Abstract
Expression of ALK protein by lymphoid cells and the description of variant anaplastic lymphoma kinase (ALK) translocations have typically been restricted to cases of T-cell and null anaplastic large-cell lymphoma (ALCL). All such cases result from a novel fusion created by the ALK gene on chromosome 2p23 and NPM on 5q35 or other variant translocation partners. A rare variant of diffuse large B-cell lymphoma (DLBCL), originally described in 1997, was thought to overexpress full-length ALK in contrast to a chimeric protein characteristic of ALCL. However, full-length ALK protein lacks tyrosine kinase activity and thus the mechanism of oncogenesis has remained elusive. We describe 6 cases of ALK+ DLBCL characterized by a simple or complex t(2;17)(p23;q23) involving the clathrin gene (CLTC) at chromosome band 17q23 and the ALK gene at chromosome band 2p23. All cases were studied using fluorescence in situ hybridization (FISH), complemented in one case with standard cytogenetic analysis, multicolor karyotyping (M-FISH), and reverse transcriptase-polymerase chain reaction. These results clearly demonstrate that most cases of ALK+ DLBCL share the same mechanism of deregulated ALK expression. Moreover, these results demonstrate the presence of CLTC-ALK fusions in these tumors and extend the list of diseases associated with this genetic abnormality to include classical T-cell or null ALCL, ALK+ DLBCL, and inflammatory myofibroblastic tumors. (Blood. 2003;102:2568-2573)
Introduction
Anaplastic large cell lymphoma (ALCL) is a well-defined subtype of non-Hodgkin lymphoma (NHL) with characteristic morphologic features, immunophenotype, and molecular genetic alterations.1 ALCL is of T-cell or null phenotype, positive for CD30, frequently expresses cytotoxic granule proteins, and in most cases, ALK protein.2,3 Most cases are characterized by the t(2;5)(p23;q35), which results in a fusion between the anaplastic lymphoma kinase (ALK) gene at chromosome band 2p23 and nucleophosmin (NPM) at chromosome band 5q35.4 Several variant translocations now have been described, including the t(2;17)(p23;q23) involving the clathrin gene (CLTC) at 17q23.5,6 The initial report of this variant translocation in ALCL identified the clathrin-like gene (CLTCL) at band 22q11 rather than the CLTC gene as the ALK partner, due to an error in GenBank. Subsequently it was established that the clathrin-like sequence was in fact clathrin, corresponding to a t(2;17)(p23;q23) rather than the previously assumed t(2;22)(p23; q11). The clathrin gene encodes a coated vesicle protein involved in the intracellular transport of various molecules.
In 1997, a series of 7 cases of an unusual variant of diffuse large B-cell lymphoma (DLBCL) that appeared to express full-length ALK protein, rather than the typical NPM-ALK fusion, was described.7 The results of immunohistochemistry with an antibody to the extracellular domain of ALK, together with Western blotting in one case, suggested the expression of full-length ALK protein. One of these cases had a clone identified by standard cytogenetic analysis, but no breakpoint involving 2p23 was identified. With the exception of this earlier case, karyotypic findings in ALK+ DLBCL have not been reported.
In reviewing a series of ALK+ DLBCL cases, we became aware of a single case with a granular cytoplasmic ALK staining pattern and a clonal karyotype that harbored a breakpoint involving chromosome band 2p23. The karyotype was complex, but the use of locus-specific fluorescence in situ hybridization (FISH) and multicolor karyotyping revealed the presence of a t(2;17)(p23;q23). On the strength of this observation and the similar ALK staining pattern of virtually all other cases of ALK+ DLBCL, we reasoned that CLTC-ALK fusions might be present in some of these cases. To test this hypothesis, we performed reverse transcriptase-polymerase chain reaction (RT-PCR) and/or FISH studies on all cases with available material. The present report details the results of these investigations.
Since the original description of deregulated ALK expression in ALCL, a number of other unrelated tumors have been shown to express the ALK protein, including neuroblastoma, rhabdomyosarcoma, and inflammatory myofibroblastic tumors (IMTs).8,9 Although most ALK rearrangements have been described only in ALCL, 4 (TPM3-ALK, TPM4-ALK, RANBP2-ALK, and CLTCALK) have been implicated in the pathogenesis of IMTs.5,10 In this report we describe the presence of the CLTC-ALK fusion in a series of 6 cases of ALK+ DLBCL, extending the list of diseases with this translocation involving the ALK gene. Although this study cannot exclude a role for deregulated expression of full-length ALK in some cases, it firmly establishes a role for the CLTC-ALK fusion in the pathogenesis of ALK+ DLBCL.
Patients, materials, and methods
Study population
In total, 11 patients of ALK+ DLBCL were available for study from the consultative files of 4 of the authors (G.D., H.K.M.-H., N.L.H., and R.D.G.). Five patients were excluded, 3 because the unstained sections had been stored for several years and produced weak signals with FISH. These were considered not evaluable. Two other patients had only Bouin-fixed sections and were excluded from the analysis. The remaining 6 patients form the basis of this study. Details of the clinical characteristics for all 6 patients were gathered from chart reviews where available. Stage of disease was determined using the Ann Arbor staging system. Approval for these studies was obtained from the British Columbia Cancer Agency and Centre Hospitalier Universitaire (CHU)-Purpan institutional review boards. Informed consent was provided according to the Declaration of Helsinki.
