Abstract
The expression of ferroportin1 (FPN1) in reticuloendothelial macrophages supports the hypothesis that this iron-export protein participates in iron recycling from senescent erythrocytes. To gain insight into FPN1's role in macrophage iron metabolism, we examined the effect of iron status and erythrophagocytosis on FPN1 expression in J774 macrophages. Northern analysis indicated that FPN1 mRNA levels decreased with iron depletion and increased on iron loading. The iron-induced induction of FPN1 mRNA was blocked by actinomycin D, suggesting that transcriptional control was responsible for this effect. After erythrophagocytosis, FPN1 mRNA levels were also up-regulated, increasing 8-fold after 4 hours and returning to basal levels by 16 hours. Western analysis indicated corresponding increases in FPN1 protein levels, with maximal induction after 10 hours. Iron chelation suppressed FPN1 mRNA and protein induction after erythrophagocytosis, suggesting that FPN1 induction results from erythrocyte-derived iron. Comparative Northern analyses of iron-related genes after erythrophagocytosis revealed a 16-fold increase in FPN1 levels after 3 hours, a 10-fold increase in heme oxygenase-1 (HO-1) after 3 hours, a 2-fold increase in natural resistance macrophage-associated protein 1 (Nramp1) levels after 6 hours, but no change in divalent metal ion transporter 1 (DMT1) levels. The rapid and strong induction of FPN1 expression after erythrophagocytosis suggests that FPN1 plays a role in iron recycling. (Blood. 2003;102:4191-4197)
Introduction
Macrophages of the reticuloendothelial system (RES) play a major role in iron metabolism by recycling iron from red blood cells. Iron recycling is achieved through the phagocytosis of damaged or senescent erythrocytes, with the subsequent return of iron to the circulation and ultimately to the erythron, where the metal is incorporated into heme during red cell maturation. Each day, the RES ingests an estimated 360 billion senescent erythrocytes,1 recycling approximately 25 mg iron into the circulation.2 Although recycling of iron by the RES represents the largest flux of iron within the body, the precise mechanisms of intracellular iron handling and iron release from the macrophage remain poorly defined.3
Abnormal iron metabolism in the RES is a hallmark feature of common disorders such as hereditary hemochromatosis (HH) and the anemia of chronic disease (because of prolonged inflammation, infection, or other underlying diseases). In HH, macrophages of the liver, spleen, and bone marrow contain little iron compared with those of subjects with secondary iron overload,4,5 suggesting that altered RE iron metabolism might be a primary defect of this disease. The paradoxically low iron stores in the RES of patients with HH appear to result from increased iron release. After intravenous injection of 59Fe-labeled heat-damaged red blood cells, patients with HH were shown to release elevated amounts of 59Fe.6 Similarly, isolated monocytes from patients with HH were found to release twice as much 59Fe in a low molecular weight form as control cells after ingesting 59Fe-labeled erythrocytes.7 The anemia of chronic disease, in contrast, is characterized by decreased macrophage iron release and increased RE iron stores.8 Impaired RE iron release from 59Fe-labeled red cells has been documented in rat models of acute infection9 and inflammation,10 as well as in patients with inflammation.6 The inhibition of iron release in vitro has also been observed in inflammatory mouse peritoneal macrophages11 and J774 macrophages treated with lipopolysaccharide (LPS).12
Our understanding of iron metabolism has been greatly advanced by the recent identification and characterization of transmembrane iron transport proteins such as natural resistance macrophage-associated protein 1 (Nramp1; also known as Slc11a1), divalent metal ion transporter 1 (DMT1; also known as Nramp2, divalent cation transporter 1 [DCT1], or Slc11a2), and ferroportin 1 (FPN1; also known as metal transport protein 1 [MTP1], iron-regulated transporter 1 [IREG1], or Slc11a3). Nramp1, which is expressed exclusively in monocytes and macrophages, localizes to lysosomes and late endosomes and is rapidly recruited to membranes of maturing phagosomes.13 The main function of Nramp1 appears to be the transport of iron and other metals into and/or out of the phagosome.14 DMT1, the apical membrane iron transporter of the duodenum,15 also localizes to recycling endosomes where it transports iron from transferrin into the cytosol.16,17 Like Nramp1, DMT1 has been shown to become recruited to the phagolysosome in J774 cells,18 suggesting that this protein plays a role in intracellular iron handling in the macrophage as well. However, the potential roles Nramp1 and DMT1 play in iron recycling remain unknown.
FPN1 is the first cellular iron exporter to be identified.19-21 Efflux of iron mediated by FPN1 has been demonstrated by increased iron export in iron-loaded Xenopus oocytes expressing FPN1,20,21 and by cellular iron depletion in HEK293T cells transiently transfected with the full-length FPN1 cDNA.19 In duodenal epithelial cells, FPN1 localizes to the basolateral membrane where it is believed to export iron from the duodenum into the portal blood circulation.19-21 The observation that FPN1 is particularly abundant in RE macrophages of the liver, spleen, and bone marrow22 supports the hypothesis that this protein may also function to export from the RES iron derived from senescent erythrocytes. Consistent with this possibility are recent clinical reports of iron accumulation in the RES of patients with mutations in FPN1.23-27 The purpose of the present study was to explore the role of FPN1 in iron recycling by examining the effect of iron status on FPN1 expression in the murine J774 macrophage cell line, particularly after phagocytosis of red cells, when iron liberated from heme significantly expands the cellular iron pool.
Materials and methods
Cell culture
The mouse macrophage cell line J774, clone E,28 was kindly provided by Dr Philip Stahl (Washington University Medical School, St Louis, MO) and grown in alpha-minimum essential medium (MEM; Mediatech, Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Norcross, GA), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2.
