Abstract
A 6-year-old male with vertebral-basilar artery thrombosis was recognized to have high-molecular-weight kininogen (HK) deficiency. The propositus had no HK procoagulant activity and antigen (< 1%). Using monoclonal antibodies (Mabs) to kininogen domain 3, the propositus, family members, and Fitzgerald plasma were determined to have detectable low-molecular-weight kininogen. Mabs to HK domains 5 and 6 do not detect HK antigen in the propositus' plasma. The propositus has a single base pair (bp) deletion in cDNA position 1492 of exon 10 affecting amino acid 480 of the mature protein and resulting in a frameshift and a premature stop codon at position 1597 (amino acid 532). Unexpectedly, Mabs to the heavy chain and domain 5 of HK detect a 92-kDa form of HK in Fitzgerald plasma, the first HK-deficient plasma. The 92-kDa Fitzgerald HK has amino acid residues through 502, corresponding to domains 1 through 5, but lacks epitopes of domain 6 (positions 543 to 595). Fitzgerald DNA has a normal exon 10, but a 17-bp mutation in intron 9. These combined results indicate that mutations in the kininogen gene may differentially affect biosynthesis, processing, and/or secretion of HK.
Introduction
Characterization of the molecular defect of deficient proteins can indicate the relation of structure to function of a protein. Hereditary high-molecular-weight kininogen (HK) deficiency is a rare entity. Williams trait has a molecular defect of C to T transition at nucleotide 586 resulting in a stop codon in exon 5 which prematurely stops biosynthesis of HK after amino acid position 195 (corresponding to position 177 of the mature protein).1,2 Williams trait is associated with an absence of both HK and low-molecular-weight kininogen (LK) in plasma.1,3 Alternatively, Fitzgerald trait, the first HK-deficient plasma recognized, has absent functional HK activity and 40% normal levels of LK antigen though plasma bradykinin levels were found to be normal.4,5
A cohesive hypothesis has recently been elaborated for the assembly, activation, and physiologic activity of HK and the plasma kallikrein/kinin system.6 The pivotal protein in the assembly and activation of this system is plasma HK. The plasma kininogens, HK and LK, are multidomain proteins whose major purpose is to provide the biologically active peptide, bradykinin. However, as part of kinin deliverance, the kininogens have other activities recognized by structure and function analysis. The identical heavy chain of both HK and LK has the ability to bind to cell membranes, inhibit cellular cysteine proteases, and interfere with thrombin activation of protease activated receptor 1.7, 8, 9, 10 The unique light chain domain of HK, produced by alternative splicing of the kininogen RNA, has the ability to bind to cell membranes to bring prekallikrein (PK) and factor XI to cell and artificial surfaces for activation.11, 12, 13 When PK bound to HK assembles on its multiprotein receptor complex on endothelial cells or cell matrix, PK is rapidly activated to kallikrein by the endothelial cell enzyme, prolylcarboxypeptidase.3,14, 15, 16 Since prolylcarboxypeptidase is also an angiotensin II degrading enzyme, the plasma kallikrein-kinin system may counterbalance the renin-angiotensin system.6,16, 17, 18 Formed plasma kallikrein liberates bradykinin from HK, stimulating nitric oxide synthesis and the residual cleaved HK expresses new antiproliferative and antiangiogenic activity.19, 20, 21 Thus the plasma kallikrein-kinin system is intimately involved in regulation of vascular biology.
In this report, we examined the plasma and DNA of a 6-year-old boy with a cerebral artery thrombosis who was found to have HK deficiency and Fitzgerald trait. These investigations indicate that truncation or frameshift at or before position 480 of the mature HK prevents biosynthesis, processing, and/or the secretion of HK into plasma whereas defects in intron 9 of the kininogen gene may result in the absence of synthesis of domain 6 of HK.
Patients, materials, and methods
Case report
A 6-year-old male with no previous medical history and whose parents are first cousins presented with cephalgia and vomiting occurring 10 days after moderate cervical trauma followed by a loss of consciousness and subsequent visual impairment. On computerized axial tomography (CAT) and angiography, the patient had an extensive left vertebral-basilar artery thrombosis and a left vertebral artery dissection. The patient had a prolonged activated partial thromboplastin time (APTT) and received 45 mL/kg fresh frozen plasma prior to arteriography and then 10 mL/kg per day for 8 days which resulted in the normalization of the APTT and resolution of neurologic symptoms. On day 8 when the APTT returned to admission values, the patient had a neurologic relapse with new areas of cerebral ischemia on CAT. The patient was anticoagulated with warfarin and one month later there was a new headache and vomiting despite warfarin anticoagulation with an international normalized ratio of 2.0. In time, however, there was full neurologic recovery with warfarin therapy for 6 months. There has been no recurrence after 2 years of follow-up. After full informed consent according the Declaration of Helsinki, both plasma and cells for the preparation of DNA were collected from the patient and his immediate family.