Histology and immunohistochemistry
Routine hematoxylin and eosin (H&E) sections were prepared using standard techniques from both formalin-fixed and Bouin-fixed paraffin blocks. Immunohistochemical analyses included a large panel of monoclonal antibodies including CD20, CD79a, CD45, kappa, lambda, IgM, IgG, IgA, Bcl-2, Bcl-6, perforin, CD3, CD4, CD5, CD30, CD43, CD57, CD138, and epithelial membrane antigen (EMA) antigens. The detection of ALK protein was performed on paraffin sections fixed in buffered formalin using monoclonal antibodies directed against fixative-resistant epitopes of the cytoplasmic portion of ALK (antibodies ALK1 and ALKc, generous gifts from Dr K. Pulford [Oxford, United Kingdom] and B. Falini [Perugia, Italy], respectively).
Cytogenetic analysis, locus-specific FISH, and multicolor FISH
The lymph node in case 1 was mechanically disaggregated, cultured for 24 hours, harvested, and chromosome preparations G-banded according to standard techniques. The karyotype was described according to the International System for Human Cytogenetic Nomenclature (ISCN 1995). Locus-specific (Vysis, Downers Grove IL) and multicolor karyotyping (M-FISH) (Metasystems, Altlussheim, Germany) were performed on metaphase spreads as previously described.11
Using bioinformatic resources available at the University of California at Santa Cruz (http://genome.ucsc.edu), BAC clones RP11-316J18 and CTD-2601E3 were selected as telomeric and centromeric probes flanking the CLTC locus, respectively. For the dual-color FISH assay flanking the CLTC locus, clones were differentially labeled with Texas Red (TR, telomeric) and Spectrum Green (SG, centromeric). As break-apart assay for the ALK locus, the commercially available LSI-ALK probe was applied (ALK telomeric in Spectrum Orange and ALK centromeric in SG; Vysis, Downers Grove, IL). For the multicolor interphase assay flanking both ALK and CLTC, the commercial LSI-ALK probe was combined with the CLTC clones, the latter being labeled with TR (telomeric) and diethylaminocoumarin (DEAC, centromeric). Bacterial cultivation, BAC DNA isolation, and labeling and probe preparation were performed as previously described.12
Locus-specific interphase FISH was performed on paraffin-embedded tissue sections. Briefly, deparaffinized slides were pretreated by pressure cooking for 2 minutes 40 seconds in EDTA (ethylenediaminetetraacetic acid) buffer (1 mM, pH 8.0) and subsequent incubation in pepsin solution for 5 minutes at 37°C to increase DNA accessibility. Then, slides were fixed in 1% paraformaldehyde for 2 minutes, dehydrated through increasing ethanol series, and air dried. The appropriate probe mix (1.5 μL) was applied to the tissue section and covered with a round 10-mm coverslip. Both probe and target DNA were simultaneously denatured at 70°C for 12 minutes and incubated overnight at 37°C. Posthybridization washes were performed according to the “rapid-wash protocol” provided by Vysis. Slides were counterstained with 4′6-diamidino-2-phenylindole 2HCl (DAPI) and mounted in Vectashield antifade solution (Vector Laboratories, Burlingame, CA). Image acquisition and processing was performed as previously described.12
Cell lines, RNA extraction, and RT-PCR
The SU-DHL-1 and RH30 cell lines were kindly provided by Dr M. L. Cleary (Stanford University Medical Center, Stanford, CA) and Dr S. W. Morris (St Jude Children Research Hospital, Memphis TN), respectively. The COST cell line has been established in the laboratory of one of the authors (G.D.) from a t(2;5)-positive ALCL. We also have used as positive controls for RT-PCR studies NIH3T3 cell lines transfected by the ALK-EC cDNA (encoding the ALK extracellular domain) in the pcDNA3 expression vector and by the full-length CLTC-ALK cDNA. Total RNA was extracted from frozen lymph node samples using the SV total RNA isolation kit (Promega, Charbonnieres, France) following the manufacturer's recommendations. Synthesis of the first cDNA strand was performed as previously described, using an ALK specific primer.6 CLTC-ALK transcripts were detected by RT-PCR as previously described, using the primers ALK1 and CLA1 in the first round, followed by a nested PCR with the inner primer pair ALK2/CLA3, yielding a 270-bp product.6
ALK transcripts corresponding to the ALK extracellular domain were detected by 2 rounds of RT-PCR. The first step consisted of an ALK gene-specific reverse transcription at 48°C for 45 minutes, followed by 30 cycles comprising a denaturation step at 94°C for 45 seconds, an annealing step at 68°C for 45 seconds, and an elongation step at 72°C for 45 seconds. The first round of PCR was performed using ALK-EC1 (5′-CCATCTCCTTCTCCTGATTATTTT-3′) and ALK-EC2 (5′-CACTGCAGACAAGCTGCGGTT-3′) primers. The second round was performed on a 2-μL aliquot from the first amplification, using nested primers ALK-ECint1 (5′-ATGACCTCAGGAACCAGAGCTGGTCC-3′) and ALK-EC int2 (5′-CCAACCATGCTTCCCTGGAGTGG -3′), yielding a 317-base pair (bp) product.
