Abstract
Monocyte-derived dendritic cells (DCs) and macrophages (Mϕs) generated in vitro from the same individual blood donors were exposed to 5 different pathogens, and gene expression profiles were assessed by microarray analysis. Responses to Mycobacterium tuberculosis and to phylogenetically distinct protozoan (Leishmania major, Leishmania donovani, Toxoplasma gondii) and helminth (Brugia malayi) parasites were examined, each of which produces chronic infections in humans yet vary considerably in the nature of the immune responses they trigger. In the absence of microbial stimulation, DCs and Mϕs constitutively expressed approximately 4000 genes, 96% of which were shared between the 2 cell types. In contrast, the genes altered transcriptionally in DCs and Mϕs following pathogen exposure were largely cell specific. Profiling of the gene expression data led to the identification of sets of tightly coregulated genes across all experimental conditions tested. A newly devised literature-based clustering algorithm enabled the identification of functionally and transcriptionally homogenous groups of genes. A comparison of the responses induced by the individual pathogens by means of this strategy revealed major differences in the functionally related gene profiles associated with each infectious agent. Although the intracellular pathogens induced responses clearly distinct from the extracellular B malayi, they each displayed a unique pattern of gene expression that would not necessarily be predicted on the basis of their phylogenetic relationship. The association of characteristic functional clusters with each infectious agent is consistent with the concept that antigen-presenting cells have prewired signaling patterns for use in the response to different pathogens.
Introduction
In addition to their function in antigen presentation, macrophages (Mϕs) and dendritic cells (DCs) play an important role in sensing pathogens and delivering both start-up and class differentiation signals to the adaptive immune system. Although the ability of both cells to produce interleukin-12 (IL-12) makes them capable of directing Th1 responses, the ability of Mϕs to initiate an inflammatory response to infectious agents through the production of proinflammatory molecules such as interleukin 8 (IL-8), tumor necrosis factor α (TNF-α), and IL-1β and their microbicidal effector functions distinguishes them from DCs. In contrast, DCs have the unique capacity to capture antigen from the periphery and deliver it to secondary lymphoid organs. Precursor DCs that encounter pathogenic organisms induce the production and release of chemokines and cytokines that, in turn, can attract and/or activate other cell types such as eosinophils, Mϕs, and natural killer cells. Moreover, DCs themselves are important producers of type I interferons (IFNs), TNF-α, and IL-1β, mediators that can enhance DC activity in an autocrine manner. Triggering of both DC and Mϕ function by pathogens is thought to occur as a consequence of ligation of distinct pattern recognition receptors on the surface of these cells.
An important question concerns whether recognition of distinct pathogens by Mϕs and DCs leads to distinct intracellular signals in these cells. Other studies of global transcriptional responses in human Mϕs or DCs to bacterial, influenza, and Candida infections conducted previously have identified a set of commonly induced genes.1-3 Indeed, it has been established that pathogen-derived bacterial stimuli, at least, are sampled by the members of the Toll-like receptor (TLR) family, a family that uses a shared adaptor molecule that leads to activation of a common signaling pathway. The set of genes was found to be induced by multiple bacterial pathogens identified as being regulated by nuclear factor-κB (NF-κB).3 This family of transcription factors is activated downstream of TLR signaling pathways, and expression of its members is known to be autoregulated. Furthermore, global gene expression analysis in mouse Mϕs has suggested that infection by the protozoan Leishmania parasite results in general suppression of gene expression.4
In the present study, the responses of DCs and Mϕs derived from the same monocyte precursor to a wide range of phylogenetically diverse parasites has been detailed at a time when these responses are maximal. We chose the infective stages of both intracellular (Toxoplasma gondii, Leishmania major, and Leishmania donovani) and extracellular (Brugia malayi) parasites because, in contrast with previously published data, the pathogens used in this study are characterized by their capacity to establish chronic infection and induce a broad range of immunologic responses. As such, intracellular pathogens such as T gondii and Mycobacterium tuberculosis promote a strong protective Th1 response characterized by the production of IFN-γ,5,6 whereas Leishmania infections, depending on the species, may or may not drive a Th1 response.7,8 In contrast, infection with extracellular parasites such as the helminth B malayi typically leads to a prototypical Th2 response characterized by high levels of IL-4/IL-5 and immunoglobulin E (IgE) (reviewed in King9 ).
With the use of a new filtering strategy (data on Blood website; see the Supplemental Figures link at the top of the online article) that allows for a comprehensive analysis of gene regulation in response to infection with diverse pathogens, our data demonstrate that Mϕs and DCs share a vast majority of the genes expressed at baseline (96% of the 4057 genes found to be transcribed in either cell type) but that each cell type was found to respond very distinctly to pathogen exposure. Changes in gene expression were more profound and diversified in DCs; unexpectedly, however, Mϕs were found to respond to a wider range of pathogens. In-depth gene expression and functional mining of the data obtained in this study revealed the existence of functionally homogeneous groups of genes that are coordinately regulated in both cell types. This approach allowed us to pinpoint for the first time the participation of many known and newly described genes in the context of innate immunity to infection.
Materials and methods
In vitro generation of DCs and Mϕs
CD14+ peripheral blood–derived monocytes were isolated from leukopacks from healthy donors by counterflow centrifugal elutriation under protocols approved by the institutional review boards of both the National Institute of Allergy and Infectious Diseases and the Department of Transfusion Medicine of the National Institutes of Health for these studies. Informed consent was provided according to the Declaration of Helsinki.10 Fresh monocytes were cultured in 6-well tissue-culture plates at 2 to 3 × 106/mL (no. 3596 Costar; Fisher Scientific, Suwannee, GA) in complete RPMI 1640 (BioWhittaker, Walkersville, MD) supplemented with 20 mM glutamine (BioWhittaker), 10% heat-inactivated human AB serum (Gemini Bioproducts, Woodland, CA), 100 μg/mL penicillin, and 100 g/mL streptomycin (Biosource International, Rockville, MD). For generation of DCs, recombinant human (rh) IL-4 and recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF; PeproTech, Rocky Hill, NJ) were added to the culture at 50 ng/mL on days 1, 3, and 5 of culture. For generation of Mϕs, rhM-CSF (PeproTech) was added at 1000 μ/mL on the same days. Both cell types were exposed to the pathogens on day 8 of culture, and 16 hours later the cells were harvested and used for RNA preparation. DCs harvested at day 8 were repeatedly shown to be CD1a+, HLA-DR+, CD86+, CD40+, CD3-, CD14-/lo, CD19-, and CD56- by flow cytometry (FACSCalibur; Becton Dickinson, Sunnyvale, CA).