Treatment of cells
To deplete J774 cells of iron, the chelators desferrioxamine mesylate (DFO) or salicylaldehyde isonicotinoyl hydrazone (SIH) were added to the cell culture media to achieve final concentrations of 0 to 200 μM, and cells were incubated for 20 hours. DFO (Novartis Pharma, Basel, Switzerland) was prepared as a 100-mM stock solution in water, and SIH (a generous gift from Dr Prem Ponka, Lady Davis Institute for Medical Research, Sir Mortimer B. Davis Jewish General Hospital, McGill University, Quebec, Canada) was prepared as a 100-mM stock in dimethyl sulfoxide (DMSO; Sigma-Aldrich, St Louis, MO). Sterile water and DMSO were used as controls for studies using DFO and SIH, respectively.
Iron loading of macrophages was achieved by incubating cells for 20 hours in the presence of Fe-nitrilotriacetate (Fe-NTA; 0-400 μM). Fe-NTA (molar ratio of 1:4) was prepared as a 20-mM stock from NTA and ferric chloride hexahydrate29 (Sigma-Aldrich). To inhibit transcription, cells were treated for 8 to 16 hours with actinomycin D (1 μg/mL; Sigma-Aldrich) in the presence or absence of 200 μM Fe-NTA.
Erythrophagocytosis by J774 macrophages
Preparation of erythrocytes. Rat and human erythrocytes were used in this study. Rat blood (gift from Dr Ramon Molina, Harvard School of Public Health) was collected sterilely and mixed 1:1 in Alsever solution (Sigma-Aldrich). Human venous blood collected from a healthy volunteer was mixed 1:1 in Alsever solution. After centrifugation at 1000g for 5 minutes at 4°C, plasma and buffy coat were removed, and erythrocytes were washed 3 times in Alsever solution. Collection and use of human red blood cells was approved by the Institutional Review Board of the Harvard School of Public Health.
Opsonization of erythrocytes. Rat erythrocytes were opsonized by incubating 1 × 109 red cells with rabbit antirat red blood cells, immunoglobulin G (IgG) fraction (1:20; Research Diganostics, Flanders, NJ). Human erythrocytes were opsonized by incubating 1 × 109 red cells with goat antihuman red blood cell, IgG fraction (1:10; Strategic Biosolutions, Newark, DE). Erythrocytes were incubated with IgG for 15 minutes at 37°C and then washed twice in 20 volumes of Alsever solution.
Phagocytosis. Opsonized erythrocytes were added to the J774 cell monolayer and incubated for 1.5 to 2 hours at 37°C (details in legends to Figures). Noningested opsonized erythrocytes were removed by performing a 10-second hypotonic lysis with water (double processed, tissue-culture water; Sigma-Aldrich), followed by 2 washes with Dulbecco phosphate-buffered saline (PBS; Sigma-Aldrich). Control cells were subjected to the same lysis and washing steps as cells treated with erythrocytes. All solutions used were sterile filtered and endotoxin tested. At various times after erythrophagocytosis, cells were collected for Northern or Western analysis as described.
Northern blot analysis
Cell monolayers were washed once in PBS, and RNA was isolated by using RNABee (TelTest, Friendswood, TX) according to the manufacturer's protocol. Northern blot analysis was performed as described previously.30 The following plasmids were digested with restriction enzymes as indicated, and the cDNA fragments produced were used as templates to synthesize 32P-labeled probes for Northern blot analysis: pCMV-SPORT6-mouse ferroportin (GenBank accession no. BE554084, ATCC no. 5382233; ATCC, Manassas, VA) was cut with SalI and NotI; pBluescriptSK(-)-human β-actin (GenBank accession no. AA173249, ATCC no. 769559; ATCC) was cut with EcoRI and XhoI; pT7T3D-Pac-rat transferrin receptor 1 (TfR1; GenBank accession no. AI454017; Resgen, Huntsville, AL) was cut with EcoR1 and NotI; pT7T3D-pac-rat DMT1 (GenBank accession no. AA819678, ATCC no. 3811001; ATCC) was cut with EcoR1 and NotI; pBluescriptSK(-)-mouse Nramp1 (GenBank accession no. AI265129, ATCC no. 1966896; ATCC); pBSK-heme oxygenase-1 (HO-1)31 (a kind gift from Dr Stella Kourembanas, Brigham and Women's Hospital, Boston, MA) was cut with HindIII. All cDNAs used for random priming were verified by sequencing.
Western blot analysis
Western analysis of ferritin. Cells were harvested, pelleted by centrifugation at 800g for 5 minutes at 4°C, and washed twice in 20 volumes ice-cold PBS. Cells were lysed in 4 volumes of ice-cold lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris (tris(hydroxymethyl)aminomethane), pH 8.0, phenylmethylsulfonyl fluoride [PMSF; 100 μg/mL], aprotinin [1 μg/mL], leupeptin [1 μg/mL], pepstatin [1 μg/mL]). Cell lysates were kept on ice and further disrupted manually (for approximately 20 seconds) using a polypropylene pellet pestle designed for use with 1.5-mL microcentrifuge tubes (Kimble/Kontes, Vineland, NJ). After centrifugation of lysates at 100 000g for 5 minutes at 4°C, supernatants were removed, snap frozen in liquid nitrogen, and stored at -80°C until analysis. Total protein content was determined by Bradford assay. Samples (200 μg) were mixed with Laemmli buffer (1 × final concentration), and heated at 95°C for 5 minutes, and proteins were electrophoretically separated on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel. The protein was transferred to a Protran nitrocellulose membrane (Schleicher & Schuell, Keene, NH), which was then preincubated with blocking solution (5% nonfat dry milk in Tris-buffered saline containing 0.01% Tween20 [TBST], pH 7.4) for 1 hour, followed by a 1-hour incubation with sheep antihuman ferritin (1:1000; The Binding Site, Birmingham, United Kingdom) primary antibody. After several washes with TBST, the blot was incubated with a 1:10 000 dilution of secondary antibody (horseradish peroxidase-linked rabbit antisheep antibody; Pierce, Rockford, IL) for 45 minutes. All incubations and wash steps were performed at room temperature. Cross-reactivity was visualized by using enhanced chemiluminescence (SuperSignal WestPico; Pierce) and quantified using QuantityOne application software (Bio-Rad, Hercules, CA).
