 previous papers.14,15 All the LBLs were cytoplasmic CD3+ and nuclear TdT+ (terminal deoxynucleotidyl transferase). All the PTCLs had a monoclonal TCR rearrangement.24
RT-PCR for CD94
The cDNA sequence of CD94 was taken from Genbank U30610.25 For reverse transcription, an antisense primer, 5′-acaaactgtgttcactagcACCCATTTTTCTTGGCAAGAACAGCA-3′ (281-256) was used. The capital letters correspond to the antisense sequence of CD94 cDNA from position 281 to 256. The small letters constitute a random sequence for increased PCR specificity and incorporation of fluorescent labels.
Specifically, reverse transcription was done in a 20-μL reaction mixture at 40°C for 50 minutes. The mixture included 0.1 μg RNA purified from formalin-fixed, paraffin-embedded tissue blocks, 0.5 μM antisense primer, 200 U reverse transcriptase, 50 mM Tris (tris(hydroxymethyl)aminomethane)-HCl at pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT (dithiothreitol), and 200 μM of each dNTP (deoxynucleoside triphosphate). A reaction for β-actin was run as a positive control, and a reaction under the same condition except for the omission of reverse transcriptase was run as a negative control.
The cDNA from 2 μL RT reaction mixture was then PCR-amplified by a forward primer, 5′-TTGTTGAAAAATTCTTTTACTAAACT-3′ (172-197) and a fluorescence-labeled primer of the random sequence, 5′-tetcttctgacaaactgtgttcactagc-3′. Specifically, a 20-μL PCR mixture included cDNA, 0.3 μM of each primer, 15 mM Tris-HCl at pH 8.0, 1.5 mM MgCl2, 50 mM KCl, 200 μM of each dNTP, and 1 U Taq polymerase. The reaction mixture was subjected to 35 cycles of PCR. Each cycle consisted of denaturation at 94°C for 45 seconds, annealing at 45°C for 45 seconds, and extension at 72°C for 45 seconds.
At the end of 35 cycles, a portion of the PCR product was loaded on and separated by a high-resolution electrophoretic system (ABI377 equipped with GeneScan software; Perkin-Elmer, Foster City, CA). The size for the RT-PCR product of CD94 is 135 nucleotides (nt).
Sequence alignment and RT-PCR for the NKG2 family
The NKG2 family, with Genbank accession numbers in parentheses, includes NKG2A (X54867) and its splicing variant 2B, NKG2C (X54869), NKG2D (X54870), NKG2E (L14542), and NKG2F (AJ001683).26-30 Multiple sequence alignment was performed with the MAP program from Baylor College of Medicine, Houston, TX.
In the top part of Figure 1, the consensus structure of the NKG2 family is shown.
Structure of the NKG2 family and the RT-PCR strategy. The upper part shows the consensus structure of the NKG2 family, including a 5′ untranslated region (UT), an intracytoplasmic domain (cyt), a transmembrane domain (TM), an extracellular carbohydrate recognizing domain (CRD), and a 3′ UT. The middle part shows alignment of the cDNA sequences, in which the exon boundaries are marked by the vertical bars. The lower part shows sequences of the TM-CRD junction used for RT-PCR. The exon encompassing the TM-CRD junction spans nucleotides 448 to 501 (A), 294 to 338 (C), 580 to 619 (D), and 469 to 522 (F). The sequences used for the forward primers are in italicized letters. The consensus sequence, 5′-TGTGGCCCTTGTCCTGAAAAGTGGAT-3′, is used for the binding of reverse primer. The figure is drawn to show relative position only but is not proportional to exact sizes.