Materials
Williams plasma, which is total kininogen-deficient plasma, was generously donated to this laboratory by the late Mayme Williams of Philadelphia.1 Fitzgerald plasma was generously provided by Dr Guillermo Scicli (Henry Ford Hospital, Detroit, MI) in 1979, and more recently, by Dr Oscar Carretero (Henry Ford Hospital).4,5 These plasmas have been continuously frozen at -70°C since freezing at collection time. Prekallikrein-deficient plasma was directly donated to this laboratory. Normal human pooled plasma was purchased from George King, Overland Park, KS. Purified HK was purchased from Enzyme Research Laboratories, South Bend, IN. Polyclonal antisera to the light chain of HK,3 monoclonal antibodies HKH14, HKH15, HKL10, HKL13, HKL14, HKL16, HKL12, HKL24, and HKL2522 (Table 1), and antisera to the HK domain 5 peptide HKH20 was prepared as previously reported23 (Table 1).
Coagulant assays
Platelet-poor plasma was prepared from blood anticoagulated with 3.2 g/dL sodium citrate by centrifugation at 2000g at room temperature for 20 minutes. If not used immediately, it was stored -20°C until use. Some of the plasma from the patient and his family was lyophilized for shipment to the United States. The APTT (PTTA Stago, Stago, Asnieres, France), prothrombin time (Innovin Dade Behring, Marburg, Germany), thrombin time (Thrombin IS, Dade Behring), fibrinogen (Fibriprest, Stago), factor II (Deficient II Stago), factors VII+X (Deficient VII+X Stago), factor V (Deficient V, Stago) were measured using an automatic analyzer STA (Stago). Antithrombin heparin cofactor activity (Thrombin, Dade Behring and S-2238 Chromogenix, Stockholm, Sweden), protein C activity (Staclot Protein C, Stago), plasminogen amidolytic activity (Stachrom Plasminogen Stago), and lupus anticoagulant detection (Staclot LA Stago) were measured using a semiautomatic analyzer ST888 (Diagnostica Stago) according to the procedures of the manufacturers. Free protein S antigen (Asserachrom Protein S Stago) was measured by an enzyme-linked immunosorbent assay (ELISA) using a EL 312 e Bio-kinetics Reader. Factors VIII, IX, XI, XII (Deficient plasmas, Stago), prekallikrein, and HK clotting activity (HMWK Deficient factor, Immuno AG, Vienna, Austria) were measured on a semiautomatic analyzer KC10 Amelung (Sigma, St Louis, MO). Von Willebrand factor ristocetin cofactor activity (von Willebrand reagent Dade Behring) was measured using an Affibio aggregometer. All these parameters were expressed in U/mL using a pool of plasmas from 25 healthy volunteers as a calibrated control. One U/mL was defined as the amount of protein assayed in 1 mL pooled normal human plasma. HK procoagulant activity on resuspended lyophilized patient plasma also was measured by one stage APTT-based assay using automated APTT reagent (Organon Teknika, Durham, NC) and total kininogen-deficient (Williams) plasma in an Amelung K4 Micro Coagulation Analyzer (Sigma).1 Samples were compared against standard curve from pooled normal human plasma diluted in 0.01 M Tris, 0.15 M NaCl, pH 7.4 (TBS). One U/mL HK was defined as that amount of procoagulant activity in a 1/10 dilution of pooled normal plasma.
PK chromogenic activity
Kallikrein activity was measured by utilizing chromogenic substrate H-D-Pro-Phe-Arg-pNA (S-2302; Diapharma, Franklin, OH) as previously reported.24 Normal pooled human plasma (50 μL) for the standard curve or patient plasma samples were acid treated to neutralize plasma protease inhibitors by incubating them with 50 μL of 1/6 N HCl for 15 minutes at room temperature followed by 50 μL of 0.1 M sodium phosphate, pH 7.6, 0.15 M NaCl, 1 mM EDTA (ethylenediaminetetraacetic acid), and 50 μLof 0.17 N NaOH. The samples then were diluted in 350 μL of 50 mM Tris, 0.15 M NaCl, pH 7.9, containing 0.1% polyetheylene glycol. Residual PK was determined by taking 50 μL of the acidified, prediluted plasma and incubating it with 50 μL of plasma prekallikrein activator (Chromogenix) for 5 minutes. After incubation, 50 μL of substrate S-2302 (0.4 mM final concentration) was added to the activated plasma and hydrolysis proceeded for 10 minutes. The reaction was stopped with 100 μL 50% acetic acid. The absorbance was read at 405 nm. The units of kallikrein activity were determined by comparison with that produced by an equal amount of pooled normal human plasma.