Western analysis of FPN1. Cell monolayers were washed once in ice-cold PBS and lysed in ice-cold SDS lysis buffer (170 mM Tris-HCl [pH 6.8], 2% SDS, 5% glycerol, 5% β-mercaptoethanol, and protease inhibitors). Protease inhibitors (Complete, Mini protease inhibitor cocktail; Roche, Indianapolis, IN) were used as recommended by the manufacturer for samples with high proteolytic activity. Lysis buffer (200 μL) was used to lyse cells in each well of a 6-well plate. Cell lysates were kept on ice, sonicated for 10 seconds, snap frozen in liquid nitrogen, and stored at -80°C until analysis. Protein content was determined using the RC DC Protein Assay (Bio-Rad). Samples (100 μg) were mixed with bromophenol blue (0.01%), and without prior heating, proteins were electrophoretically separated on a 10% SDS-polyacrylamide gel. Molecular weight standards (Precision Plus Protein standards, All Blue; Bio-Rad) were run in parallel. The protein was transferred onto a Protran or Optitran nitrocellulose membrane (Schleicher & Schuell), which was then preincubated with blocking solution (described earlier), followed by overnight incubation at 4°C with a 1:2500 dilution of affinity-purified rabbit anti-FPN1 antibody.19 After washing with TBST, the blot was incubated at room temperature with a 1:2000 dilution of secondary antibody (horseradish peroxidase-linked donkey antirabbit IgG antibody; Amersham Biosciences UK Limited, Buckinghamshire, England) for 45 minutes. Cross-reactivity was visualized by using enhanced chemiluminescence (SuperSignal WestPico; Pierce) and quantified using QuantityOne application software (Bio-Rad).
Cell transfections
To overexpress FPN1, HEK293T cells at 60% confluency were transfected with either pSPORT2 cytomegalovirus (CMV) or pSPORT2 CMV containing full-length mouse FPN1 cDNA19 using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. Forty-eight hours after transfection, cell lysates were prepared and analyzed for FPN1 protein levels by Western analysis as described earlier.
Nonheme iron determination
J774 cells from a nearly confluent plate were harvested, weighed, and then digested in 2 mL of 3 M HCl and 10% trichloroacetic acid for 20 hours at 65°C. Nonheme iron levels were determined colorimetrically using the method of Torrance and Bothwell.32
Results
FPN1 mRNA levels decrease in response to iron depletion
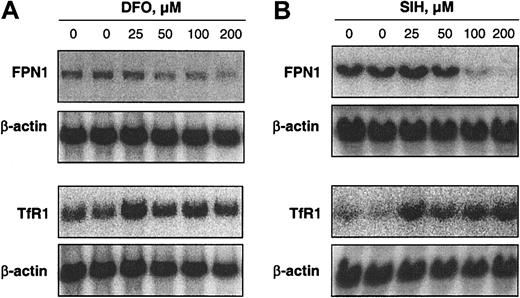
Treatment of J774 cells for 20 hours with the iron chelator DFO (0-200 μM) decreased FPN1 mRNA levels in a dose-dependent manner (Figure 1A). To determine whether this decrease in FPN1 mRNA levels in DFO-treated cells was statistically significant, another experiment was performed in which cells were treated for 20 hours with either 100 μM DFO or sterile water as a control. The mean FPN1 mRNA level in DFO-treated cells was 23% ± 5% (± SE, n = 4) lower than control (P = .012). For comparison, the level of TfR1 mRNA, which is stabilized under low-iron conditions,33 was also determined. The increase in TfR1 mRNA levels in response to DFO is consistent with other studies using this same cell line34 and confirms iron depletion by this chelator. TfR1 mRNA levels in DFO-treated cells in Figure 1A increased by an average of 87% (range, 40%-116%). A limitation of DFO, however, is that it crosses cell membranes inefficiently and, thus, has relatively poor access to the intracellular chelatable iron pool.35 Therefore, the effects of a lipophilic iron chelator, SIH, were further examined. SIH penetrates cells very efficiently and extracts iron from cultured cells more potently than DFO.35 As shown in Figure 1B, FPN1 mRNA levels were markedly decreased after 20 hours of treatment with 100 to 200 μM SIH. The more pronounced decrease in FPN1 mRNA levels by SIH corresponds with a greater increase in TfR1 mRNA levels (average increase of 294%; range, 265%-338%), supporting the idea that a more significant decrease in cellular iron was induced relative to DFO. The suppression of FPN1 mRNA levels by iron chelation is consistent with the diminished FPN1 expression observed in iron-deficient mouse liver.19
Iron depletion decreases FPN1 mRNA levels in J774 macrophages. Northern analysis of FPN1 and TfR1 mRNA expression in J774 cells incubated for 20 hours in the presence of DFO (A) or SIH (B) at the indicated concentrations. Total RNA was size-fractionated in a 1% agarose gel and transferred to a Nytran N membrane. The Northern blot was hybridized with either 32P-labeled murine FPN1 or rat TfR1 probe and then stripped and re-probed for β-actin. Hybridized probes were detected by PhosphorImaging (Quantity One Software; Bio-Rad). Results shown in each panel are representative of 2 independent experiments.