Structure of the NKG2 family and the RT-PCR strategy. The upper part shows the consensus structure of the NKG2 family, including a 5′ untranslated region (UT), an intracytoplasmic domain (cyt), a transmembrane domain (TM), an extracellular carbohydrate recognizing domain (CRD), and a 3′ UT. The middle part shows alignment of the cDNA sequences, in which the exon boundaries are marked by the vertical bars. The lower part shows sequences of the TM-CRD junction used for RT-PCR. The exon encompassing the TM-CRD junction spans nucleotides 448 to 501 (A), 294 to 338 (C), 580 to 619 (D), and 469 to 522 (F). The sequences used for the forward primers are in italicized letters. The consensus sequence, 5′-TGTGGCCCTTGTCCTGAAAAGTGGAT-3′, is used for the binding of reverse primer. The figure is drawn to show relative position only but is not proportional to exact sizes.
Below the consensus sequence, in the middle part of Figure 1, individual cDNAs were aligned with exon boundaries indicated by the vertical bars. Note the correspondence between the domains and the exon structure. NKG2B is a splicing variant of NKG2A without the consensus TM-CRD junction. NKG2C and NKG2E are highly homologous and have identical TM-CRD junctions. NKG2F also has homology with C and E and is a truncated variant at the 3′ end. Finally, NKG2D is only distantly related to the others.
In the lower part of Figure 1, the highly homologous sequence at the TM-CRD junction is shown. This portion is used for the design of an efficient RT-PCR approach by a consensus reverse primer and 4 specific forward primers. The consensus reverse primer is 5′-acaaactgtgttcac tagcATCCACTTTTCAGGACAAGGGCCACA-3′. The specific forward primers, and their cDNA positions in parentheses, were 5′-TTGTTATTCCCTCTACATTAA-3′ for 2A/2B (436-456); 5′-ATAGTTCTTATTCCTTTCCT-3′ for 2C/2E (278-297 of 2C or 316-335 of 2E); 5′-G AGTGCTGTATTCCTAAACT-3′ for 2D (560-579); and 5′-AATAGTTCTTATTCCTTGTAT TGG-3′ for 2F (452-475).
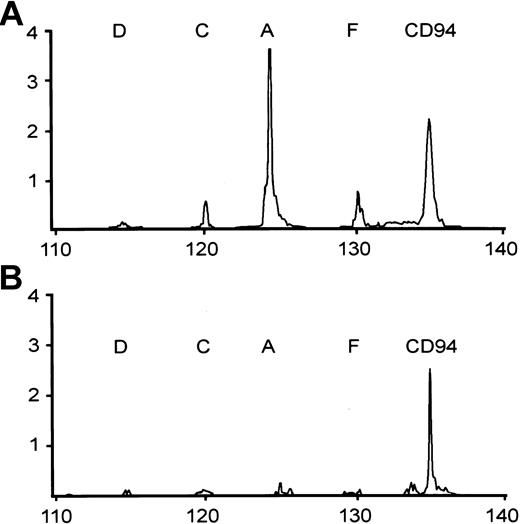
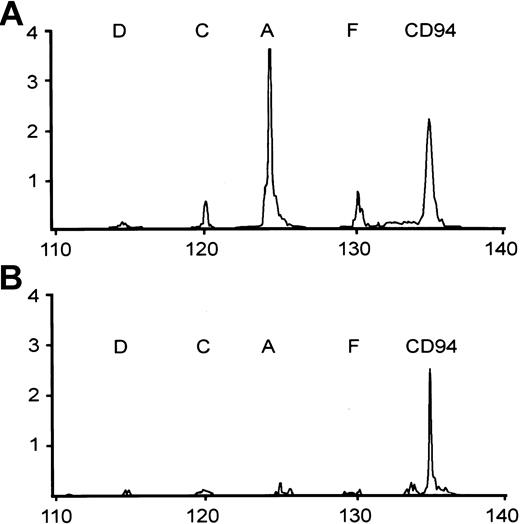
The conditions for RT-PCR were the same as that for CD94, except for the primers. The PCR products were also separated by the GeneScan system. The expected size for the RT-PCR products is 125 nt for A, 120 nt for C/E, 115 nt for D, and 130 nt for F. NKG2B, which does not have the correct TM-CRD junction, is not amplified. Illustrative patterns of the CD94/NKG2 analysis are shown in Figure 2.