Parasite and mycobacteria preparation
B malayi live L3s isolated from the proboscises of infected Aedes aegypti were provided by Dr John McCall (University of Georgia, Athens). The L3s were then incubated in a 6-well plate in RPMI plus penicillin/streptomycin/amphotericin B, 200 mM l-glutamine, and gentamicin for 1 hour, following which the L3s were isolated individually, washed twice in the same media, counted, and replated in a 6-well plate prior to incubation with DCs or Mϕs.
Leishmania parasites were cultured at 22°C without CO2 in 199 medium supplemented with 20% heat-inactivated fetal calf serum (HyClone Laboratories, Logan, UT), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, 40 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (BioWhittaker), 0.1 mM adenine (in 50 mM HEPES), 5 mg/mL hemin (in 50% triethanolamine), and 1 mg 6-biotin/mL. Infective-stage metacyclic promastigotes were isolated from stationary culture (4 to 5 days old) by negative selection using peanut agglutinin1 (Vector Laboratories, Burlingame, CA) for L major (NIH Friedlin V1 strain MHOM/IL/80/FN) or monoclonal antibody (MoAb) MG-12 for L donovani (strain MHOM/IN/95/9515). Before infection, parasites were opsonized with 5% normal human serum by incubation at 37°C for 30 minutes. Parasites tested negative for Mycoplasma and below the detection limits for endotoxin.
Tachyzoites of the RH strain of T gondii were cultivated in human foreskin fibroblasts maintained in Dulbecco modified Eagle medium (DMEM; Life Technologies, Gaithersburg, MD) supplemented with 10% fetal calf serum (FCS; HyClone) and antibiotics.
Mycobacteria stocks from logarithmically growing cultures were frozen in phosphate-buffered saline (PBS) containing 10% glycerol, and representative vials were thawed and enumerated for viable colony-forming unit on Middlebrook 7H11 plates. Before in vitro infection, cryopreserved M tuberculosis aliquots were thawed and suspended in culture medium by vortexing briefly. M tuberculosis bacilli were sonicated to disrupt small aggregates of bacteria.
Exposure/infection of DCs and Mϕs
DCs and Mϕs from 7 donors were cultured at 2 to 5 × 106 cells/2 mL in 6-well tissue-culture plates and were separately exposed to the parasites for 16 hours. Infected cells were harvested at the conclusion of the experiment, cytospins were prepared and Wright-Giemsa stained, and infections were monitored by light microscopy. For B malayi, 2 to 5 × 106 DCs were exposed to 50 or 5 live L3s. For T gondii, cells were infected at a 3:1 parasite-to-cell ratio. For L major and L donovani, cells were infected at a 5:1 parasite-to-cell ratio. All the cells compared had similar infection ratios, ranging from 200 to 600 parasites/100 cells. For M tuberculosis, cells were infected at a bacteria-to-MOI (multiplicity of infection) ratio of 10:1.
RNA preparation and microarray hybridization
The RNA from each of the 7 donors was extracted independently, pooled, and used for subsequent labeling and hybridization. Total RNA was prepared using the RNAeasy mini kit (Qiagen, Valencia, CA), and used to generate cRNA probes. Preparation of cRNA, hybridization, and scanning of the HU95 arrays were performed according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). Labeled cRNA was hybridized to the Affymetrix HU95A microarray, and fluorescence was measured at 570 nm in an Affymetrix scanner. This entire procedure was repeated a second time, so that there were a total of 2 independent cRNA preparation and microarray hybridizations for each condition.
Microarray data processing
We chose a filtering strategy based on a fixed change in average difference (Ad) intensity values (IAd). This value represents a difference between the Ad intensities of a gene on one chip compared with another. This filtering approach was used in combination with a fixed 5-fold change cutoff. Details about the filtering method used in this study are available as supplemental material on the Blood website.
An algorithm used for mining the biomedical literature is described in detail elsewhere.11 Briefly, term occurrence frequencies in relevant abstracts are determined for each gene using a statistical package (Simstat/Wordstat; Provalis Research, Montreal, QC, Canada), and frequently occurring terms identified for each gene are selected without intervention of the investigator with the use of a standard but arbitrary filtering criterion. Occurrence patterns are analyzed for the resulting subset of terms by hierarchic clustering.
Real-time quantitative RT-PCR (Taqman)
The same pooled RNA used for microarray was used for real-time reverse transcriptase–polymerase chain reaction (RT-PCR). Predeveloped assay reagent (PDAR) probes, reagents, and equipment were used as recommended by the manufacturer (Applied Biosystem, Fullerton, CA).
Results
Constitutive gene expression in human monocyte-derived DCs and Mϕs
DCs and Mϕs were exposed to the pathogens on day 8 of culture, and 16 hours later the cells were harvested, lysed, and stored for subsequent RNApreparation. Because of donor-to-donor differences, human experimental systems can be prone to considerable variability. Therefore, cells from a total of 7 donors were prepared and infected independently. The RNA from all 7 donors was extracted separately; equivalent amounts of RNA from each donor were pooled and used for subsequent labeling and hybridization. We assessed the variability associated with the preparation of targets and processing of microarrays by replicating this procedure with another set of RNA from the same donors and a second set of microarrays (total of 2 chips for each condition; Supplemental Figure S1A). Raw signal values as well as fold-change values for this entire data set are publicly available (NCBI GEO accession no. GSE360).
The validity of our experimental design was tested further by comparing the basal expression values for immature (uninfected) DCs obtained in this study (7-donor pool performed twice) with another independent experiment using uninfected DCs from a separate pool of 4 donors (Supplemental Figure S1B). Furthermore, for a limited number of genes, microarray expression data were confirmed using real-time quantitative RT-PCR (Supplemental Figure S2).