Iron depletion decreases FPN1 mRNA levels in J774 macrophages. Northern analysis of FPN1 and TfR1 mRNA expression in J774 cells incubated for 20 hours in the presence of DFO (A) or SIH (B) at the indicated concentrations. Total RNA was size-fractionated in a 1% agarose gel and transferred to a Nytran N membrane. The Northern blot was hybridized with either 32P-labeled murine FPN1 or rat TfR1 probe and then stripped and re-probed for β-actin. Hybridized probes were detected by PhosphorImaging (Quantity One Software; Bio-Rad). Results shown in each panel are representative of 2 independent experiments.
Iron loading increases FPN1 mRNA levels in J774 macrophages in a transcription-dependent manner
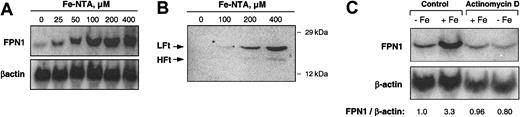
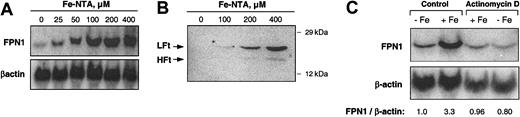
To investigate the effect of iron loading on FPN1 mRNA levels, J774 cells were treated with Fe-NTA(0-400 μM) for 20 hours, and total RNA was collected for Northern analysis. As shown in Figure 2A, a dose-dependent increase in FPN1 mRNA levels was observed. When normalized to β-actin, FPN1 mRNA levels after treatment with 400 μM Fe-NTA were 17-fold higher than basal levels. An increase in cellular iron content subsequent to Fe-NTA treatment was confirmed by the observed increase in ferritin protein levels (Figure 2B). Use of the simple iron chelate ferric ammonium citrate, which is more commonly used to load macrophages with iron,34,37-39 also increased FPN1 mRNA levels but to a much lesser degree (data not shown). To determine whether iron loading induces transcription, J774 cells were exposed for 8 hours to 200 μM Fe-NTA in the presence or absence of 1 μg/mL actinomycin D, an inhibitor of RNA synthesis. As shown by the results of Northern analysis (Figure 2C), cotreatment with actinomycin D completely blocked the iron-induced increase in FPN1 mRNA.
Iron loading increases FPN1 mRNA levels in J774 macrophages. (A) Northern analysis of FPN1 mRNA expression in J774 cells incubated with the indicated concentrations of Fe-NTA for 20 hours. Results shown are representative of 2 independent dose-response experiments. (B) Western analysis of ferritin in J774 cells incubated with the indicated concentrations of Fe-NTA for 20 hours (LFt indicates ferritin light chain; HFt, ferritin heavy chain). Proteins (200 μg) from cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE), blotted, and immunoblotted using antiferritin antibody as described in “Materials and methods.” The positions and masses of molecular weight markers (carbonic anhydrase and cytochrome c, 29 and 12 kDa, respectively) are indicated. Data are from a single experiment performed as a control to confirm previous results reported in the literature.36 (C) Northern analysis of FPN1 expression in cells treated for 8 hours in the presence (+Fe) or absence (-Fe) of 200 μM Fe-NTA and actinomycin D (1 μg/mL). The ratio of FPN1 signal intensity relative to β-actin is shown. Results shown are representative of 3 similar experiments.
Iron loading increases FPN1 mRNA levels in J774 macrophages. (A) Northern analysis of FPN1 mRNA expression in J774 cells incubated with the indicated concentrations of Fe-NTA for 20 hours. Results shown are representative of 2 independent dose-response experiments. (B) Western analysis of ferritin in J774 cells incubated with the indicated concentrations of Fe-NTA for 20 hours (LFt indicates ferritin light chain; HFt, ferritin heavy chain). Proteins (200 μg) from cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE), blotted, and immunoblotted using antiferritin antibody as described in “Materials and methods.” The positions and masses of molecular weight markers (carbonic anhydrase and cytochrome c, 29 and 12 kDa, respectively) are indicated. Data are from a single experiment performed as a control to confirm previous results reported in the literature.36 (C) Northern analysis of FPN1 expression in cells treated for 8 hours in the presence (+Fe) or absence (-Fe) of 200 μM Fe-NTA and actinomycin D (1 μg/mL). The ratio of FPN1 signal intensity relative to β-actin is shown. Results shown are representative of 3 similar experiments.