Two representative patterns of RT-PCR analysis of CD94 and NKG2 family, including NKG2A (A), NKG2C (C), NKG2D (D), and NKG2F (F). CD94+ NTENL expresses NKG2A strongly, and NKG2C and 2F weakly (A); a second NTENL expresses CD94 only (B).
Two representative patterns of RT-PCR analysis of CD94 and NKG2 family, including NKG2A (A), NKG2C (C), NKG2D (D), and NKG2F (F). CD94+ NTENL expresses NKG2A strongly, and NKG2C and 2F weakly (A); a second NTENL expresses CD94 only (B).
Group-specific RT-PCR for KIRs
We have used a group-specific RT-PCR to demonstrate a restricted KIR repertoire as a clonality test for NK cell lymphomas.15 Briefly, the KIRs can be put into 1 of 3 splicing groups, KIR2DL4, KIR2D, or KIR3D. Three pairs of group-specific primers were designed to target the unique junctions created by splicing. They are 5′-GCTGAGAGAGAAG GTTCTCATAT-3′ and 5′-CACTCCCCCACTGGGTGGTCGGC-3′ for KIR 2DL4, 5′-TGGGTGGGCCAGAGGAAGGTTT-3′ and 5′-CATGGCGTGTGTTGGGTTCTTCTTG-3′ for KIR2D, and 5′-TGGGTGGGCCAGGAGGAAGGTTT-3′ and 5′-CACTCCCCCCACT GGGTGGTCGGC-3′ for KIR3D.
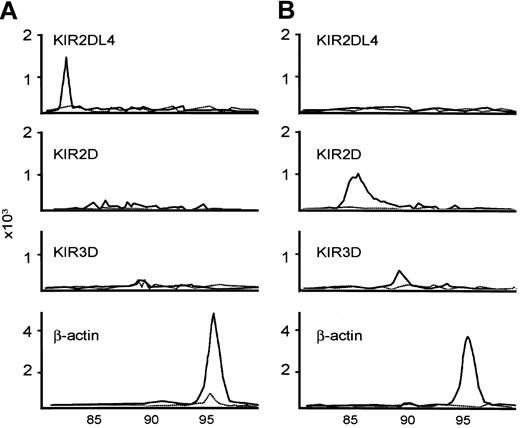
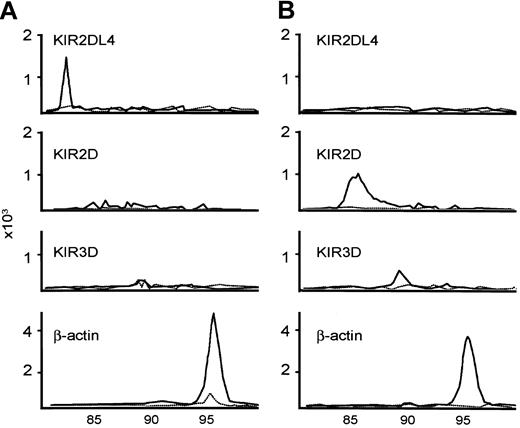
The RT-PCR conditions were the same as those for CD94 and the NKG2 family, except that fluorescence-labeled deoxycytidine triphosphate (dCTP) was used at a concentration of 2 μM 6-carboxytetramethylrhoda-mine [TAMRA]-dCTP during the PCRs. The sizes for the PCR products were 83, 85, and 90 for KIR2DL4, KIR2D, and KIR3D, respectively. Two common patterns of KIR repertoires are shown in Figure 3.
Examples of KIR repertoire by RT-PCR. KIR2DL4+KIR2D-KIR3D-NTENL is shown (A); another KIR2DL4-KIR2D+KIR3D+ NTENL is shown (B). The expected sizes of the PCR products for KIR2DL4, KIR2D, KIR3D, and β-actin are 83, 86, 90, and 96, respectively. The solid line is RT-PCR with reverse transcriptase; the dashed line is RT-PCR without reverse transcriptase, as negative control.