Results
Clinical, histological, and immunophenotypic findings
The incidence of ALK+ DLBCL is unknown and cannot be estimated with confidence from this study. The reported cases represent uncommon DLBCL cases from the consultative files of several of the authors and thus are heavily influenced by both selection and referral bias. Nonetheless, a small study of ALK expression using a tissue microarray of 120 de novo DLBCL patients from British Columbia failed to detect any ALK-positive cases, suggesting a frequency of less than 1% of DLBCL (R.D.G. and G.D., unpublished observations, October 2002). The details of all 6 cases included in the study are described in Table 1. The median age was 48.5 years, and there was a clear male predominance (5:1); 4 of the 6 patients had advanced-stage disease at diagnosis, 3 of the patients for whom follow-up information was available have either progressive disease or have died of lymphoma. No information regarding the stage at diagnosis, bone marrow status, treatment, or follow-up was available for case 2. Case 6 was treated with rituximab in addition to cyclophosphamide + adriamycin + vincristine + prednisone (CHOP) chemotherapy, despite the absence of CD20 expression by the tumor cells.
The histologic findings in all 6 cases were similar. A distinct sinusoidal growth pattern was evident, and a mixture of large immunoblasts and numerous plasmablastic cells characterized the infiltrates. Plasmablastic cells were defined as having eccentric nuclei, amphophilic cytoplasm, prominent Golgi zones, and lacking the expression of mature pan-B-cell antigens. Case 6 was characterized by prominent necrosis, but this was not a feature of the other 5 cases. No hallmark cells, typical of T-cell or null ALCL were apparent. These features were identical to those described in the original report.7
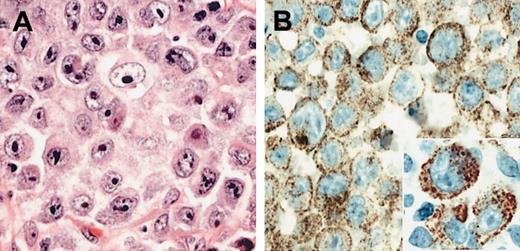
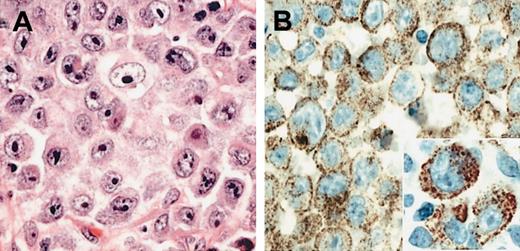
The immunophenotypic results in all 6 cases were also similar. All 6 cases lacked expression of pan-B-cell antigens CD20 and CD79a. CD138 was strongly positive in all cases, as was EMA (Table 1). Four cases showed light-chain restriction with expression of lambda, and 3 of these also expressed IgA heavy chains. In 2 cases immunoglobulin expression was technically not interpretable, however, one of these (case 1) showed a distinct clonal IGH rearrangement using consensus PCR strategies for framework one and J-region gene segments (data not shown). CD30 was negative in all 6 cases, highlighting the distinction from ALCL. The expression of Bcl-2 protein was assessed in 4 cases with all 4 being negative. Weak focal expression of CD4 was seen in 2 cases and CD57 in a single case, but CD3 was negative in all cases. The pattern of ALK expression was identical in all 6 cases. ALK protein expression was restricted to the cytoplasm and showed a fine granular pattern in all cells (Figure 1). In addition, some cases showed membrane reinforcement of ALK staining.
Morphology of ALK and DLBCL. (A) H&E-stained section of the diagnostic lymph node biopsy from case 1 showing neoplastic cells with immunoblastic and plasmablastic features. (B) The ALK-1 stain reveals fine granular cytoplasmic localization of the ALK protein (inset). Original magnifications: ×400 (A-B); ×800 (inset).
Morphology of ALK and DLBCL. (A) H&E-stained section of the diagnostic lymph node biopsy from case 1 showing neoplastic cells with immunoblastic and plasmablastic features. (B) The ALK-1 stain reveals fine granular cytoplasmic localization of the ALK protein (inset). Original magnifications: ×400 (A-B); ×800 (inset).