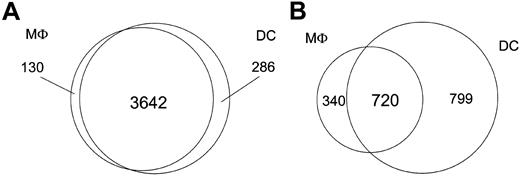
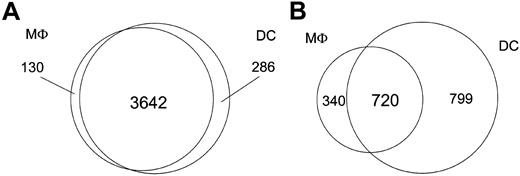
Although the focus of this study was to compare the transcriptional responses of DCs and Mϕs with both intracellular and extracellular eukaryotic parasites, we were able to gain insight into the gene expression profiles of these cell types in their steady state because both the DCs and Mϕs were derived from the same human monocyte donors. As such, DCs and Mϕs expressed comparable (96% similar) numbers of genes basally (3928 genes in DCs; 3772 genes in Mϕs). In addition to the 3642 genes that were transcriptionally active in both DCs and Mϕs, 130 were unique to Mϕs, with 286 genes being specific to DCs (Figure 1); however, we observed marked differences between Mϕs and DCs in the number of genes uniquely regulated following pathogen exposure. Only 40% of the regulated genes (720 genes) were shared by both cell types, whereas 340 were unique to Mϕs and 799 to DCs (data not shown).
Transcriptional differences between DCs and Mϕs. The Venn diagrams illustrate transcriptional differences between DCs and Mϕs. (A) Number of genes expressed by either DCs or Mϕs alone, or by both cell types at baseline before pathogen exposure. (B) Number of genes altered by either DCs or Mϕs alone, or by both cell types upon pathogen exposure.
Transcriptional differences between DCs and Mϕs. The Venn diagrams illustrate transcriptional differences between DCs and Mϕs. (A) Number of genes expressed by either DCs or Mϕs alone, or by both cell types at baseline before pathogen exposure. (B) Number of genes altered by either DCs or Mϕs alone, or by both cell types upon pathogen exposure.
As expected, we found genes such as CD1a, CD1b, CD1c, CD1e, and LAMP3 to be expressed only by DCs and not by Mϕs; conversely, among those genes known to be expressed uniquely by Mϕs, CSF3R expression could easily be seen. Among the 130 Mϕ-specific genes expressed at baseline were genes known to be involved in cell adhesion (EMILIN [elastin], CSPG2 [chondroitin sulfate proteoglycan], ITGBL1 [integrin b-like 1], and ITGA6 [integrin alpha 6]). Moreover, both proapoptotic (TRAF3, IL-1β) and antiapoptotic (API5 and TOSO) genes were also expressed uniquely by Mϕs. For DCs, similarly, there was unique expression of other cell adhesion molecules (CDH2), chemotaxins (SCYA13, SCYA17, and SCYD1), and molecules involved in apoptosis (BCL10 and CASP10) (Table 1). A complete list of constitutively expressed genes by both DCs and Mϕs can be found in the supplemental data (database accession numbers to come).
Pathogen-induced gene expression patterns in human monocyte-derived DCs and Mϕs
Global changes in gene expression have previously been reported for either Mϕs or DCs in response to influenza virus, yeast, bacteria, and bacterial products.1 In each of these studies, common and pathogen-specific alterations in transcriptional responses to infectious agents could be identified by comparing the transcriptional responses with both intracellular and extracellular eukaryotic parasites concurrently. In contrast with previously published work, we were able to compare the transcriptional responses elicited by a variety of pathogens in both DCs and Mϕs obtained from the same donors (Figure 2). We used the infective stages of 3 protozoa (L major, L donovani, T gondii) and one helminth (B malayi); Mϕ and DC responses to infection with M tuberculosis were detailed for comparison.
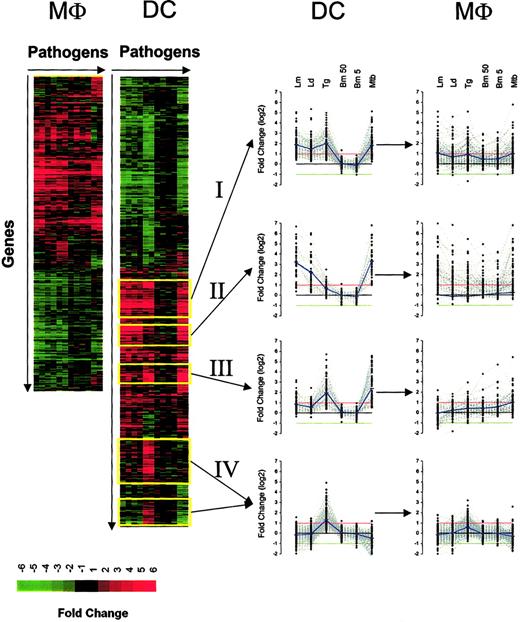
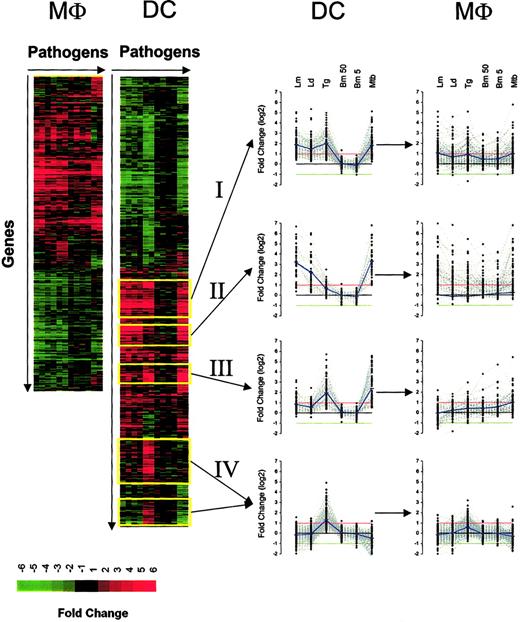
Hierarchic clustering of genes regulated in both DCs and Mϕs following pathogen exposure. Genes were arranged according to their gene expression pattern (fold change) by hierarchic clustering, with the genes arranged vertically and the pathogens (in replicate) arranged horizontally for DCs (left side) and Mϕs (right side). Red indicates gene induction; green, gene repression. Analysis was done for DCs and Mϕs independently. The expression profile of 4 major clusters (yellow outline) is also plotted for both DCs and Mϕs. The red and green lines in each graph (in middle) represent the 2-fold change values; blue lines represent the average fold-change values.