Examples of KIR repertoire by RT-PCR. KIR2DL4+KIR2D-KIR3D-NTENL is shown (A); another KIR2DL4-KIR2D+KIR3D+ NTENL is shown (B). The expected sizes of the PCR products for KIR2DL4, KIR2D, KIR3D, and β-actin are 83, 86, 90, and 96, respectively. The solid line is RT-PCR with reverse transcriptase; the dashed line is RT-PCR without reverse transcriptase, as negative control.
T-cell receptor γ gene rearrangement
Immunohistochemistry and in situ hybridization
Immunohistochemical stains for the epsilon chain of CD3 (CD3ϵ, polyclonal; Dako SA, Glostrup, Denmark), membrane CD3 (CD3m, clone PS-1; Novocastra Lab, Newcastle, United Kingdom),31 and CD56 (Novocastra Lab) were done for all cases on formalin-fixed, paraffin-embedded tissue sections. Immunohistochemistry for CD94 (clone HP-3D9; BD PharMingen, San Diego, CA) and NKG2A (clone T-20; Santa Cruz Biotechnology, Santa Cruz, CA) was done on acetone-fixed fresh frozen tissue sections of 4 NTENLs that were CD94+and NKG2A+ by RT-PCR. Illustrative figures are shown in Figure 4A-E.
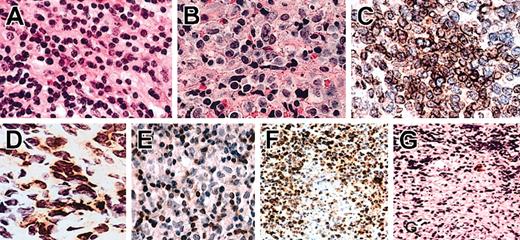
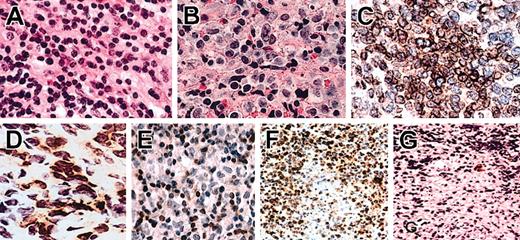
Hematoxylin and eosin (H/E) sections, immunohistochemistry, and in situ hybridization of NTENL. Representative examples of grade I (A) and grade III (B) nuclear morphologies. Note the smaller nuclei with condensed chromatin in panel A, and the larger nuclei with open chromatin in panel B. Not shown here, an intermediate morphology is grade II. By immunohistochemistry, an NK-NTENL was diffusely CD94+ (C) and focally NKG2A+ (D). Another NK-NTENL was CD3m-, with CD3m+ small T cells (E). In situ hybridization with the antisense cDNA probe demonstrated CD94+ lymphoma cells with focal necrosis (F); a corresponding area stained by the sense cDNA probe showed rare cells with nonspecific staining (G). All at magnification × 1000, H/E on paraffin sections (A-B), immunoperoxidase stains on frozen sections (C-D), or paraffin sections (E-G).
Hematoxylin and eosin (H/E) sections, immunohistochemistry, and in situ hybridization of NTENL. Representative examples of grade I (A) and grade III (B) nuclear morphologies. Note the smaller nuclei with condensed chromatin in panel A, and the larger nuclei with open chromatin in panel B. Not shown here, an intermediate morphology is grade II. By immunohistochemistry, an NK-NTENL was diffusely CD94+ (C) and focally NKG2A+ (D). Another NK-NTENL was CD3m-, with CD3m+ small T cells (E). In situ hybridization with the antisense cDNA probe demonstrated CD94+ lymphoma cells with focal necrosis (F); a corresponding area stained by the sense cDNA probe showed rare cells with nonspecific staining (G). All at magnification × 1000, H/E on paraffin sections (A-B), immunoperoxidase stains on frozen sections (C-D), or paraffin sections (E-G).