Immunoblot analysis reveals iron-induced expression of FPN1 protein
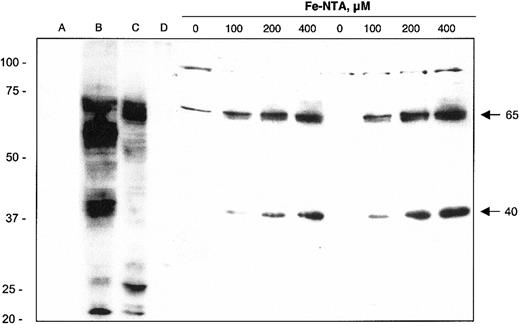
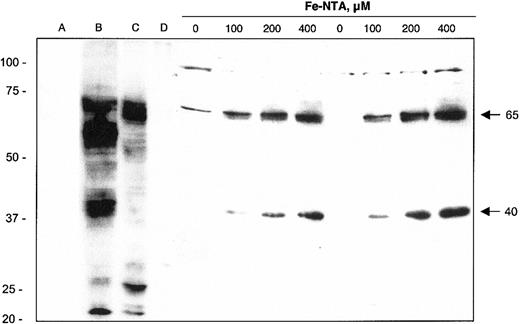
Western blot analysis of J774 macrophage cell lysates indicates that the induction of FPN1 mRNA on iron treatment is associated with increased FPN1 protein expression (Figure 3). Two immunoreactive bands with molecular masses of approximately 65 kDa and approximately 40 kDa were induced by iron treatment in a dose-dependent manner. The iron-induced 65-kDa band is close to the predicted molecular mass of FPN1 (62 kDa) and comigrates with a band detected in HEK cells transfected with full-length mouse FPN1 cDNA (lane B). Moreover, immunoreactivity of a protein of the same mass was detected in duodenum (lane C), but not ileum (lane D), of a bled rat. Alignment of the 65-kDa band with these positive control samples supports the identity of this protein as FPN1. The second immunoreactive band of 40 kDa may represent a precursor, a splice variant, a cleaved fragment, a different member of the same protein family, or a cross-reacting nonrelated protein.
Immunoblot analysis of FPN1 protein expression in J774 macrophages. Lysates were collected from HEK293T cells transfected with pSPORT2 CMV vector (lane A) or pSPORT2 CMV containing full-length mouse FPN1 cDNA (lane B). Additional control lysates were prepared from duodenal (lane C) and ileal (lane D) mucosal cells from a bled rat (material provided by Dr Ramon Molina, Harvard School of Public Health, Boston, MA). Cell lysates were also prepared from J774 cells incubated for 40 hours with the indicated concentrations of Fe-NTA. Results from 2 different samples at each Fe-NTA treatment concentration are shown. Protein samples were prepared for electrophoresis as described in “Materials and methods” and separated on a 10% SDS polyacrylamide gel; 100-μg aliquots were electrophoresed except for lane B, which contained 1 μg. Proteins were transferred to nitrocellulose for immunoblotting as detailed in “Materials and methods.” Arrows on the right identify the positions and estimated mass (kDa) of iron-responsive FPN1-immunoreactive bands. The position and mass (in kDa) of molecular weight markers (Precision Plus Protein standards, All Blue; Bio-Rad) are indicated on the left. Results shown are representative of 3 independent experiments.
Immunoblot analysis of FPN1 protein expression in J774 macrophages. Lysates were collected from HEK293T cells transfected with pSPORT2 CMV vector (lane A) or pSPORT2 CMV containing full-length mouse FPN1 cDNA (lane B). Additional control lysates were prepared from duodenal (lane C) and ileal (lane D) mucosal cells from a bled rat (material provided by Dr Ramon Molina, Harvard School of Public Health, Boston, MA). Cell lysates were also prepared from J774 cells incubated for 40 hours with the indicated concentrations of Fe-NTA. Results from 2 different samples at each Fe-NTA treatment concentration are shown. Protein samples were prepared for electrophoresis as described in “Materials and methods” and separated on a 10% SDS polyacrylamide gel; 100-μg aliquots were electrophoresed except for lane B, which contained 1 μg. Proteins were transferred to nitrocellulose for immunoblotting as detailed in “Materials and methods.” Arrows on the right identify the positions and estimated mass (kDa) of iron-responsive FPN1-immunoreactive bands. The position and mass (in kDa) of molecular weight markers (Precision Plus Protein standards, All Blue; Bio-Rad) are indicated on the left. Results shown are representative of 3 independent experiments.
FPN1 mRNA and protein levels increase transiently after erythrophagocytosis
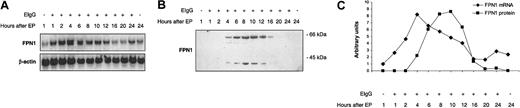
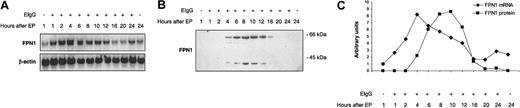
Because macrophages in vivo obtain most of their iron by ingesting senescent erythrocytes, the effect of erythrophagocytosis on FPN1 expression was investigated. As shown in Figure 4A and C, FPN1 mRNA levels increased transiently after erythrophagocytosis, reaching maximal levels (8-fold higher than basal levels) after 4 hours and returning to basal levels by 16 hours. FPN1 protein expression, as indicated by the band at 65 kDa, increased to maximal levels by 10 hours and then decreased (Figure 4B-C). No immunoreactive bands at 40 or 65 kDa were detected in cell lysates from opsonized erythrocytes alone (data not shown). Cell viability was more than 95% when assessed by trypan blue exclusion 24 hours after erythrophagocytosis. Induction of FPN1 mRNA after erythrophagocytosis was also observed in IC-21 cells (a cell line derived by transformation of normal mouse peritoneal macrophages with SV40), indicating that the effect of erythrophagocytosis on FPN1 expression is not limited to J774 cells (data not shown).
FPN1 mRNA and protein levels increase transiently after erythrophagocytosis (EP) by J774 macrophages. J774 cells (approximately 1 × 106) were incubated in the presence of 15 × 106 opsonized human erythrocytes (EIgG) for 1.5 hours. Noningested erythrocytes were lysed and removed, and incubation continued for the times indicated. (A) Northern analysis of FPN1. Results are from a single experiment using human erythrocytes and were similar to data from experiments using sheep erythrocytes (n = 2) and rat erythrocytes (n = 2). (B) Western blot analysis of FPN1. Results shown are representative of 2 independent experiments performed using human erythrocytes. (C) FPN1 mRNA levels normalized to β-actin (♦), and protein levels of the 65-kDa FPN1 band (▪) plotted as a function of time after EP.