Radial immunodiffusion for HK and PK
Radial immunodiffusion to quantitate the amount of HK and PK antigen was performed in 1% agarose, as previously reported.1,25 HK antigen was determined using a polyclonal antibody to the light chain of HK.3 Plasma PK antigen was measured using a monospecific goat polyclonal antiserum as previously reported.25 The amount of HK and PK in plasma was determined by comparison with the known amount of each of these proteins in a characterized pool of normal human plasma.3,25
Gel electrophoresis and Western blotting
Plasma from family members and normal human pooled plasma was diluted 1:20 and Williams and Fitzgerald plasmas were diluted 1:10 with TBS and sample buffer. The samples were then reduced with 2% β-mercaptoethanol and boiled for 5 minutes followed by application to a 7% polyacrylamide gel electrophoresis in the presence of 0.1% (wt/vol) sodium dodecyl sulfate (SDS-PAGE) at 30 mA for 60 minutes. Molecular mass markers (Bio-Rad Laboratories, Richmond, CA) were myosin (217 kDa), β-galactosidase (123 kDa), bovine serum albumin (71 kDa), ovalbumin (48 kDa), phosphorylase B (112 kDa), carbonic anhydrase (36.2 kDa), soybean trypsin inhibitor (29.9 kDa), and lysozyme (21.3 kDa). The electrophoresed proteins were then transferred to nitrocellulose at 9 mA overnight. The membranes were blocked with TBS containing 5% (wt/vol) dry milk powder and 0.05% (vol/vol) Tween 20 (Blotto) as previously reported.25 The immunoblot of the transferred proteins was performed by incubating the primary antibody in Blotto. Bound antibody was detected by a horseradish peroxidase–coupled secondary antibody, followed by the chemiluminescence detection system (Amersham, Arlington Heights, IL).
PCR amplification and DNA sequencing
DNA from the patient and family were prepared from leukocytes. DNA from 2 mL to 10 mL thawed Fitzgerald and Williams trait plasmas were extracted from a 15 000g centrifugation pellet. Factor V Gln506 and factor II G20210 mutations were screened using established techniques.26 Since immunoblot studies indicated that the defects in the patient's and Fitzgerald plasma HK were in domains 5 or 6 and this region is coded by exon 10 of HK, 2 sets of PCR primers were prepared to amplify exon 10 of HK (GenBank accession no. M11437.1).2,11 The 5′ sense primer of the first set corresponded to nucleotides 5′-AGGCCTCCAGGTTTTTCACCTTTCCGA-3′ (nucleotide positions 26 to 52 of exon 10) and the antisense primer 5′-AGAAAGGCCATCAGTGAGATCGAAATA-3′ (nucleotides 791 to 817). The first set of primers produced a 791-bp DNA fragment. A second set of PCR primers closer to the exon 10 deletion site was designed. The sense primer was 5′-CTTGATGATGATCTTGAACACCAAGGG-3′ (nucleotides 320 to 346 of exon 10) and the antisense primer was 5′-ATTGTGCTTTCCATTCTTTTTGCCTTT-3 (nucleotides 410-436). This second set of primers produced a 117-bp DNA fragment. Each cycle of polymerase chain reaction (PCR) consisted of 2 minutes of denaturation at 94°C and 30 cycle repetitions of denaturing for 1 minute at 94°C, annealing for 1 minute at primer-specific temperature, and extension for 2 minutes at 72°C. A Perkin-Elmer 9700 thermal cycler was used for these amplifications. PCR products were analyzed by electrophoresis in 1% agarose submarine gels for size (Gibco BRL, Life Technologies, Grand Island, NY) in 1× TBE buffer. DNA fragments were purified with the Qiaquick DNA extraction kit (Qiagen, Hilden, Germany). DNA sequences were determined in an ABI Model 3700 sequencer at the University of Michigan's DNA Sequencing Core, Ann Arbor, MI.
Additional studies were performed to sequence exon 5 of HK from DNA from Williams trait plasma and normal leukocytes. PCR primers were prepared for HK exon 5 (GenBank accession no. M11524). The sense primer, 5′-ATTGTTTCAGGTGGTGGCTG-3′, was prepared corresponding to nucleotide positions 1 to 20 of exon 5. The antisense primer, 5′-ACGCCTACTTACACCATTCC-3′, was derived from nucleotides 110 to 128 of exon 5. Further studies were performed to sequence kininogen's 2.1 kb intron 9. A deductive PCR sequencing approach was employed starting with a sense primer from the 3′ end of exon 9 (GenBank accession no. M11528) and an antisense primer from the 5′ end of exon 10. The sequential set of sense and antisense primers for sequencing intron 9 are shown in Table 2 and Table 3.