Hierarchic clustering of genes regulated in both DCs and Mϕs following pathogen exposure. Genes were arranged according to their gene expression pattern (fold change) by hierarchic clustering, with the genes arranged vertically and the pathogens (in replicate) arranged horizontally for DCs (left side) and Mϕs (right side). Red indicates gene induction; green, gene repression. Analysis was done for DCs and Mϕs independently. The expression profile of 4 major clusters (yellow outline) is also plotted for both DCs and Mϕs. The red and green lines in each graph (in middle) represent the 2-fold change values; blue lines represent the average fold-change values.
Despite DCs and Mϕs having more than 96% similarity in basal gene expression, here we found that their responses to various pathogens differed dramatically. With the use of expression data from genes that exceeded the defined filtering criteria for at least one condition and in replicate, we found that the number of transcriptionally altered genes was greater in DCs (1519 genes) than in Mϕs (1060 genes) (Figure 2). Furthermore, the responses to each of the pathogens were more complex and diversified in DCs for both induced and repressed genes. For example, on the basis of hierarchic clustering algorithms, there were approximately 12 major clusters in DCs for induced genes and only half as many in Mϕs; for the repressed genes, there were only 3 major clusters in Mϕs compared with 7 to 8 in DCs. Because DCs demonstrated the most diverse changes on exposure to the pathogens, we focused our attention on 4 distinct clusters (Figure 2): cluster I, genes induced by all intracellular pathogens; cluster II, genes induced by Leishmania and M tuberculosis (MTB); cluster III, genes induced by T gondii and M tuberculosis; and cluster IV, genes only induced by T gondii. It should be noted that, in general, the extracellular filarial parasite (B malayi) induced expression of very few genes (between 7 and 14 genes, depending on the number of L3s) in DCs.
Although most of the induced genes noted among the 4 major clusters were found to be regulated coordinately in both DCs and Mϕs, the expression pattern seen in each of these 2 cell types differed dramatically (Figure 2). Those genes induced by each of the intracellular pathogens in DCs (cluster I) were uniformly regulated (neither on average significantly up- or down-regulated). Similarly, for those sets of genes induced only by Leishmania spp and MTB in DCs (cluster II) for the Mϕs, at least, each of the pathogens (including extracellular B malayi) induced the same set of genes. This finding of DC-specific responses not being recapitulated in Mϕs held true for the other induced clusters as well.
Functional assignments of coordinately expressed gene profiles
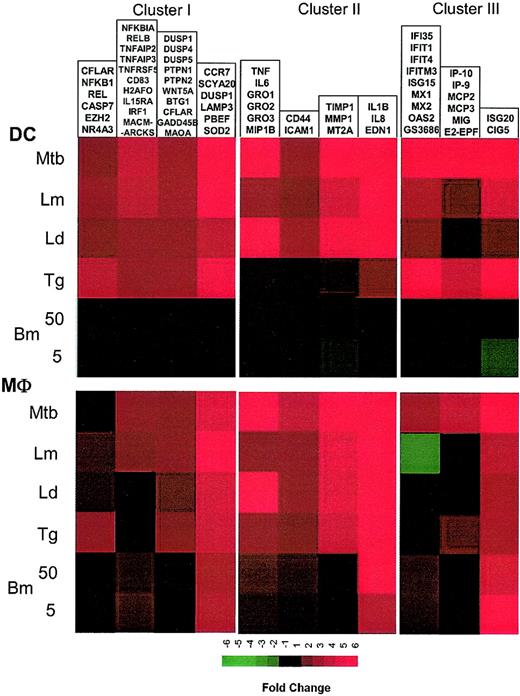
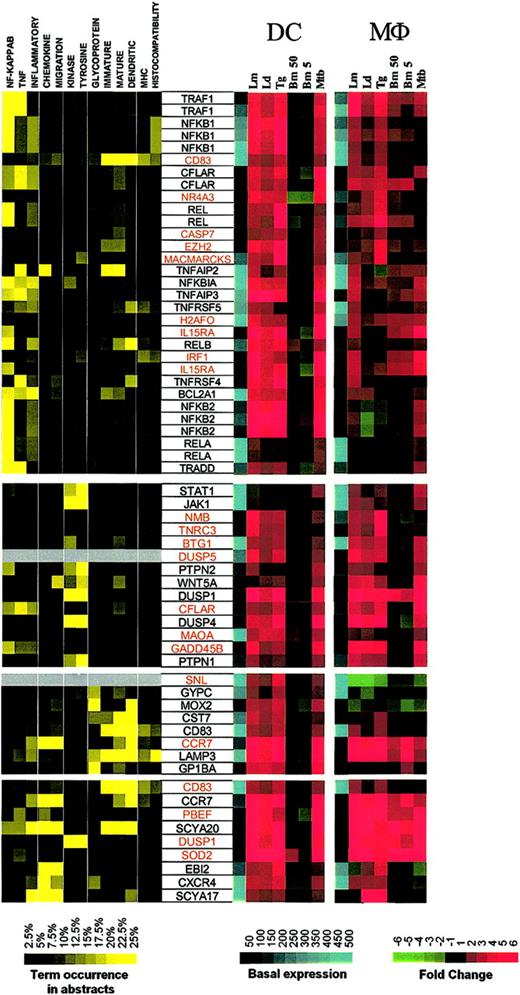
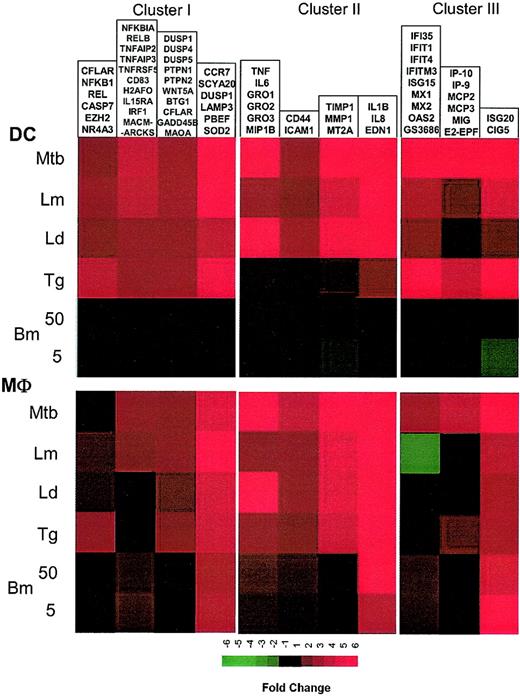
To identify the relationships among these coordinately expressed genes, a literature-mining algorithm was used to help order the data with respect to possible function among the 4 major induced clusters identified with the use of standard hierarchic clustering. The application of this algorithm to those genes induced in DCs by each of the intracellular pathogens (but not extracellular B malayi) demonstrated that cluster I (Figure 3; Supplemental Figure S3) included a large family of genes belonging to the NF-κB signaling pathway (NFKB1, NFKB2, REL, RELA, RELB, NFKBIA) as well as apoptosis regulators (TRAF1, CFLAR, BCL2A1, TRADD), and TNF-related molecules (TNFSF4/OX40, TNFSF5/CD40, TNFAIP2, TNFAIP3). A second group consists of kinases involved in cell growth and cytokine production (JAK1 [Janus kinase 1], STAT1 [signal transducer and activator of transcription 1], PTPN2 [protein tyrosine phosphatase, nonreceptor type 2], WNT5A [wingless-type MMTV integration site family, member 5A], DUSP1 [dual specificity phosphatase 1], DUSP4, PTPN1). A third and probably functionally unrelated group includes glycoproteins, 2 of which are known to be expressed primarily in mature DCs (CD83 and LAMP3). Of interest, another DC maturation marker, CCR7, was found to be associated with LAMP3 according to patterns of gene expression.