Cytogenetic, M-FISH, and dual/multicolor interphase FISH
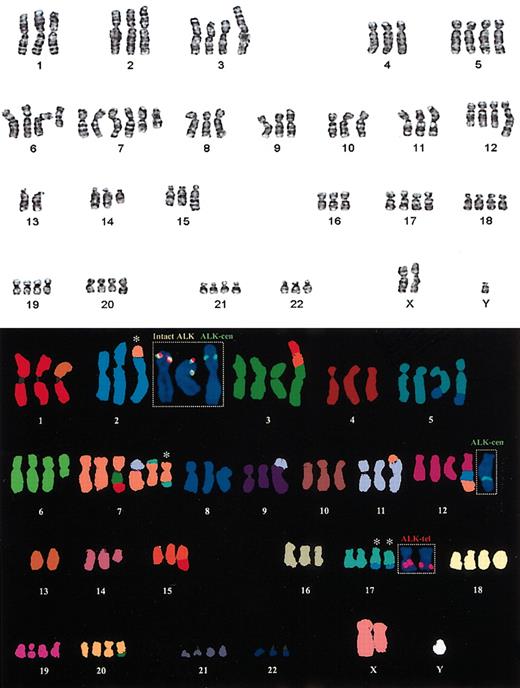
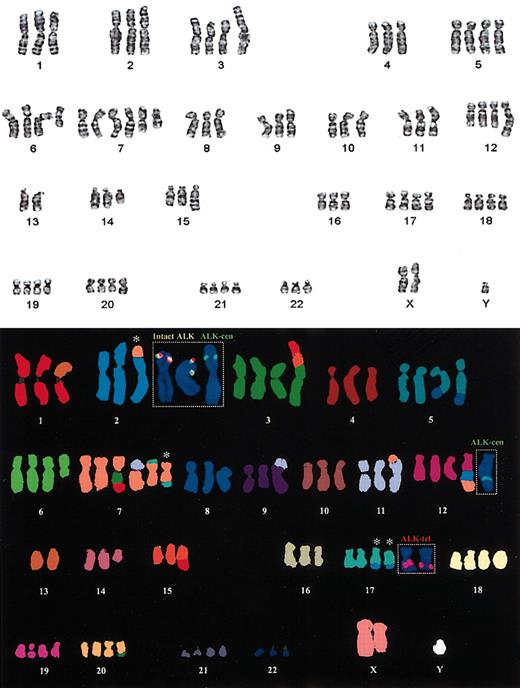
Cytogenetic analysis was available for case 1 only. This showed a complex karyotype with an add (2) (p23) and add (17) (q23). Locus-specific FISH with the dual colored ALK probe confirmed a breakpoint resulting in splitting apart of the ALK gene flanking probes (Figure 2, insert). The M-FISH confirmed a 3-way balanced t(2;17;7)(p23;q23; q?22) with duplication of the der(17)t(2;17;7)(p23;q23;q?22) and a partial duplication of the der(2)t(2;17;7)(p23;q23;q?22), whereby a duplicated portion of the derivative p-arm with the CLTC-ALK fusion was translocated to chromosome 12. These results were confirmed by dual and multicolor interphase FISH (Figure 2B and Figure 3G).
Standard cytogenetic analysis showing the complex karyotype from case 1 and M-FISH with dual-color FISH confirming the t(2;17;7)(p23;q23;q?22). The inserted locus-specific FISH images show the location of the multiple copies of the ALK gene. The colocalization of the green and red probes (yellow signal) is seen on the 2 copies of the normal chromosome 2. The centromeric ALK probe (green) is seen on the der(2) and on the derivative chromosome 12. The telomeric ALK probe (red) is localized to the 2 copies of the der(17). The white asterisks denote the chromosomes involved in the 3-way translocation. The combined G-band and M-FISH karyotype interpretation was as follows: 80, XXY, der(1)t(1;13)(q10;q10), t(2;17;7)(p23;q23; q22), del(3)(p25), +der(3)(3qter-> 3p21::15q11-> 15q21::1q25-> 1qter), der(5)t(5;8)(q33; q22), +der(5)t(5;8)(q33;q22),+del(6)(q?21),+der(7)(7pter-> 7q11::3?-> 3?::1q25::1qter), +der(7)t(7;11)(q11;p11),+der7t(7;17)(?;?),der9t(9;11)(p23;?q23),der11t(7;11)(p13;p14),+der12-(12pter-> 12q21::2p1?3-> p23::7q22-> 7qter), -13, del(14)(q?13q24), der(15)t(1;15)(q25; q22),+der(17)t(2;17;7)(p23;q23;q22), +18,+19,+der(20)t(3;20)(?p25;q12),+21.
Standard cytogenetic analysis showing the complex karyotype from case 1 and M-FISH with dual-color FISH confirming the t(2;17;7)(p23;q23;q?22). The inserted locus-specific FISH images show the location of the multiple copies of the ALK gene. The colocalization of the green and red probes (yellow signal) is seen on the 2 copies of the normal chromosome 2. The centromeric ALK probe (green) is seen on the der(2) and on the derivative chromosome 12. The telomeric ALK probe (red) is localized to the 2 copies of the der(17). The white asterisks denote the chromosomes involved in the 3-way translocation. The combined G-band and M-FISH karyotype interpretation was as follows: 80, XXY, der(1)t(1;13)(q10;q10), t(2;17;7)(p23;q23; q22), del(3)(p25), +der(3)(3qter-> 3p21::15q11-> 15q21::1q25-> 1qter), der(5)t(5;8)(q33; q22), +der(5)t(5;8)(q33;q22),+del(6)(q?21),+der(7)(7pter-> 7q11::3?-> 3?::1q25::1qter), +der(7)t(7;11)(q11;p11),+der7t(7;17)(?;?),der9t(9;11)(p23;?q23),der11t(7;11)(p13;p14),+der12-(12pter-> 12q21::2p1?3-> p23::7q22-> 7qter), -13, del(14)(q?13q24), der(15)t(1;15)(q25; q22),+der(17)t(2;17;7)(p23;q23;q22), +18,+19,+der(20)t(3;20)(?p25;q12),+21.