To confirm the positive CD94 results of RT-PCR, in situ hybridization for CD94 was done for all the 10 ENLNTs that were CD94+ by RT-PCR. The vector pCMV-sports6 with a CD94 cDNA insert (GenBank BC028009) was purchased from InVitrogen (Carlsbad, CA). A biotin-labeled antisense cDNA probe, corresponding to the nucleotides 271 to 55 of CD94, was obtained by single-strand PCR. A sense cDNA probe was similarly obtained as a negative control. The probe was used at a concentration of 100 ng/mL. The hybridization buffer, per 100 mL, was composed of 10 g dextran sulfate, 40 mL deionized formamide, 20 mL 20 × standard saline citrate (SSC), 1 mL 2 M Tris at pH 7.4, 1 mL 100 × Denhardt solution, 1 mL 2 mg/mL salmon DNA, and 37.5 mL water. Hybridization was done at 45°C overnight. Posthybridization wash at 45°C for 15 minutes was done twice with 1 × SSC. The slide was then stained by the ABC method with tyramine amplification (Dako SA). Illustrative staining is shown in Figure 4F.
Survival analysis
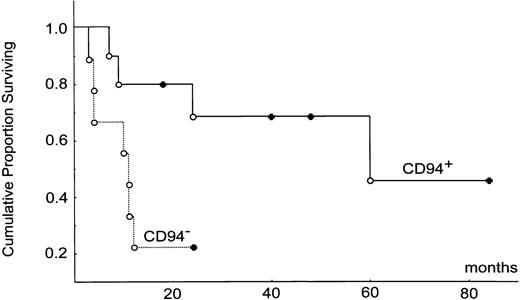
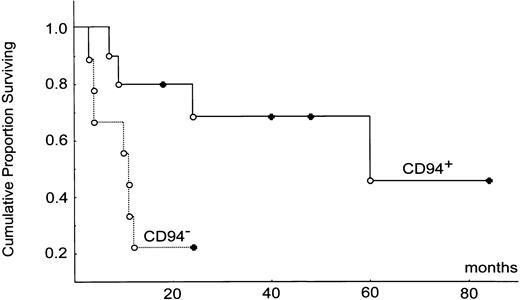
Kaplan-Meier survival analysis based on the product-limit method was done by the statistical software Statistica (StatSoft, Tulsa, OK). Two-sample comparison between the CD94+ versus CD94- subtypes was done by the Cox F test. Survival curves for the CD94+ and CD94- subtypes are shown in Figure 5.
Kaplan-Meier survival analysis. CD94+ and CD94-subtypes are shown, complete follow-up (○) and censored case (•). The median survival times for the 2 groups were 60 months versus 10 months (P = .026).
Kaplan-Meier survival analysis. CD94+ and CD94-subtypes are shown, complete follow-up (○) and censored case (•). The median survival times for the 2 groups were 60 months versus 10 months (P = .026).
Results
Restricted KIR repertoire without a monoclonal TCR rearrangement supports an NK lineage for a subset of NTENLs
Mature NK cells express clonotypical KIRs. It has been estimated that the total KIR repertoire of a polyclonal population of NK cells, such as those from the peripheral blood, consisted of about 6 to 9 KIRs. In contrast, a monoclonal population of NK cells, such as those from NK lymphoma/leukemias, had a restricted KIR repertoire of usually 1 but sometimes 2 KIRs by immunohistochemistry.32-35 Such findings suggested that a restricted KIR repertoire might be used as a clonality test for NK cell lymphomas.
On the basis of these observations, we developed a simplified but systematic analysis of KIR repertoires by RT-PCR. In this approach, a restricted KIR repertoire without a monoclonal TCR rearrangement was taken as a criterion for the NK lineage.15 Conversely, a monoclonal TCR rearrangement without a restricted KIR repertoire was used for the T lineage. In addition, a restricted KIR repertoire plus a monoclonal TCR rearrangement was used for mixed NK/T differentiation.16 Applying these criteria to Table 1, we assigned 19 of the 25 NTENLs to the NK lineage (NK-NTENL) and the remaining 6 to the mixed NK/T lineage (NK/T-NTENL). It is interesting to note that, although all NTENLs were CD3ϵ+, the NK-NTENLs were CD3m-, whereas the NK/T-NTENLs might or might not be CD3m+.