Cluster I: genes up-regulated by intracellular pathogens. Gene expression and literature profiles are shown for both DCs and Mϕs. Color intensity correlates with basal gene expression values (blue), fold-change values (green and red), and term occurrence in abstracts (yellow). Genes listed in red were not part of the original functional cluster and were integrated on the basis of their expression profile similarity to other genes in this cluster. Names for the genes abbreviated in this figure can be found in supplemental Table 1.
Cluster I: genes up-regulated by intracellular pathogens. Gene expression and literature profiles are shown for both DCs and Mϕs. Color intensity correlates with basal gene expression values (blue), fold-change values (green and red), and term occurrence in abstracts (yellow). Genes listed in red were not part of the original functional cluster and were integrated on the basis of their expression profile similarity to other genes in this cluster. Names for the genes abbreviated in this figure can be found in supplemental Table 1.
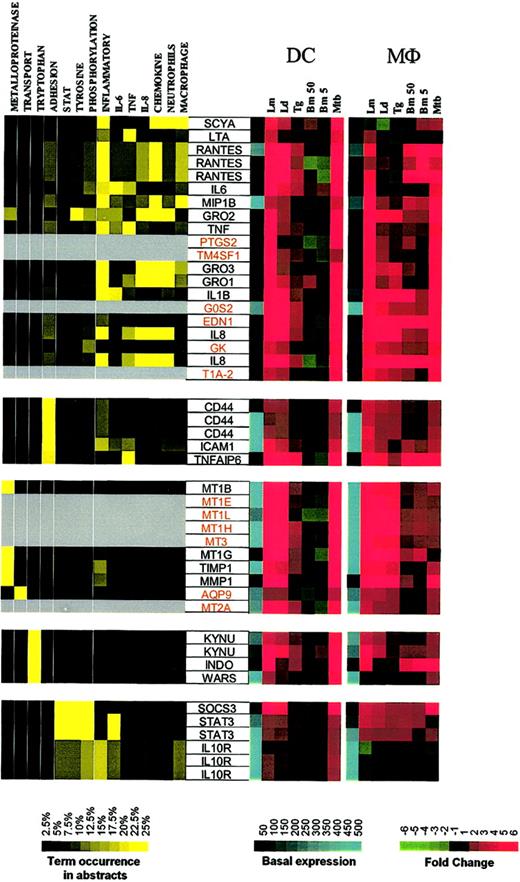
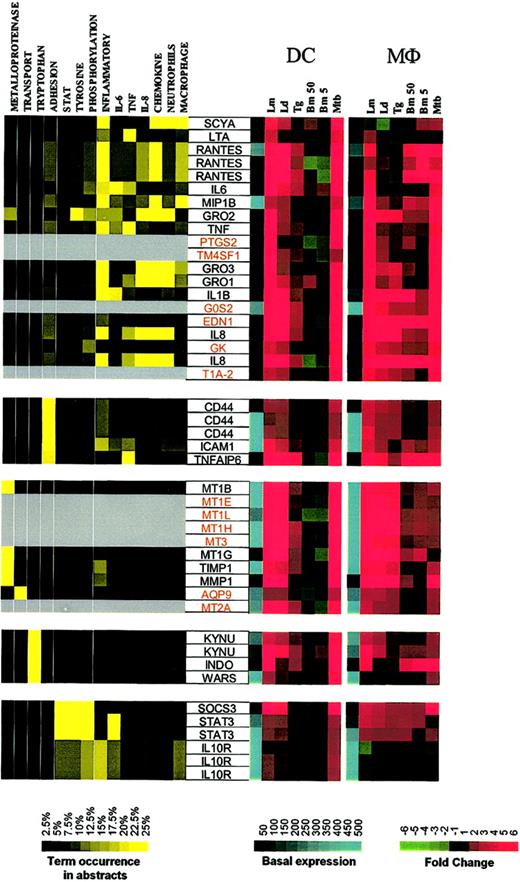
The second major expression cluster consists of genes in DCs whose expression is induced primarily by Leishmania spp and M tuberculosis and can be subgrouped on the basis of function (Figure 4; Supplemental Figure S4). The first subgroup consists of inflammatory mediators (RANTES, MIP-1β, Gro1-3, IL-8, IL-6, IL-1β, TNF, LTA). Some of these genes are clearly related to inflammatory responses, with PTGS2 (prostaglandin-endoperoxide synthase 2) being a key enzyme in prostaglandin biosynthesis, while EDN1 (endothelin 1) has been described as a proinflammatory cytokine.12 Moreover, we demonstrate a strong induction of EDN1 expression in both DCs and Mϕs, suggesting that both of these cell types are an important but heretofore unrecognized source of endothelin. Although not formally linked to inflammatory processes, the other genes in this major subgroup include TM4SF1 (transmembrane 4 superfamily member 1), a tumor-associated cell surface protein involved in the regulation of cell development, activation, growth, and motility; GK (glycerol kinase); G0S2; and T1A-2, a mucin-type lung type-I cell membrane–associated glycoprotein previously found to be expressed only by endothelial and epithelial cells. The present data suggest that perhaps these (and other coordinately regulated) genes may have an unrecognized role in DC- and Mϕ-mediated inflammation.