Dual and multicolor interphase FISH for the detection of breakpoints in the ALK and CLTC loci. (A) Ideograms of chromosomes 2 and 17 showing the ALK(2p23) and CLTC(17q23) flanking probes labeled with different dyes. The schematic interphase nucleus displays a FISH signal pattern indicating intact ALK and CLTC loci. (B) Scheme of chromosomes 2 and 17 containing the t(2;17)(p23;q23) reciprocal translocation as well as an interphase nucleus harboring this translocation. Derivative chromosomes 2 and 17 show a colocalization of green/pink and pale blue/red signals, respectively. The latter points to the CLTC-ALK fusion leading to activation of the ALK tyrosine kinase. (C) Interphase FISH in case 2 hybridized with the LSI ALK probe. Arrows point to tumor cells with isolated red and green signals indicating the presence of a breakpoint in the ALK locus. Arrowheads point to nontumoral cells containing the signal constellation for 2 intact ALK loci. (D-E) Interphase cells from case 3 (D) and case 4 (E) hybridized with the dual-color CLTC break-apart probe. Both cases show a split of the red and green signals pointing to a breakpoint in the CLTC locus. (F-G) False color display of interphase cells from case 2 (F) and case 1 (G) hybridized with the multicolor break-apart probe for ALK and CLTC (described in panels A-B). Both cells contain red/pale blue colocalizations, which indicate the fusion of the 5′-end of CLTC with the 3′-end of ALK leading to activation of the ALK tyrosine kinase. Besides, the presence of isolated green and pink signals indicates that the reciprocal 5′ALK-3′CLTC fusion is missing, probably due to complex chromosomal aberrations. The lack of an isolated red 3′ALK signal renders an additional fusion other than CLTC-ALK highly unlikely.
Dual and multicolor interphase FISH for the detection of breakpoints in the ALK and CLTC loci. (A) Ideograms of chromosomes 2 and 17 showing the ALK(2p23) and CLTC(17q23) flanking probes labeled with different dyes. The schematic interphase nucleus displays a FISH signal pattern indicating intact ALK and CLTC loci. (B) Scheme of chromosomes 2 and 17 containing the t(2;17)(p23;q23) reciprocal translocation as well as an interphase nucleus harboring this translocation. Derivative chromosomes 2 and 17 show a colocalization of green/pink and pale blue/red signals, respectively. The latter points to the CLTC-ALK fusion leading to activation of the ALK tyrosine kinase. (C) Interphase FISH in case 2 hybridized with the LSI ALK probe. Arrows point to tumor cells with isolated red and green signals indicating the presence of a breakpoint in the ALK locus. Arrowheads point to nontumoral cells containing the signal constellation for 2 intact ALK loci. (D-E) Interphase cells from case 3 (D) and case 4 (E) hybridized with the dual-color CLTC break-apart probe. Both cases show a split of the red and green signals pointing to a breakpoint in the CLTC locus. (F-G) False color display of interphase cells from case 2 (F) and case 1 (G) hybridized with the multicolor break-apart probe for ALK and CLTC (described in panels A-B). Both cells contain red/pale blue colocalizations, which indicate the fusion of the 5′-end of CLTC with the 3′-end of ALK leading to activation of the ALK tyrosine kinase. Besides, the presence of isolated green and pink signals indicates that the reciprocal 5′ALK-3′CLTC fusion is missing, probably due to complex chromosomal aberrations. The lack of an isolated red 3′ALK signal renders an additional fusion other than CLTC-ALK highly unlikely.
Interphase cytogenetic analyses with dual-color probes flanking the ALK and CLTC loci were performed on formalin-fixed, paraffin-embedded tissue sections of the 5 evaluable DLBCL cases. In case 5, only old unstained sections were available, resulting in poor probe hybridization that prevented reliable evaluation. This case was not studied by FISH but was analyzed using RT-PCR (Table 1). The other 5 cases displayed a split of the signals flanking ALK and CLTC, respectively, indicating a chromosomal breakpoint within both loci (Figure 3). In order to confirm the presence of a t(2;17)(p23;q23) translocation leading to CLTC-ALK fusion, a 4-color interphase FISH assay was applied (Figure 3). In 3 patients (cases 1, 2, and 6), fusion of the CLTC-centromeric (5′) and ALK-telomeric (3′) signals were observed in all evaluated cells with a breakpoint in the ALK locus indicating the presence of the CLTC-ALK oncogenic fusion gene. Remarkably, the ALK-centromeric (5′) and CLTC-telomeric (3′) signals appeared isolated in the cell nucleus. This finding suggests the presence of a 3-way or more complex translocation in which the reciprocal ALK-CLTC fusion is lost. In case 1, this finding from interphase FISH is in line with the results of the metaphase analyses and M-FISH studies showing the presence of a t(2;17;7)(p23;q23;q?22) 3-way translocation. In the 2 additional patients (cases 3 and 4), the presence of the CLTC-ALK fusion could not be formally proven due to the lack of reliably evaluable signals from the CLTC-centromeric (5′) probe labeled with DEAC. Nevertheless, there was clearly a fusion of signals for 3′ CLTC and 5′ ALK in the evaluated cells with an ALK break-apart pattern. Considering the breakpoints affecting ALK and CLTC, along with the typical ALK protein expression pattern, this finding strongly suggests the presence of an oncogenic CLTC-ALK fusion in these 2 ALK+ DLBCL cases. In case 5, FISH studies were not successful, however, RT-PCR evaluation revealed the CLTCALK fusion (Figure 4B).