FPN1 mRNA and protein levels increase transiently after erythrophagocytosis (EP) by J774 macrophages. J774 cells (approximately 1 × 106) were incubated in the presence of 15 × 106 opsonized human erythrocytes (EIgG) for 1.5 hours. Noningested erythrocytes were lysed and removed, and incubation continued for the times indicated. (A) Northern analysis of FPN1. Results are from a single experiment using human erythrocytes and were similar to data from experiments using sheep erythrocytes (n = 2) and rat erythrocytes (n = 2). (B) Western blot analysis of FPN1. Results shown are representative of 2 independent experiments performed using human erythrocytes. (C) FPN1 mRNA levels normalized to β-actin (♦), and protein levels of the 65-kDa FPN1 band (▪) plotted as a function of time after EP.
Iron chelation suppresses FPN1 induction after erythrophagocytosis
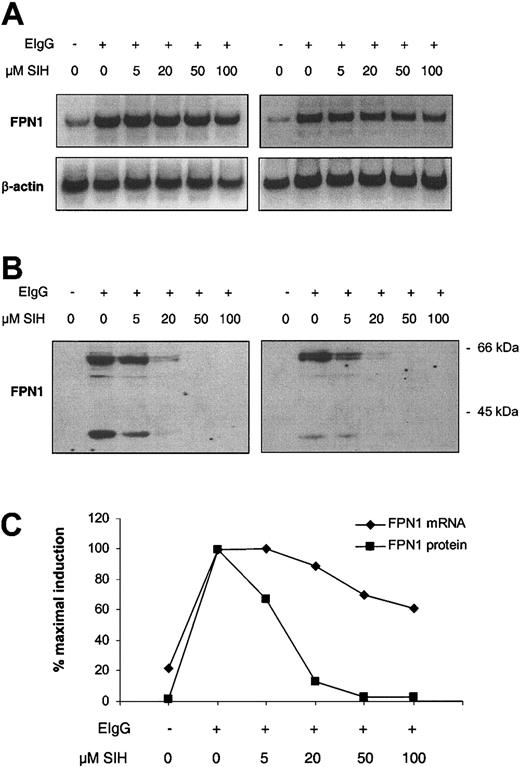
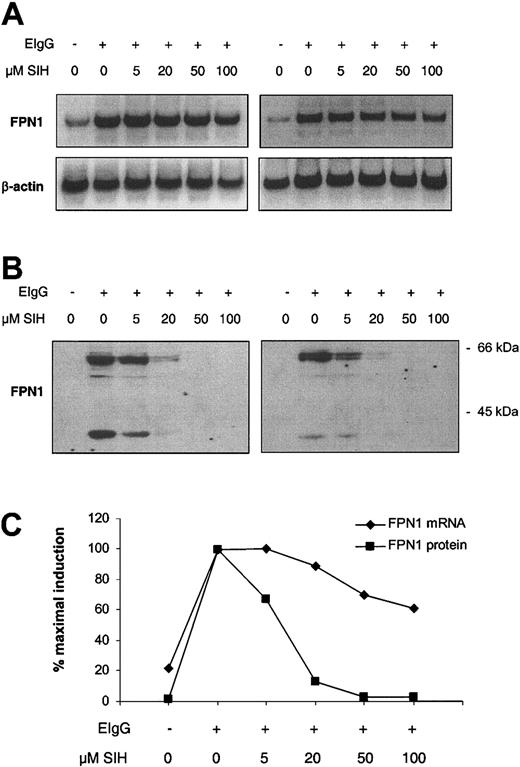
To test whether the increase in FPN1 expression after erythrophagocytosis was due to increased cellular iron, the lipophilic chelator SIH was added to cells after ingestion of opsonized red cells, and FPN1 mRNA and protein levels were analyzed 6 hours later. SIH added immediately after the erythrophagocytosis period resulted in a pronounced dose-dependent suppression of FPN1 induction at the protein level, with no detectable expression following the addition of 50 or 100 μM SIH (Figure 5B-C).
Iron chelation suppresses the induction of FPN1 after erythrophagocytosis by J774 macrophages. J774 cells (approximately 1 × 106) were incubated in the presence (+) or absence (-) of 15 × 106 opsonized human erythrocytes (EIgG) for 1.5 hours. Noningested erythrocytes were lysed and removed, SIH was added at the indicated concentrations, and incubation was continued for 6 hours. (A) Northern analysis of FPN1. (B) Western blot analysis of FPN1. (C) FPN1 mRNA levels normalized to β-actin (♦), and protein levels of the 65-kDa FPN1 band (▪) plotted as a function of SIH concentration. A third independent experiment, in which FPN1 protein (but not mRNA) was measured, yielded similar results.
Iron chelation suppresses the induction of FPN1 after erythrophagocytosis by J774 macrophages. J774 cells (approximately 1 × 106) were incubated in the presence (+) or absence (-) of 15 × 106 opsonized human erythrocytes (EIgG) for 1.5 hours. Noningested erythrocytes were lysed and removed, SIH was added at the indicated concentrations, and incubation was continued for 6 hours. (A) Northern analysis of FPN1. (B) Western blot analysis of FPN1. (C) FPN1 mRNA levels normalized to β-actin (♦), and protein levels of the 65-kDa FPN1 band (▪) plotted as a function of SIH concentration. A third independent experiment, in which FPN1 protein (but not mRNA) was measured, yielded similar results.