Cluster II: genes up-regulated by Leishmania spp and M tuberculosis. Gene expression and literature profiles are shown for both DCs and Mϕs. Color intensity correlates with basal gene expression values (blue), fold-change values (green and red), and term occurrence in abstracts (yellow). Genes listed in red were not part of the original functional cluster and were integrated on the basis of their expression profile similarity to other genes in this cluster. Names for the genes abbreviated in this figure can be found in supplemental Table 2.
Cluster II: genes up-regulated by Leishmania spp and M tuberculosis. Gene expression and literature profiles are shown for both DCs and Mϕs. Color intensity correlates with basal gene expression values (blue), fold-change values (green and red), and term occurrence in abstracts (yellow). Genes listed in red were not part of the original functional cluster and were integrated on the basis of their expression profile similarity to other genes in this cluster. Names for the genes abbreviated in this figure can be found in supplemental Table 2.
Other functional subgroups within this cluster include adhesion molecules (CD44, ICAM1, TNFAIP6 [TNF-α–induced protein 6]), metalloproteinases and their inhibitors (metallotheineins, TIMP, MMP1), antibacterial genes involved in tryptophan metabolism and known to be IFN-γ dependent (KYNU [L-kynurenine hydrolase]), INDO (indoleamine 2,3-dioxygenase), and WARS (tryptophanyl-tRNA synthetase).
Another important expression cluster includes those genes that are coordinately but differentially regulated by each of the pathogens under study (Figure 5A; Supplemental Figure S5). This cluster consists of genes highly induced by T gondii and M tuberculosis, moderately by L major, slightly by L donovani, or with no change by B malayi L3. The induced genes can be subcategorized into 3 functional subgroups, the most important of which consists of interferon-related genes involved in signaling (STAT-1, STAT-4, IRF4 and IRF7, NMI, IFP35), antiviral activities (OAS2, OASL, MX1, MX2, ISG15, ISG20), or proliferation (IFITM1). Associated with this subgroup as well is MyD88, a finding that may suggest that the Toll/IL-1R family of receptors13 and other interferon-induced genes in this subfamily are under common transcriptional control. Of interest, most of the genes in this subgroup were also found to be transcriptionally regulated in Mϕs as well, although the pattern of regulation was quite different than that seen in DCs (Figure 5B).
Cluster III: genes up-regulated by T gondii and M tuberculosis. (A) Gene expression and literature profiles are shown for both DCs and Mϕs. The color intensity correlates with basal gene expression values (blue), fold-change values (green and red), and term occurrence in abstracts (yellow). Genes listed in red were not part of the original functional cluster and were integrated on the basis of their expression profile similarity to other genes in this cluster. (B-C) Expression profiles are shown for groups of genes that belong to cluster II and are coordinately regulated in both cell types. The red and green lines in each profile represent the 2-fold change values; blue lines represent the average fold-change values. Names for the genes abbreviated in this figure can be found in supplemental Table 3.
Cluster III: genes up-regulated by T gondii and M tuberculosis. (A) Gene expression and literature profiles are shown for both DCs and Mϕs. The color intensity correlates with basal gene expression values (blue), fold-change values (green and red), and term occurrence in abstracts (yellow). Genes listed in red were not part of the original functional cluster and were integrated on the basis of their expression profile similarity to other genes in this cluster. (B-C) Expression profiles are shown for groups of genes that belong to cluster II and are coordinately regulated in both cell types. The red and green lines in each profile represent the 2-fold change values; blue lines represent the average fold-change values. Names for the genes abbreviated in this figure can be found in supplemental Table 3.
A second subgroup in this DC cluster includes the CXC chemokines (IP9, IP10, MIG) and the CC chemokines (MCP2, MCP3). MCP2, MIG, IP9, and IP10 are also transcriptionally coregulated in Mϕs. Within this subgroup, however, the chemokine expression pattern for DCs and Mϕs are shown to be similar (Figure 5C), a finding that contrasts with what was seen with the interferon-induced genes mentioned earlier. In addition, E2-EPF, a ubiquitin-conjugating enzyme, was found to share the same profile as these chemokines.
The third and very important subgroup in this expression cluster consists of genes involved in major histocompatibility complex (MHC) class I antigen processing and presentation. These genes are linked with protein degradation (PSMA2, PSMA3), MHC-peptide assembly and transport (B2M, TAPBP), and antigen presentation (HLA-A, -C, -F, -G, and -J). As shown in Figure 5A, these genes are highly expressed in unexposed DCs and Mϕs, but nonetheless they are up-regulated in response to T gondii and M tuberculosis in DCs only.
An additional cluster consisting of genes specifically induced by T gondii in DCs was also identified and consisted of a large number of functional subcategories involved in cellular and subcellular processing (data not shown). These subcategories included groups of genes involved in cell structure, protein and vesicle trafficking, cell-cycle regulation, transcriptional and translational machinery, and apoptosis. Genes belonging to the TNF superfamily (eg, TNFSF4-OX40L, TNFRSF9-4-1BB) were also regulated coordinately and specifically by T gondii.