Further investigations were performed to sequence the heavy chain of PK from the propositus and his family. PCR was performed to amplify exons 3 to 6 and 8 to 10 corresponding to PK's Apple domains 1, 2, and 4 using the primers and PCR conditions as described by Yu et al.27
Results
Investigations on plasma HK and PK levels in the patient and family
The propositus had a markedly prolonged APTT with a normal prothrombin time, thrombin time, and clottable fibrinogen (Table 4). On a 1:1 mixing study, there was complete correction of the APTT. Investigations for most of the established prothrombotic risk factors were negative (Table 4). The patient had a factor XII coagulant activity of 0.35 U/mL. The patient's prolonged APTT was mostly due to an HK deficiency. The propositus had less than 0.01 U/mL HK procoagulant activity when compared with pooled normal human plasma (Table 5). The HK procoagulant activities of his father, mother, and sister on a fresh sample upon presentation in France were 0.38 U/mL, 0.59 U/mL, and 0.65 U/mL, respectively. PK activity and antigen levels also were reduced in the propositus and family members (Table 5). Some reduction of the PK values could have resulted from lyophilization, shipment to, and resuspension of the plasmas in Ann Arbor, MI. Reduced HK values in family members were noted in the resuspended lyophilized plasma samples. However, low plasma PK values also have previously been reported in HK-deficient patients.28 Reconstitution of their plasma with up to 1.2 U/mL purified HK did not restore the PK amidolytic or coagulant activity to a normal level (data not shown), as previously reported in other HK-deficient patients.27 The Apple domains 1, 2, and 4 of the heavy chain of the prekallikrein gene of the propositus and family members were sequenced looking for a polymorphism that might result in interference with HK binding to PK.29,30 In exon 5 that codes for Apple domain 2 of the prekallikrein gene, there was a polymorphism at bp 180 changing an adenine to guanidine that changed an asparagine to serine in the propositus and other family members.27
Immunoblot investigations
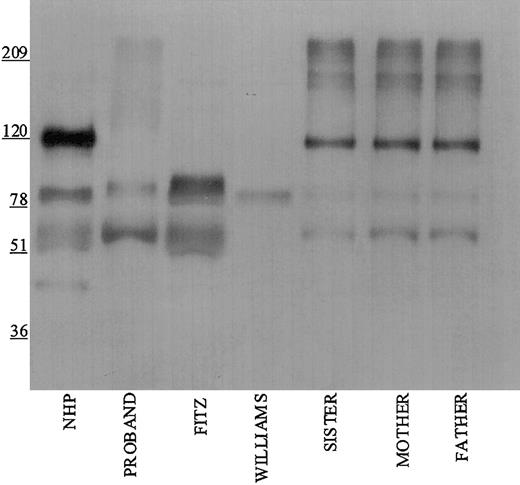
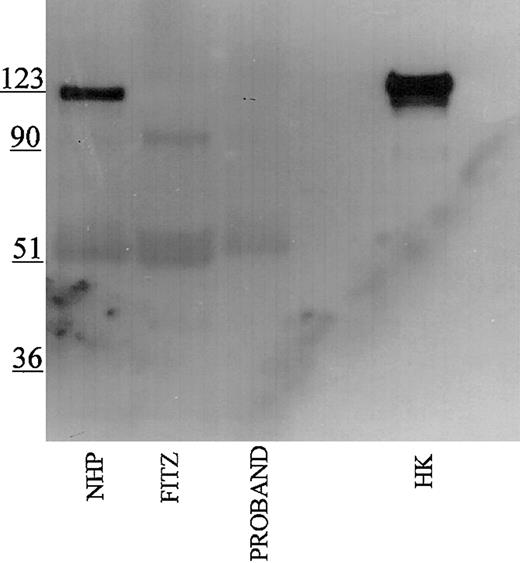
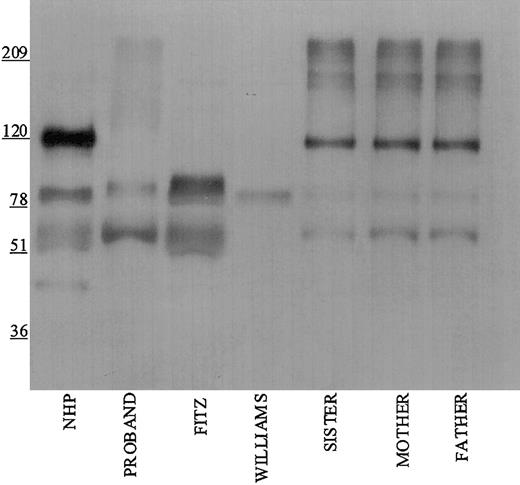
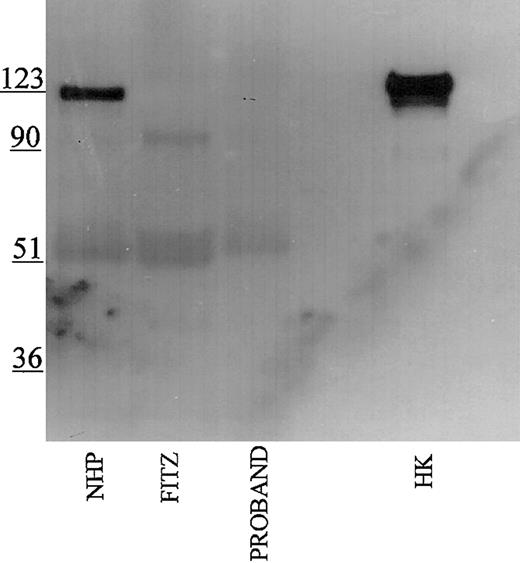