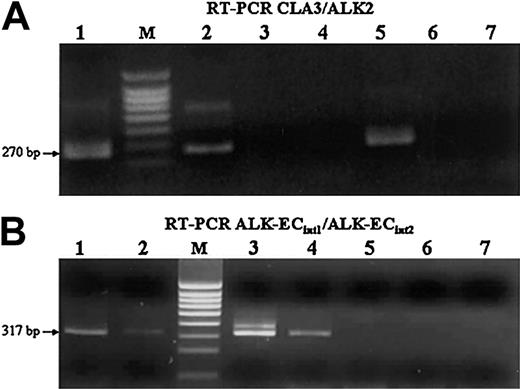
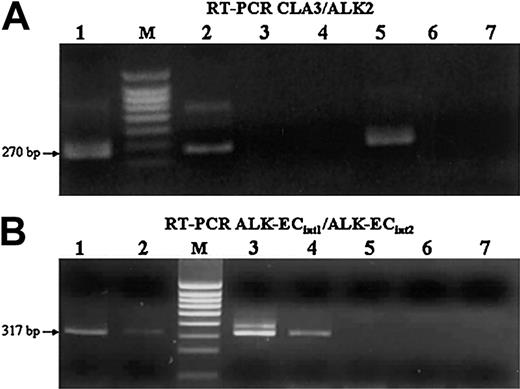
RT-PCR analysis of CLTC-ALK and ALK extracellular domain transcripts. (A) Cases 1 and 5 (lanes 1-2) and the NIH3T3 cell line transfected with CLTC-ALK cDNA, used as positive control (lane 5), show the expected 270-bp CLTC-ALK RT-PCR product (M indicates size marker). No band could be amplified from the Rh30 (lane 3), NIH3T3 cell line transfected with ALK-EC cDNA (lane 4), SU-DHL1 (lane 6), and COST (lane 7) cell lines, used as negative controls. (B) Transcripts encoding the extracellular portion of ALK also were present in the cases expressing CLTC-ALK transcripts (lanes 1-2), as in the Rh30 (lane 3) cell line and in the NIH3T3 cell line transfected with ALK-EC cDNA (lane 4), used as positive controls. As expected, no signal was present in either of the t(2;5)-positive SU-DHL1 (lane 6) and COST (lane 7) cell lines, nor in the NIH3T3 cell line transfected with CLTC-ALK cDNA (lane 5).
RT-PCR analysis of CLTC-ALK and ALK extracellular domain transcripts. (A) Cases 1 and 5 (lanes 1-2) and the NIH3T3 cell line transfected with CLTC-ALK cDNA, used as positive control (lane 5), show the expected 270-bp CLTC-ALK RT-PCR product (M indicates size marker). No band could be amplified from the Rh30 (lane 3), NIH3T3 cell line transfected with ALK-EC cDNA (lane 4), SU-DHL1 (lane 6), and COST (lane 7) cell lines, used as negative controls. (B) Transcripts encoding the extracellular portion of ALK also were present in the cases expressing CLTC-ALK transcripts (lanes 1-2), as in the Rh30 (lane 3) cell line and in the NIH3T3 cell line transfected with ALK-EC cDNA (lane 4), used as positive controls. As expected, no signal was present in either of the t(2;5)-positive SU-DHL1 (lane 6) and COST (lane 7) cell lines, nor in the NIH3T3 cell line transfected with CLTC-ALK cDNA (lane 5).
RT-PCR
Detection of hybrid CLTC-ALK transcripts. Our RT-PCR technique showed that a 270-bp CLTC-ALK band could be amplified from 2 patients (Figure 4A, cases 1 and 5, lanes 1-2). Lane 5 shows the positive control, representing the NIH3T3 cell line transfected with CLTC-ALK cDNA.
Detection of ALK transcripts. Analysis by RT-PCR showed that the extracellular portion of ALK transcripts also was present in the cases expressing CLTC-ALK transcripts (Figure 4B, cases 1 and 5, lanes 1-2). As expected, the 317-bp ALK-EC band could be amplified from RH30 cell line (Figure 4B, lane 3) and the other positive control cell line (NIH3T3 cell line transfected with ALK-EC) shown in lane 4. No signal was evident from the SU-DHL-1 and COST cell lines (Figure 4B, lanes 6-7) or in lane 5 (NIH3T3 cell line transfected with CLTC-ALK cDNA) used as negative controls. The sequences of full-length ALK and the CLTC-ALK fusions for cases 1 and 5 are as follows:
Full-length ALK (primers ALK EC int1 and ALK EC int2), case 1: CCGAGGAGGCCTCCCNGATGGACTTGCTGGATGGGCCTGGGGCAGAGCGTTCTAAGGAGATGCCCAGAGGCTCCTTTCTCCTTCTCAACACCTCAGCTGACTCCAANCACACCATCCTGAGTCCGTGGATGAGGAGCAGCAGTGAGCACTGCACACTGGCCGTCTCGGTGCACAGGCACCTGCAGCCCTCTGGAAGGTACATTGCCCAGCTGCTG.