Pathogen-specific signatures in DCs and Mϕs
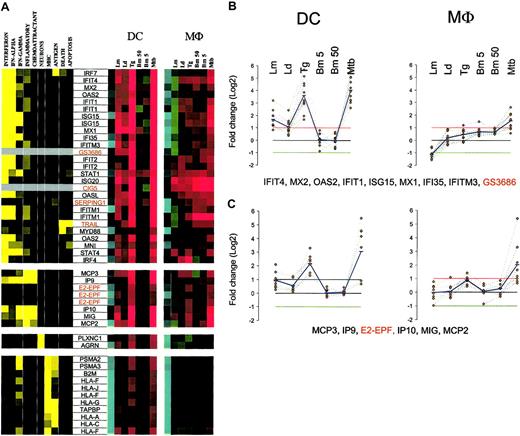
By averaging the changes in mRNA expression among genes sharing similar expression profiles for both DCs and Mϕs, pathogen-specific signatures can be clearly noted in either DCs or Mϕs (Figure 6A-B). By using the set of interferon-induced and “inflammatory” genes, unique and pathogen-specific expression profiles can be constructed for both DCs and Mϕs. Even in Mϕs, which have previously been shown to have relatively uniform expression/induction of proinflammatory genes by bacteria and their products,2,3 we show here that Mϕs have quite unique responses following exposure/infection to different protozoan and helminth parasites (and in contrast to an intracellular mycobacteria). For example, genes known to be expressed in response to IFN differ dramatically when the stimulus for expression is infection with different pathogens (M tuberculosis, Leishmania versus T gondii versus B malayi). Indeed, these differences extended even to different species of the same genus (L major compared with L donovani). This complex but pathogen-specific pattern of gene expression was equally evident in DCs (Figure 6A). Moreover, in contrast to all the other pathogens studied, the transcriptional response to Brugia was almost silent.
Coregulated transcriptional components of the innate immune response to infection. Expression values of groups of genes coordinately regulated in both cell types are averaged and are represented based on color intensity. Data derive from clusters I, II, and III.
Coregulated transcriptional components of the innate immune response to infection. Expression values of groups of genes coordinately regulated in both cell types are averaged and are represented based on color intensity. Data derive from clusters I, II, and III.
Discussion
Infections with helminth and protozoan parasites have prototypical and distinct outcomes that can often be directly related to the nature of the innate responses that occur at the initiation of infection. The power of the present study was in its ability to evaluate gene expression in 2 of the major effector cells of the innate response (DCs and Mϕs derived from the same donors) both in their steady state and in response to organisms that were not only phylogenetically diverse but that also have differing cell type requirements, intracellular localization, and mechanisms of cell entry. Moreover, all the pathogens used in this study are characterized by their capacity to establish chronic infection in humans and to induce a distinct but broad range of immunologic responses.
Having the ability to assess DCs and Mϕs derived from identical donors in parallel allowed us to minimize individual-to-individual variability and to demonstrate that, of the 12 000 genes expressed on a high-density oligonucleotide microarray, approximately 4000 genes are expressed in both DCs and Mϕs at baseline, with only 130 genes being Mϕ specific and 286 being uniquely expressed by DCs (Figure 1). Proteomic approaches or less dense DNA microarrays have also been used to compare the baseline transcriptome of human CD14+ blood monocytes and their derived DCs.14 With the use of DCs from 3 different times during their maturation process (days 1, 7, and 14), 40% of the 6300 genes sampled were shown to be expressed in DCs, with approximately 4% having altered regulation during the process of DC differentiation. These regulated genes were, in large part, related to cell adhesion and motility, cell growth, and broadly to the regulation of the immune response. In another study specifically examining expression of the NF-κB/Rel pathway during DC and Mϕ differentiation from monocyte precursors, it has been demonstrated that Mϕ- and lipopolysaccharide (LPS)–matured DCs differ from each other both at baseline and in response to at least one differentiation signal.15
The differential responsiveness by both DCs and Mϕs to the parasite pathogens was perhaps the hallmark of the present study. Furthermore, it is in the DC and Mϕ responses to each of the infectious parasitic microorganisms that the diversity of the cellular responses among the varied parasites could be detailed and compared more broadly. Indeed, we were able to identify discrete expression signatures induced by individual sets of pathogens for both cell types (DC or Mϕ) studied. Notably, although there were sets of genes coordinately induced by a given pathogen, the pattern of induction often differed between Mϕs and DCs (Figures 2 and 6). This difference in cell type–specific coordinate regulation of gene expression was most notable for interferon-induced genes (Figure 5B).
Many factors have been shown to influence the differentiation and development of DCs from their precursor cells. These factors include a wide range of stimuli such as corticosteroids,16,17 IFN-α, and IFN-β.18 Pathogen-specific molecules such as LPS, peptidoglycans, and lipoproteins trigger DC activation and maturation through Toll-like receptors (TLR)19 and induce production of IL-12 and other cytokines as well as responsiveness to chemokines that promote migration of DCs from peripheral tissues to lymphoid organs. The process of DC activation following infection with parasites is less well understood. Murine fetal skin–derived DCs,20,21 murine splenic DCs,22,23 and blood-derived human DCs24 produce IL-12 following infection with Leishmania. Of interest, soluble Toxoplasma antigen (STAg) stimulates IL-12 production by splenic DCs as well as inducing maturation and migration of DCs from the red pulp and marginal zones of the spleen into the T-cell region.25 Live Toxoplasma infection has been shown to induce prototypic DC maturation26 ; however, STAg has no effect on the production of IL-12 in monocyte-derived DCs.27 Furthermore, M tuberculosis infection of human DCs resulted in increased surface expression of costimulatory molecules as well as secretion of IL-12 by these cells.28 Other stages of the B malayi life cycle have also been shown to increase the expression of some costimulatory molecules but not lead to production of IL-12 by these cells.27 Although all intracellular parasites have been found to induce DC maturation as measured by a limited set of parameters, this study demonstrates major differences at the level of global gene expression.
To simulate physiologic conditions more precisely, all cells and pathogens were cultured in the presence of human heat-inactivated AB serum (rather than fetal calf serum). Therefore, it is possible that macrophages and dendritic cells respond to opsonic ligands as well as to microbial molecules, although the control cells (not exposed to pathogens) to which all of the infected/exposed cells were compared, were cultured in AB serum as well. Preopsonization occurred only with the leishmanial organisms, as this is how leishmanial infections have commonly (and historically) been performed because it both resembles the normal physiologic state and because, for macrophages, at least, entry occurs through a complement receptor–mediated process.