Studies were performed to immunophenotype the defect in the HK of the patient and his family members. Initial investigations were performed with a polyclonal antibody directed to the light chain of HK (Figure 1). Using this antibody, normal human plasma showed a prominent 120-kDa band for HK on reduced SDS-PAGE. Each member of the propositus' family also had a similar band, although at reduced quantity of antigen. No bands at 120 kDa were seen in the propositus, Fitzgerald, or Williams plasmas. Additional nonspecific bands both above and below the 120 kDa band were detected in all the plasmas using this antisera.
Immunoblotting of plasma with polyclonal antibody directed to the light chain of HK. Pooled normal human plasma (NHP) and plasmas from the family members were diluted 1:20 in TBS prior to the addition of an equal volume of sample buffer. Plasmas from the propositus, Fitzgerald (FITZ), and Williams were diluted 1:10. All samples were reduced with 2% β-mercaptoethanol and boiling prior to being applied to a 7% SDS-PAGE. After electrophoresis, the samples were electroblotted onto nitrocellulose and incubated with polyclonal antibody AHMWK3 at 1:500 in Blotto.23 Bound antibody was detected by peroxidase-conjugated secondary antibody to goat immunoglobulin G (IgG) followed by chemiluminescence. The figure is a representative immunoblot of 3. In this and subsequent figures, the numbers on the left represent molecular mass standards in kilodaltons.
Immunoblotting of plasma with polyclonal antibody directed to the light chain of HK. Pooled normal human plasma (NHP) and plasmas from the family members were diluted 1:20 in TBS prior to the addition of an equal volume of sample buffer. Plasmas from the propositus, Fitzgerald (FITZ), and Williams were diluted 1:10. All samples were reduced with 2% β-mercaptoethanol and boiling prior to being applied to a 7% SDS-PAGE. After electrophoresis, the samples were electroblotted onto nitrocellulose and incubated with polyclonal antibody AHMWK3 at 1:500 in Blotto.23 Bound antibody was detected by peroxidase-conjugated secondary antibody to goat immunoglobulin G (IgG) followed by chemiluminescence. The figure is a representative immunoblot of 3. In this and subsequent figures, the numbers on the left represent molecular mass standards in kilodaltons.