Increased ferritin and nonheme iron levels after erythrophagocytosis are associated with up-regulation of FPN1, HO-1, and Nramp1 but not DMT1 mRNA levels
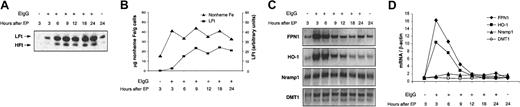
To further confirm increased iron content in J774 cells after erythrophagocytosis, levels of ferritin and nonheme iron were measured. As shown in Figure 6A-B, ferritin levels, particularly light-chain ferritin (LFt), increased over the first 6 hours after erythrophagocytosis and remained elevated during the next 18 hours. The time course of changes in cellular nonheme iron levels indicates that red cell iron was liberated from heme primarily during the first 3 hours after erythrophagocytosis (Figure 6B).
Changes in ferritin, nonheme Fe, and mRNA levels of FPN1, HO-1, Nramp1, and DMT1 after erythrophagocytosis (EP) by J774 macrophages. J774 cells (approximately 4 × 106) were incubated in the presence of 100 × 106 opsonized rat erythrocytes (EIgG) for 2 hours. Noningested erythrocytes were lysed and removed, and incubation was continued for the times shown. (A) Western analysis of ferritin (LFt indicates ferritin light chain; HFt, ferritin heavy chain). The position of murine LFt above that of HFt was determined previously in studies of MEL cells stably transfected with HFt.40 (B) Nonheme Fe (▴), and LFt (▪) levels plotted as a function of time after EP. (C) Northern analysis of FPN1, HO-1, Nramp1, and DMT1. (D) mRNA levels of FPN1 (♦), HO-1 (▪), Nramp1 (▴), DMT1 (○), all normalized to β-actin, plotted as a function of time after EP. Results shown in panels A-B are from a single experiment and confirm previously published data.7,41-43,45 Results shown in panels C-D are representative of at least 2 similar experiments.
Changes in ferritin, nonheme Fe, and mRNA levels of FPN1, HO-1, Nramp1, and DMT1 after erythrophagocytosis (EP) by J774 macrophages. J774 cells (approximately 4 × 106) were incubated in the presence of 100 × 106 opsonized rat erythrocytes (EIgG) for 2 hours. Noningested erythrocytes were lysed and removed, and incubation was continued for the times shown. (A) Western analysis of ferritin (LFt indicates ferritin light chain; HFt, ferritin heavy chain). The position of murine LFt above that of HFt was determined previously in studies of MEL cells stably transfected with HFt.40 (B) Nonheme Fe (▴), and LFt (▪) levels plotted as a function of time after EP. (C) Northern analysis of FPN1, HO-1, Nramp1, and DMT1. (D) mRNA levels of FPN1 (♦), HO-1 (▪), Nramp1 (▴), DMT1 (○), all normalized to β-actin, plotted as a function of time after EP. Results shown in panels A-B are from a single experiment and confirm previously published data.7,41-43,45 Results shown in panels C-D are representative of at least 2 similar experiments.
Other iron-induced effects were measured after erythrophagocytosis (Figure 6C-D). As has been shown previously,43 HO-1 mRNA levels were markedly increased immediately after erythrophagocytosis and paralleled changes in FPN1 transcript levels, with maximal induction (10-fold and 16-fold, respectively) observed 3 hours after erythrophagocytosis. A small but reproducible 2-fold increase in Nramp1 mRNA levels occurred 6 to 8 hours after erythrophagocytosis (n = 3). An increase in Nramp1 mRNA levels in bone marrow macrophages treated with hemin has been reported previously.44 Among the iron-related genes examined, only levels of DMT1 mRNA did not change during the 24-hour period after erythrophagocytosis, consistent with another report that DMT1 mRNA levels in J774 cells do not respond to changes in cellular iron status.34
Discussion
RES macrophages of the liver, spleen, and bone marrow acquire iron as a result of erythrophagocytosis during the normal process of removing effete or damaged red cells from the circulation. After erythrophagocytosis, hydrolytic enzymes present in the phagolysosome degrade the red blood cell. Proteolytic digestion of hemoglobin subsequently liberates heme, which is assumed to cross the phagolysosomal membrane either by diffusion or by a specific transporter to reach membrane-bound HO-1 in the endoplasmic reticulum. Oxidation of the heme ring by HO-1 produces biliverdin, carbon monoxide, and free iron, the latter of which is then either released from the cell or stored in ferritin. With each red cell ingested, the macrophage accrues an estimated 109 iron atoms. Studies in erythrophagocytosing rat Kupffer cells show that within 24 hours, about half of the acquired iron is purged from the macrophage, either bound to ferritin or transferrin, or in a form readily available for binding to transferrin.45 The rate of iron release is greatest during the first 8 hours after erythrophagocytosis, then declines slowly and progressively thereafter.45
To investigate the potential role of FPN1 in erythrocyte iron recycling, changes in FPN1 mRNA and protein levels in J774 macrophages after erythrophagocytosis were assessed. Both increased substantially during the first 10 hours after erythrophagocytosis and then returned to basal levels. Transcript levels of HO-1, which is involved in heme degradation, paralleled the changes observed for FPN1, whereas mRNA pools for Nramp1 and DMT1, 2 other iron transport proteins, were only modestly enhanced or remain unaffected, respectively. The correlation between the time course of FPN1 expression in J774 macrophages after erythrophagocytosis and iron release by erythrophagocytosing rat Kupffer cells45 is consistent with the hypothesis that FPN1's transport activity is integral to iron export. The observation that iron chelation can suppress FPN1 induction after erythrophagocytosis provides strong evidence that increased levels of this protein result from iron liberated from the degradation of red cell heme. It is, therefore, probable that the progressive decreases in FPN1 protein levels (after the initial rapid induction after erythrophagocytosis) reflect decreased levels of chelatable