Case 5: AGGAGGCCTCCCAGATGGACTTGCTGGATGGGCCTGGGGCAGAGCGTTCTAAGGAGATGCCCAGAGGCTCCTTTNTCCTTCTCAACACCTCAGCTGACTCCAAGCACACCATCCTGAGTCCGTGGATGAGGAGCAGCAGTGAGCACTGCACACTGGCCGTCTCGGTGCACAGGCACCTGCAGCCCTCTGGAAGGTACATTGCCCAGCTGCTGCCCCACAA.
CLTC-ALK fusion (primers CLA3 and ALK2, the ALK sequence is in italics), case 1: TTAAGGCCAGATGTCGTCCTAGAAACTGCATGGAGGCACAATATCATGGATTTTGCCATGCCCTATTTNATCCAGGTCATGAAGGAGTACTTGACAAAGGTGGATAAATTAGATGCTTCAGAATCACTGAGAAAAGAAGAAGAACAAGCTACAGAGACACAACCCATTGTTTATGTGTACCGCCGGAAGCACCAGGAGCTGCAAGCCATGCAGATG.
Case 5: CAGATGTCGTCCTAGAAACTGCATGGAGGCACAATATCATGGATTTTGCCATGCCCTATTTCATCCAGGTCATGAAGGAGTACTTGACAAAGGTGGATAAATTAGATGCTTCAGAATCACTGAGAAAAGAAGAAGAACAAGCTACAGAGACACAACCCATTGTTTATGTGTACCGCCGGAAGCACCAGGAGCTGCAAGCCATGCAGATGGAGCT.
Discussion
In the initial report describing ALK+ DLBCL, full-length ALK protein was implicated in the pathogenesis of this unusual tumor based on the results of RT-PCR, Western blot, and immunohistochemistry. A systematic analysis of ALK fusions was not performed, but NPM-ALK was for the most part excluded as a pathogenic mechanism. In the present report, however, we have definitively shown that 2 of 6 studied cases of ALK+ DLBCL harbor a translocation creating a fusion gene, CLTC-ALK, identical to that previously described in typical T-cell/null ALCL and more recently in IMTs.6,13 In the remaining 4 cases, FISH data, together with the results of characteristic granular cytoplasmic ALK immunostaining, strongly suggest a role for CTCL-ALK fusions. Thus, previous hypotheses suggesting that ALK+ DLBCL arises by different mechanisms that leads to overexpression of the full-length ALK protein may not be true in most cases of this entity.7 Moreover, this finding now expands the list of diseases associated with the CLTC-ALK alteration. In 2 of the 6 cases included in this report, we were able to demonstrate by RT-PCR the concomitant expression of full-length ALK transcripts. These results remain largely unexplained, as the ALK gene is normally silent in cells committed to the B-cell lineage. More important, previous studies had failed to document any tyrosine kinase activity associated with full-length ALK protein (G.D., unpublished observations, July 2002). Thus, the results of the current study suggest a prominent role for CLTC-ALK fusion in the pathogenesis of ALK+ DLBCL but leave open the role of full-length ALK protein. Interestingly, a 200-kDa protein consistent with native ALK also was found to be expressed in IMTs associated with TPM4-ALK and TPM3-ALK.14 In agreement with our current findings, these authors noted that native ALK protein lacked in vitro kinase activity and tyrosyl phophorylation.14 Case 5 in the current study was previously reported in 1997 (case 1 in that series), including Western blot analysis. At that time a band of approximately 200 kDa was detected, which was believed to represent full-length ALK protein. PCR studies of this same case in the current report indicate both full-length ALK and CLTC-ALK fusion transcripts. The CLTCALK fusion protein on Western blotting would have an expected molecular weight of about 220 kDa. We cannot exclude the possibility that the previously reported band of about 200 kDa was CLTC-ALK, as the resolution of Western blotting in the high molecular weight range is often poor. Unfortunately, the lack of any additional frozen material precludes additional Western blot analyses.