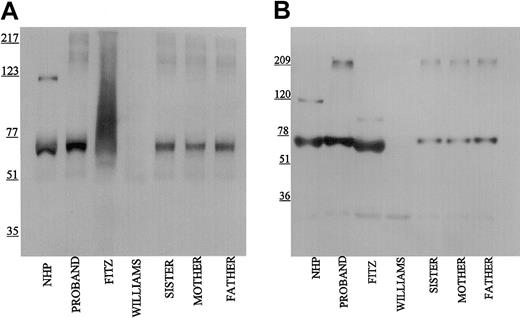
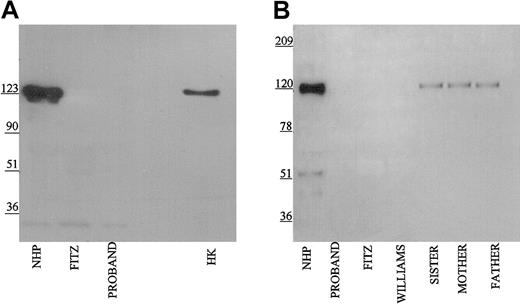
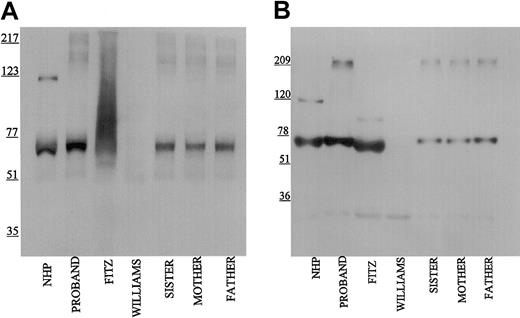
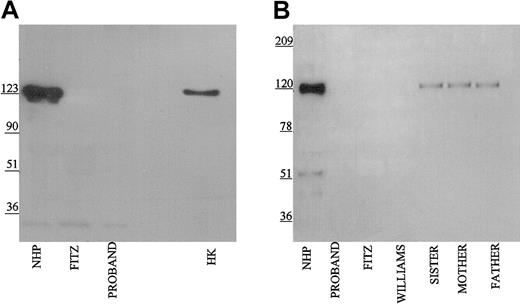
Using the Mab HKH14 to kininogens' domain 3, LK was detected in the propositus's plasma similar to normal human plasma and the patient's family (Figure 2A). In other studies not shown, the apparent plasma concentration of LK in the propositus on immunoblot was similar to other members of its family. This finding indicated that the kininogen gene was present in the propositus to direct the expression of LK of apparent normal size. Mab HKH14 also recognized the heavy chain of HK at 120 kDa in NHP. No band was seen in Williams plasma and any kininogen antigen in Fitzgerald plasma appeared as a broad smear (Figure 2A). Mab HKH15 recognized the same epitopes in all the plasmas except Williams plasma and recognized a discrete LK band in Fitzgerald plasma (Figure 2B). HKH15 also detected the heavy chain of HK in normal human plasma and faintly in the plasmas of the patient's family. Remarkably, Mab HKH15 also recognized a faint though distinct band in Fitzgerald plasma at 92 kDa (Figure 2B).
Immunoblot of plasma with monoclonal antibodies HKH 14 and HKH 15. Pooled normal human plasma (NHP), plasmas from the family members (1:20 in TBS), and plasmas from the propositus, Fitzgerald (FITZ), and Williams (1:10) were electrophoresed on a reducing 7% SDS-PAGE, followed by electroblotting onto nitrocellulose and incubation with monoclonal antibodies HKH14 (panel A) and HKH15 (panel B) at 2 μg/mL in Blotto.23 Bound antibody was detected by peroxidaseconjugated secondary antibody to mouse IgG followed by chemiluminescence.
Immunoblot of plasma with monoclonal antibodies HKH 14 and HKH 15. Pooled normal human plasma (NHP), plasmas from the family members (1:20 in TBS), and plasmas from the propositus, Fitzgerald (FITZ), and Williams (1:10) were electrophoresed on a reducing 7% SDS-PAGE, followed by electroblotting onto nitrocellulose and incubation with monoclonal antibodies HKH14 (panel A) and HKH15 (panel B) at 2 μg/mL in Blotto.23 Bound antibody was detected by peroxidaseconjugated secondary antibody to mouse IgG followed by chemiluminescence.
When Mab HKL13 directed to amino acids 402 to 419 of domain 5 of the HK light chain was used in immunoblot studies, no protein was detected in propositus and Williams plasmas (Figure 3). However, this Mab to the light chain of HK detected the 120-kDa band of reduced HK in normal plasma and the patient's family. Interestingly, a strong 92-kDa protein band was also detected in Fitzgerald plasma. Similar findings were seen when Mab HKL10, which is directed to the same epitope on domain 5 as HKL13 (Table 1), was used for immunoblot (data not shown). These data indicated for the first time that Fitzgerald plasma contained a smaller-sized form of HK.
Immunoblot of plasma with monoclonal antibody HKL13. Samples were processed as detailed in the legend to Figure 1 except that monoclonal antibody HKL13 was used at 2 μg/mL.
Immunoblot of plasma with monoclonal antibody HKL13. Samples were processed as detailed in the legend to Figure 1 except that monoclonal antibody HKL13 was used at 2 μg/mL.