iron, either through export from the cell or sequestration in ferritin. The observation that nonheme iron levels remain elevated after erythrophagocytosis (Figure 6) suggests that a large fraction of ingested iron remains in the cell. On the basis of the estimate that approximately 40% of erythrocytes are ingested during the 2-hour incubation period, about 50% of red cell iron is found in the nonheme iron pool 3 hours after erythrophagocytosis. Our preliminary measurements of iron release indicate that only 10% to 15% of ingested 59Fe is released after phagocytosis of 59Fe-labeled rat erythrocytes (data not shown), consistent with studies using 59Fe-labeled erythroblasts and cultured rat bone marrow macrophages.46 Thus, it is the rapid and strong induction of ferritin, particularly the L-isoform (Figure 6), that appears to more substantially reduce the chelatable free iron pool, perhaps even competing for its export by FPN1. Indeed, studies characterizing the intracellular forms of iron in cultured monocytes after erythrophagocytosis indicate that approximately 25% of ingested 59Fe is incorporated into ferritin within the first 4 hours, whereas less than 5% is in the low molecular weight pool.7
Previous studies using in situ hybridization have shown an increase in FPN1 mRNA levels in freshly isolated human alveolar macrophages treated with ferric ammonium citrate.37 In the present study, we demonstrate by Northern analysis that FPN1 mRNA levels in murine J774 macrophages increased on loading with Fe-NTA as well as decreased on iron depletion using 2 different iron chelators. The demonstration that actinomycin D completely blocked the induction by iron loading suggests that regulation of FPN1 gene expression occurs at the level of gene transcription. The increase in macrophage FPN1 mRNA levels on iron loading was similar to that observed for ferritin mRNA. Treatment of human monocyte-macrophages with ferric ammonium citrate has been shown to result in increased ferritin mRNA levels.38,47 The more pronounced increases in FPN1 mRNA levels in the present study likely reflect the use of highly bioavailable sources of iron such as Fe-NTA and red cell heme. Indeed, hemin has been shown to be a much more potent inducer of H-ferritin mRNA levels than ferric ammonium citrate in Friend erythroleukemic cells.48 The presence of a 5′ iron-responsive element in FPN1 mRNA suggests that, like ferritin,49 FPN1 expression is also translationally regulated. This idea is strongly supported by transient transfection studies of reporter genes driven by the FPN1 promoter in human COS7 cells19 and murine RAW264.7 macrophages.50 The fact that the addition of SIH after erythrophagocytosis completely blocked the induction of FPN1 protein despite the presence of FPN1 mRNA levels that are 3-fold greater than basal further indicates that translational control does play a major role in the regulation of the iron export protein expression. It remains to be determined to what degree transcriptional and posttranscriptional regulatory mechanisms contribute to FPN1 protein expression in the macrophage. Finally, although an iron-export protein would be expected to reside exclusively at the plasma membrane, the available immunofluorescence data in Kupffer cells indicate an intracellular distribution.19 It is possible that the cytolocalization of macrophage FPN1 is affected by phagocytosis, as has been observed for the other transmembrane iron transporters Nramp113 and DMT1.18 For example, when exposed to high cellular copper concentrations, the transport protein MNK rapidly translocates from the trans-Golgi network to the plasma membrane where it exports copper from cells.51 Perhaps an analogous redistribution of subcellular FPN1 occurs after exposure to iron, providing yet another layer of regulatory control over its function. A key question that remains is how the regulation of macrophage FPN1 is altered in conditions like HH and the anemia of chronic disease, which are both characterized by perturbations in iron release from the RES.3
The potential involvement of FPN1 in iron release from cells of the RES has been the subject of 2 other recent investigations. Yang et al22 have reported diminished macrophage FPN1 immunostaining in sections of spleen, liver, and bone marrow from LPS-treated mice. Time course experiments, which show that LPS-induced reductions in serum iron levels precede the down-regulation of splenic FPN1 protein levels, suggest that down-regulation of FPN1 may serve to maintain the hypoferremia rather than induce it. Ludwiczek et al52 have shown that THP-1 cells stimulated with interferon γ (IFN-γ) and LPS have decreased levels of FPN1 mRNA by reverse transcriptase-polymerase chain reaction (RT-PCR) and also observed decreased immunohistochemical staining for FPN1 protein. These inflammatory stimuli also significantly reduced cellular iron efflux from THP-1 cells, as measured by the release of 59Fe after incubating cells with 59Fe-labeled transferrin and 59Fe-ferric citrate. Because macrophages handle iron differently if it is obtained via phagocytosis or via endocytosis of transferrin,12,53,54 future studies need to determine the effects of inflammatory stimuli on the release of iron acquired via erythrophagocytosis. Our results clearly demonstrate that macrophage FPN1 transcript levels are modulated in response to cellular iron status, and that iron liberated by erythrophagocytosis up-regulates both protein and mRNA expression. These novel observations help to identify a significant role for FPN1 in iron recycling by the RES and further our understanding of how this process can be regulated.
Prepublished online as Blood First Edition Paper, August 7, 2003; DOI 10.1182/blood-2003-04-1250.
Supported by grants from the National Institutes of Health (grant DK56160) (M.W.-R.), (DK09998) (M.D.K.), and (DK59429) (D.J.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.