Restricted KIR repertoire without TCR rearrangement indicates a possible differentiation toward NK cell in a subset of LBLs
Most LBLs were traditionally attributed to the T lineage, but a true NK lineage was also mentioned in the literature.36 We found that 5 among 9 cases of LBLs had a monoclonal TCR rearrangement; 3 of these 5 cases were CD3m+, further supporting an origin from the T lineage. We also found 4 cases of LBLs (4 in 9, 44%) had a restricted KIR repertoire without monoclonal pattern of TCR rearrangement (Table 2).
Interpretation of the data is difficult, because, unlike mature NK cells, data on the regulation of KIRs during the development of immature NK cells are limited.37 The interpretation is further complicated by the well-known phenomenon of lineage promiscuity in primitive hematopoietic neoplasms.38 We conclude that lineage assignment based on the analysis of antigen receptors alone may not be adequate. Nonetheless, in view that a common bipotent precursor in the thymus gives rise to both T cells and NK cells,4,5 the possibility of a LBL with differentiation toward the NK lineage deserves to be further investigated.
Peripheral T-cell lymphomas rarely had transcripts of KIRs, CD94 or NKG2
We also used 10 cases of PTCLs for comparison with NTENLs. All these cases had a monoclonal TCR rearrangement, but only one case had a detectable KIR repertoire, and another case had CD94 transcripts. This finding shows that NK differentiation (acquisition of NK cell characteristics) in PTCLs is rare (Table 2).
Lineage assignments or patterns of KIR repertoires do not affect survival for NTENLs
We compared the survival times of the 25 NTENLs, as shown in the last column of Table 1. The 1-year survival rate was 0.53 (10 of 19) for the 19 NK-NTENLs and 0.67 (4 of 6) for the 6 NK/T-NTENLs. The difference was not significant (P = .69). For the 19 NKNTENLs, the presence or absence of KIR2DL4, KIR2D, and KIR3D did not affect survival significantly either. For example, the difference between the 1-year survival rate for the KIR2DL4+ NK-NTENLs (7 of 12 or 58%) and that of the KIR2DL4- NK-NTENLs (3 of 7 or 43%) was not statistically significant (P = .32). Similarly, there was no statistical significance in the comparisons KIR2D+ (3 of 7 or 43%) versus KIR2D- (7 of 12 or 58%) at a P value of .53, or KIR3D+ (5 of 7 or 71%) versus KIR3D- (5 of 12 or 42%) at a P value of .24. This information showed that patterns of KIR repertoires or the lineage assignments were not predictive of clinical outcomes.
Distinct patterns of CD94/NKG2 transcripts were identified in NTENLs
We assigned 19 of the 25 NTENLs to the NK lineage (NKNTENL) with a restricted KIR repertoire without a monoclonal TCR rearrangement. Six with a restricted KIR repertoire plus a monoclonal TCR rearrangement were classified as the mixed NK/T lineage (NK/T-NTENL). The 19 NK-NTENLs could be further separated into 10 CD94+ and 9 CD94- cases (Table 1). Among the 10 CD94+ cases, 7 cases expressed one or more NKG2 members, mostly 2A, followed by 2C and 2F. Among the 9 CD94- cases, 1 case unexpectedly expressed NKG2A, and the remaining 8 cases did not express any members of the NKG2 family. For the 6 cases of NK/T-NTENL, only 1 case expressed NKG2A, and CD94 was not expressed at all.
Correlation with morphology, immunohistochemistry (IHC), and in situ hybridization
Expressions of CD94 and NKG2A were confirmed by IHC on 4 NK-NTENLs that were CD94+/NKG2A+ by RT-PCR. As negative controls, 3 PTCLs, 3 tonsils, 3 spleens, and 3 peripheral blood smears were also tested. IHC showed strong and diffuse staining for CD94 and weaker and focal staining for NKG2A only for the 4 NK-NTENLs (Figure 4). Furthermore, in situ hybridization for CD94 was positive on all the 10 NTENLs that were CD94+ by RT-PCR, with negative staining by the sense cDNA probe. These IHC and in situ hybridization findings confirmed the data from the RT-PCR approach.