T gondii and M tuberculosis triggered the expression of many interferon-inducible genes in DCs; in contrast, these same genes were not induced by T gondii in Mϕs from the same donors. A study using microarrays to monitor global transcriptional responses to interferons in a fibroblast line has shown that both type I and II interferons (IFN-α/β and IFN-γ, respectively)29 were directly capable of inducing a set of genes similar to that found in the present study. More recently, M tuberculosis has been shown to induce type I interferons in human monocyte–derived DCs,30 with IFN-β being released early in in vitro infection and triggering an IFN-α response. The up-regulation of the interferon-inducible genes we observed 16 hours after M tuberculosis infection could, therefore, be attributed to an initial release of IFN-β. We observed a comparable transcriptional response in DCs exposed to T gondii, suggesting that a similar response might be induced by this protozoan parasite. In addition, we found several NF-κB family members (NFKB1, NFKB2, IKBA, REL, RELA, RELB) to be up-regulated in DCs by all intracellular pathogens. In contrast, many of the genes known to be under NF-κB transcriptional control were only induced following infection with Leishmania spp and M tuberculosis. That T gondii failed to induce this pathway may relate to the parasite's ability to block the translocation of NF-κB to the nucleus, as has been shown in murine Mϕs.31,32
Although most of the genes altered by exposure to Leishmania spp were common between the 2 species studied, a small number of genes were found to be differentially regulated by the 2 closely related species. Most notable was the specific down-regulation of a subset of IFN-γ–induced genes in Mϕs by L major infection. The leishmaniases are a group of diseases that result in diverse clinical manifestations. The varied clinical outcomes are attributed primarily to differences in the Leishmania species initiating the infections; for example, visceral leishmaniasis is caused by species of the L donovani complex, and infection with L major results mainly in self-limiting cutaneous lesions. It is intriguing that the genes in the differentially regulated cluster are known to be involved directly or indirectly in host defense33,34 ; it is possible that intrinsic differences between these Leishmania species in their ability to activate these genes may partially account for the evolution of healing and nonhealing forms of leishmanial disease. These genes are known to be interferon inducible35 and are transcriptionally regulated through promoter interferon stimulatory response element (ISRE) motifs.36-41 Notably, 2 other genes known to be regulated by this element, IP-10 and ISG20, were not down-regulated by L major infection. ISG20 expression was induced nearly 5-fold by L major in Mϕs and 9-fold in DCs. A unique ISRE element has been identified in the ISG20 promoter that mediates the induction of both type I and type II IFN.42
Differences between L major and L donovani also pinpointed signal transduction pathways, as these pathways appeared to be differentially used by the 2 different Leishmania species. Both Mϕs and DCs had increased expression of genes associated with an inflammatory response (Figure 6); at a quantitative level, L major was much more capable of inducing such a response than was L donovani. These results are in agreement with previous data that suggest that L major is a much stronger inducer of the inflammatory response,43 perhaps accounting for the localized containment of L major parasites to the lesion site. Moreover, the data published in mice indicate a general suppression in Mϕ gene expression after leishmanial infection4 ; however, our results indicate that human Mϕs and DCs do not behave in this manner, as similar numbers of genes were up-regulated as were down-regulated in both cell types by both Leishmania species.
Another important but perhaps expected finding was the minimal alteration in gene expression by the extracellular Brugian parasite, particularly in DCs. The role of DCs in filarial infection has not been fully understood. It has been shown that nematode infections are commonly associated with Th2 cytokines and hyporesponsive antigen-reactive T cells. We have previously shown that antigens from a different stage of the B malayi life cycle, microfilariae, impair the maturation and function of human DCs.27 In this report, we show that, in general, Brugia altered fewer genes and with lower-fold changes than each of the other intracellular pathogens. Although there are only a small number of genes (7 to 14 genes) altered by exposure of DCs to different doses of L3, approximately 90 to 130 genes were changed in Mϕs by the same doses of this parasitic stage. Our data indicate that inflammatory genes in Mϕs predominate in the response to L3. Confounding the interpretation of these data is the fact that many filarial parasites have intracellular bacteria (Wolbachia) present44 that could obviously play a role in induction of an inflammatory response. It is unlikely, however, that up-regulation of inflammatory genes in Mϕs by L3 is due to Wolbachia, because live worms were used in this study, and these intracellular bacteria are rarely released until the worm dies.44
The role of Mϕs in filarial infection has been studied in great detail.45 Mϕs can be activated to kill filarial parasites or can play a suppressor role in filarial infection by producing down-regulatory cytokines such as IL-10, which can counter the effects of IFN-γ.46 In our microarray analysis, macrophage inflammatory protein 3β (MIP-3β) was one of the genes up-regulated only by L3 in Mϕs; as a chemokine that up-regulates the production of IL-10 in monocytes,47 MIP-3β induction by the filarial parasite may provide insight into a strategy used by the parasite to modulate the host immune response.
Delineating the response profiles to various pathogenic parasitic microorganisms in the 2 cell types (Mϕs and DCs) that provide the link between the innate and adaptive immune response has identified both cell- and pathogen-specific sets of coordinately regulated genes that would not have been predicted by their phylogenetic relationships. In addition, using a newly described data analysis tool, we have been able to assign functions to many of these sets of coordinately regulated genes. By linking characteristic functional clusters with a particular pathogen studied, our data suggest that antigen-presenting cells have prewired signaling patterns for use in the response to different pathogens. This intrinsic variability in host responsiveness has broad implications for the identification of strategies used by the host to contain the pathogens and by the parasite to maintain survival and chronicity.
Although a broader selection of pathogens may be necessary to draw a more definitive conclusion, these types of comparative studies can provide valuable insights into the mechanisms governing T-cell polarization. For example, we observed a positive relationship between the capacity of pathogens to up-regulate IFN-responsive genes and induction of Th1 responses (T gondii, M tuberculosis > L major > L donovani > > > B malayi); however, such relationships were not observed within the other gene clusters.
In a more general sense, this study allows us to unravel new aspects of host/pathogen interaction. Previous studies have demonstrated that infection with intracellular pathogens such as T gondii, L major, and M tuberculosis results in up-regulation of prototypic DC maturation markers.20-26 Although our data confirm the previous findings (cluster I up-regulated by all intracellular pathogens), microarray analysis reveals major differences at a global level (clusters II, III, IV). In addition, we identified many genes that were not previously associated with innate response to infection, providing valuable leads for future studies.
Prepublished online as Blood First Edition Paper, March 27, 2003; DOI 10.1182/blood-2002-10-3232.
D.C., R.T.S., and M.A.M. contributed equally to this study.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Richard Lempicki and Jun Young for microarray hybridization, Paul Keiser for Brugia L3 preparation, Joseph Kubofcik for his help in the real-time RT-PCR experiments, and Sara Hieny for preparing Toxoplasma gondii. We also thank Brenda Rae Marshall for editorial assistance.