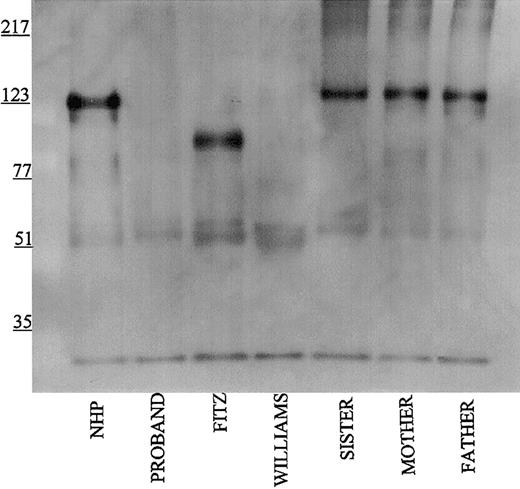
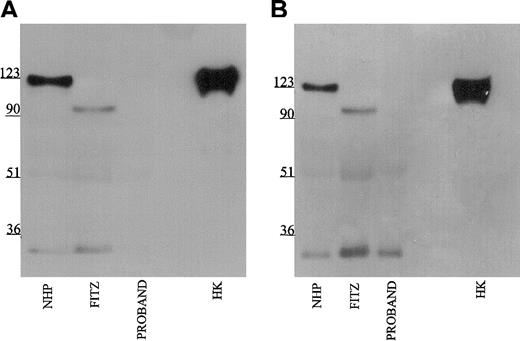
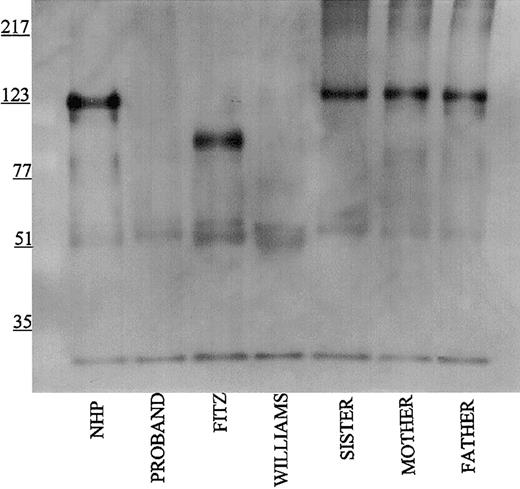
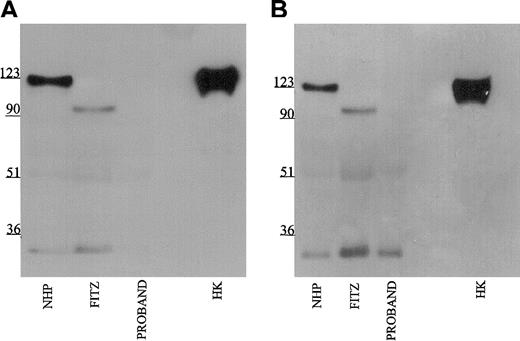
Further investigations were performed to map the 92-kDa form of HK in Fitzgerald plasma. Using monoclonal antibodies HKL12 and HKL14 which map to amino acids 440 to 458 and 420 to 502, respectively, on domain 5 of the HK light chain, the 92-kDa band in Fitzgerald plasma is detected (Figure 4A-B). Likewise, when the antipeptide antibody AHKH20, which is exclusively directed to amino acids 479 to 498 on domain 5, was used for immunoblot, the 92-kDa band of Fitzgerald HK was faintly seen (Figure 5). When Mabs HKL24 or HKL16, which are directed to amino acids 543 to 554 or 569 to 595 on domain 6, respectively, were used, no HK was detected in Fitzgerald plasma (Figure 6A-B). Similar findings were made with Mab HKL25, which has the same epitope as HKL16 (Table 1, data not shown). These combined data indicated that the protein defect in Fitzgerald HK was an absent domain 6 most probably after position 502 of mature HK.
Immunoblot of plasma with monoclonal antibodies HKL12 and HKL14. Samples were processed as detailed in the legend to Figure 1 except that monoclonal antibodies HKL12 (panel A) and HKL14 (panel B) were used at 2 μg/mL.
Immunoblot of plasma with monoclonal antibodies HKL12 and HKL14. Samples were processed as detailed in the legend to Figure 1 except that monoclonal antibodies HKL12 (panel A) and HKL14 (panel B) were used at 2 μg/mL.
Immunoblot of plasma using antibody AHKH20. Samples were processed as detailed in the legend to Figure 1 except that rabbit antipeptide antibody AHKH20 was used at a 1:500 dilution. Bound antibody was detected by peroxidaseconjugated secondary antibody to rabbit IgG followed by chemiluminescence.
Immunoblot of plasma using antibody AHKH20. Samples were processed as detailed in the legend to Figure 1 except that rabbit antipeptide antibody AHKH20 was used at a 1:500 dilution. Bound antibody was detected by peroxidaseconjugated secondary antibody to rabbit IgG followed by chemiluminescence.