As shown in Table 3, we compared 4 morphologic parameters: nuclear grade, clear-cell change, angiocentric necrosis, and reactive infiltrates of eosinophils and plasma cells. None of these parameters had statistical significance among the 3 subtypes (CD94+ NK-NTENLs, CD94- NK-NTENLs, and NK/T-NTENLs). The staining pattern of CD56, usually considered to be the characteristic marker of NK cell differentiation, did not differ significantly among the 3 subtypes either (P = .72).
Correlation with clinical and laboratory data showed CD94+ NK-NTENLs had a better prognosis than the CD94- type
The NK-NTENLs, both CD94+ and CD94- types, were associated with middle-aged men (median ages, 43 and 57 years; male-to-female sex ratio, 8:2 and 8:1), whereas the NK/T-NTENLs were associated with younger women (median age, 26 years; male-to-female sex ratio, 2:4). The differences among median ages and sex ratios were both statistically significant, P values less than .001 and equal to .047, respectively (Table 3). The 3 subtypes (CD94+ or CD94- NK-NTENLs and NK/T-NTENLs) had comparable initial clinical presentations, in terms of stage, the level of lactate dehydrogenase (LDH), and peripheral blood cell counts. However, the clinical outcomes differ significantly in terms of disease progression or 1-year survival (P = .031 for both), mainly because of the rapid progression and poor survival of the CD94- subtype.
To confirm further the significance of the 1-year survival rate, we constructed the Kaplan-Meier survival curves. Two-sample comparison between the CD94+ and CD94- subtypes of NKNTENLs showed a highly significant difference between the curves (P = .026; Figure 5). The median survival times for the CD94+ and CD94- subtypes were 60 months and 10 months, respectively. In contrast, the survival curve for the mixed NK/T subtype was between the CD94+ and the CD94- subtypes. The difference does not reach statistical significance, in part because of the small sample size. The median survival for the NK/T type was 35 months.
Disease progression in NK-NTENLs confirms an association with CD94 transcripts but not KIR repertoires
To see if other factors might also determine clinical outcomes, we assigned the 19 NK-NTENLs to 2 groups, depending on the response after treatment: nonprogressive (n = 8) and progressive (n = 11) (Table 4). The nonprogressive group consisted of patients with complete remission, partial remission, or a stable lesion after treatments. The progressive group consisted of patients with lymphoma progression, midfacial destruction, cranial nerve palsy, regional and/or systemic lymphadenopathy, or distant metastasis. Among various clinical and pathologic features evaluated in Table 4, we noticed that only CD94 had a strong association with disease progression, with a Spearman correlation coefficient of -0.59 at a P value of .007.
Among the traditional prognostic indices, such as stage, LDH, and blood counts, only the level of LDH reached marginal significance at P = .10. It is of interest to note in Table 4 that the presence or absence of KIRs had almost no correlation with disease progression (P = .96). This information confirms again that patterns of KIR repertoires do not affect survival for NK-NTENLs.
Discussion
Rearrangement of TCR or immunoglobulin genes is a developmentally regulated process for lymphocytes.39,40 It is known that TCR-β chain rearrangement takes place prior to α chain rearrangement, and immunoglobulin heavy chain rearrangement takes place prior to light chain rearrangement. This process of sequential rearrangement can be used for distinguishing immature from mature T or B cells. Despite the important role of antigen receptors, data on the expression of NK cell antigen receptors, such as KIRs, CD94, and NKG2, are quite limited, in part because of the rarity of NK cell lymphomas.32-35 To address this issue, we have collected 25 cases of NTENL, one of the largest series, for molecular analysis.