All 6 cases included in this report show a characteristic pattern of ALK staining, as evidenced by a fine granular cytoplasmic staining pattern. In the consultative files of one of the authors (G.D.), this staining pattern is seen in all cases of ALK+ DLBCL except one (G.D., unpublished data, June 2001). Additionally, this pattern is also seen in classic T-cell/null ALCL with a CLTC-ALK fusion. These results suggest that CLTC-ALK fusions likely represent the dominant oncogenic mechanism in this disease. The one exception noted above shows both nuclear and cytoplasmic ALK staining and has been shown to harbor a t(2;5) resulting in an NPM-ALK fusion.15 Taken together, these results suggest a very different distribution of ALK fusion partners in ALK-positive tumors. Firstly, classical T-cell or null ALCL is characterized by NPM-ALK fusions in the vast majority of the cases. Alternate translocation partners, although numerous in variety, are uncommon in frequency. ALK+ DLBCL appears to be mostly associated with CLTC-ALK fusions based on the results of this study, although occasional cases may harbor a t(2;5) or express full-length ALK protein. Lastly, IMTs have been shown to mostly harbor the TPM3-ALK or TPM4-ALK fusion, whereas other translocation partners, such as CLTC-ALK, are uncommon.14 IMTs associated with NPM-ALK have yet to be described.
The close association of ALK expression to pathogenesis of ALCL suggests that this molecular alteration is responsible for disease initiation. In addition, ALK is not normally expressed in lymphoid cells. NPM is the most frequently implicated fusion partner, accounting for approximately 70% of classic ALCL translocations.5 However, a number of recent studies have identified a growing list of alternative translocation partners (X-ALK) that appear also to activate ALK.16-22 All of these are characterized by changes in the subcellular localization of ALK, being located either within the nucleus, nucleolus, and cytoplasm, restricted to the cytoplasm, or in the cytoplasmic membrane. Recognition of these variant translocations suggests that nuclear and nucleolar localization of ALK, as seen in cases with the t(2;5), is not critical to pathogenesis. Moreover, retroviral-mediated gene transfer of NPM-ALK causes lymphoid malignancies in mice.23 Of importance, the mice develop CD30-negative B-cell tumors with similar morphology to the ALK+ DLBCL reported herein and elsewhere.7
Lastly, a transgenic mouse model has been recently developed using the NPM-ALK transgene under control of the murine CD4 promoter.24 These mice develop either thymic T-cell lymphomas or plasma cell tumors that express CD138. The morphologic spectrum of the plasma cell tumors is diverse, but Figure 5C from that study shows similar morphology to de novo ALK+ DLBCL.7,24 The mechanism by which the transgene is expressed in cells other than T cells is unexplained in these animals, but curiously ALK+ DLBCL commonly express weak but focal CD4 and/or CD57.7 These in vivo data provide support for our observations concerning the role of ALK fusions in B-cell tumors and strongly suggest a role for ALK in tumors of diverse cell lineages.24
Of interest is the involvement of the t(2;17)(p23;q23) with 3 distinct diseases including classic T- or null-cell ALCL, ALK+ DLBCL, and IMTs. The karyotype in case 1 was complex, leaving open the possibility that other cytogenetic alterations gave rise to the DLBCL and that the t(2;17;7) was a late event and not critical to disease initiation. However, breakpoints at 2p23 do not appear at high frequency in other lymphoma subtypes on the basis of cytogenetic analyses, and large-scale screenings of a number of lymphomas and other tumors have not disclosed ALK expression as a frequent event.25,26 Alternatively, the t(2;17) may be important to pathogenesis, but the decisions that determine cell type (myofibroblast versus lymphoid cell) and lineage (T cell versus B cell) may be heavily dependent on secondary downstream cytogenetic/molecular alterations or epigenetic changes. Thus, it seems that ALK deregulation by different mechanisms is involved in a growing number of markedly different diseases. Of note, it remains unclear whether the full-length ALK receptor is constitutively activated and whether it plays a role in a subset of ALK+ DLBCL.7
In summary, the frequency of CLTC-ALK fusions and their role in the pathogenesis of ALK+ DLBCL is further established by this report. Of note, a recent description of this same translocation in an ALK+ DLBCL Japanese patient has appeared in abstract form.27 Thus, 2 of 3 cases with a clonal karyotype thus far reported have a t(2;17) or variant.7,27 These findings, together with the characteristic restricted granular cytoplasmic staining pattern of the CLTCALK fusion in T-cell/null ALCL and its similarity to the expression of ALK protein in ALK+ DLBCL, suggest a prominent role for this translocation in this unusual subtype of DLBCL.
Prepublished online as Blood First Edition Paper, May 22, 2003; DOI 10.1182/blood-2003-03-0786.
Supported by the Ligue Nationale Contre le Cancer and the Comités du Lot, de la Haute Garone, de l'Yonne, et du Gers and the Interdisziplinäres Zentrum für klinische Krebsforschung (IZKF), Kiel, Germany; and by National Institutes of Health grant 1-UO1-CA84967-01 (R.D.G., V.S.L., and D.E.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Michel March for his excellent technical work performing immunohistochemistry and Blandine Roquefeuil and Claudia Becher for locus-specific FISH work. We also thank Andrea Moore for PCR studies, and Nicole Dastugue for assistance with cytogenetic work. We thank Miles Price for referral of case 1 and Hugh Turner for assistance with preparation of samples and histopathology of this same case. We acknowledge our fellow pathologists for referral of cases 3, 5, and 6, including Henry Y. Dong, Impath, New York; Dr T. Wang, Riddle Memorial Hospital, Media, PA; and Dr William L. Marsh, Ohio State University, Columbus, OH.