Diagnosis of NTENLs is traditionally based on the presence of pleomorphic CD56+ lymphoid cells in an angiocentric pattern and/or in association with Epstein-Barr virus (EBV) infection. The diagnosis, however, could be difficult when a low-grade CD56- lymphoid infiltrate mixed with reactive cells is found in a young patient or when extensive necrosis leaves little viable tissue for diagnosis. To resolve these difficulties, we have advocated a restricted pattern of KIRs as a diagnostic tool. Most NTENLs belong to the true NK lineage (NK-NTENL), characterized by a restricted KIR repertoire without a monoclonal TCR rearrangement. Some NTENLs belong to the mixed NK/T lineage (NK/TNTENL), characterized by both a monoclonal TCR rearrangement and a restricted KIR repertoire.15,16 Despite its diagnostic value, the pattern of the KIR repertoire has little prognostic value.
To understand better the behavior of NTENLs, we have turned to the analysis of the other NK cell antigen receptors, CD94 and NKG2. In contrast to the KIRs, which belong to the immunoglobulin superfamily, CD94/NKG2 are C-type lectins. More important, the developmental regulation of CD94/NKG2 is better defined than that of KIRs; CD94 was expressed first, followed by NKG2A, 2E, and 2C. We have shown a strong association between CD94 and NKG2 receptors. Most CD94+ cases (7 of 10) expressed one or more NKG2 members, whereas only one each of CD94- of NK- and NK/T-NTENLs cases (2 of 15) expressed NKG2A. The differential pattern of CD94 expression might further separate NK-NTENLs into a mature CD94+ subtype and a relatively immature CD94- subtype. CD94 was not expressed in NK/T-NTENLs.
Our data indicate that a division into the CD94+ and the CD94- subtypes has clinical significance, whether by the criteria of disease progression, 1-year survival, or survival curves (Tables 3-4 and Figure 5). However, a correlation between outcomes and treatments is difficult to establish in a retrospective study of this sample size. The therapeutic regimens are usually doxorubicin-based protocols such as CHOP (cyclophosphamide, hydroxydaunomycin, vincristine [Oncovin; Eli Lilly, Indianapolis, IN], prednisone) or its variation, with or without local radiotherapy. Investigations into how the biologic divisions into CD94+ and CD94- subtypes might influence the choice of therapeutic regimens are certainly of great clinical significance.
Regarding the cause of death, we noted that 7 of the 9 CD94- patients died; all of them died of rapid progression of the disease, often with distant metastases. In contrast, only 4 of the 10 CD94+ patients died; none of them had metastasis, and 2 of them died of hemophagocytic syndrome, defined clinically as fever, pancytopenia, hepatosplenomegaly, jaundice, and hemophagocytosis on bone marrow aspiration smears. Because data suggest that the hemophagocytic syndrome is often a reflection of underlying defect of NK cells,41 further investigation is required to determine whether assays of NK cell function might be useful in predicting and preventing this fatal outcome.
In conclusion, we confirmed a restricted KIR repertoire without a monoclonal TCR rearrangement in NTENLs of NK lineage and showed that the presence of CD94 transcripts is the most important prognostic factor. This finding emphasizes again the importance of antigen receptors in diagnostic hematopathology. The exact nature of the CD94- phenotype could not be ascertained at this time, but a CD94+ phenotype is consistent with that of a mature NK cell and implies a better prognosis (median survival, 60 months versus 10 months by Meier-Kaplan survival analysis). We caution that, despite being a large series of 25 cases for a rare disorder, the statistical significance is still small (P = .026 by Cox F test). Finally, we showed that the occurrence of hemophagocytosis as the cause of death is unusual even in NTENLs.
Prepublished online as Blood First Edition Paper, June 19, 2003; DOI 10.1182/blood-2003-01-0295.
Supported by grants from the National Taiwan University Hospital (NTUH) (grants NTUH-91-A12, NTUH-92-S58) (C.W.L.), the National Science Council (NSC) (grant NSC91-2320-B002-197) (C.W.L.) (NSC-91-2314-B-002-179) (S.M.H.), and the Ministry of Education, Taiwan (grant 89-B-FA01-1-4) (S.M.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mr C. C. Lu for the artwork.