Immunoblot of plasma with monoclonal antibodies HKL24 and HKL16. Samples were processed as detailed in the legend to Figure 1 except that monoclonal antibodies HKL24 (panel A) and HKL16 (panel B) were used at 2 μg/mL.
Immunoblot of plasma with monoclonal antibodies HKL24 and HKL16. Samples were processed as detailed in the legend to Figure 1 except that monoclonal antibodies HKL24 (panel A) and HKL16 (panel B) were used at 2 μg/mL.
Determination of the molecular defect in the patient and Fitzgerald DNA
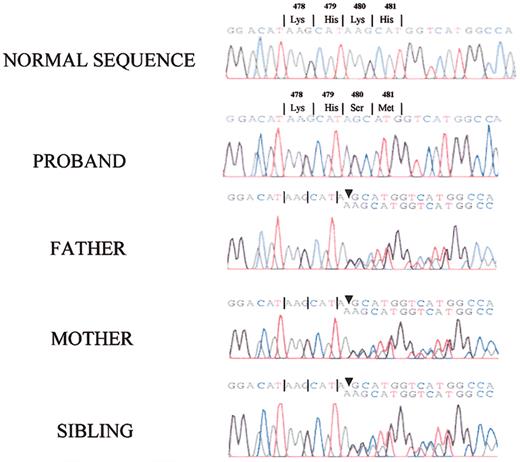
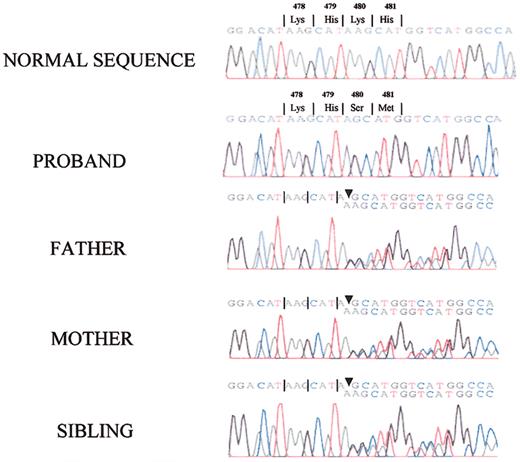
Since the patient had LK antigen encoded by exons 1 to 9 and 11 of the kininogen gene, the defect causing the absence of HK in the patient's plasma should reside in exon 10, which codes for domains 5 and 6 of HK. After patient exon 10 was prepared by PCR, a single base pair deletion was found at position 367 in exon 10 (bp 1492 of the full-length cDNA and amino acid 480 in the mature protein) that changed codon AAG for lysine to codon AGC for serine (Figure 7). This deletion resulted in an altered amino acid sequence from position 480 in the mature protein and a premature termination at bp 1597 corresponding to amino acid position 532 on the mature protein. The patient's parents and sister were heterozygous for this deletion. Also, all family members were found to have a T to C nucleotide change at bp position 627 of exon 10, corresponding to amino acid 563 in the prekallikrein/factor XI binding region of HK. This nucleotide change resulted in a codon change that produced a threonine in place of an isoleucine.
Identification of a base pair deletion within exon 10 of the HK gene. The amplified PCR fragments of exon 10 from normal human, the propositus', and all family members' DNA underwent direct nucleotide sequencing. The propositus exhibited a homozygous base pair deletion of an adenine at nucleotide position 367 in exon 10 (corresponding to position 1492 of the full-length mRNA and position 480 of the mature protein). All family members were heterozygous for the same defect in the PCR of their amplified DNA.
Identification of a base pair deletion within exon 10 of the HK gene. The amplified PCR fragments of exon 10 from normal human, the propositus', and all family members' DNA underwent direct nucleotide sequencing. The propositus exhibited a homozygous base pair deletion of an adenine at nucleotide position 367 in exon 10 (corresponding to position 1492 of the full-length mRNA and position 480 of the mature protein). All family members were heterozygous for the same defect in the PCR of their amplified DNA.
DNA amplified from authentic Fitzgerald plasma samples was found to have no changes in the sequence of exon 10. The veracity of preparing DNA from a more than 25-year-old frozen plasma specimen was confirmed by preparing DNA from frozen Williams trait plasma from the early 1980s. DNA from Williams plasma demonstrated the nucleotide 586 C to T transition in exon 5 that results in a stop codon at amino acid position 177 of the mature protein as previously described.2 Further studies sequenced the 2.1 kb intron 9 of the kininogen gene from 4 healthy individuals (GenBank accession no. AY183666) and compared this sequence to that of Fitzgerald DNA (GenBank accession no. AY206689). At nucleotide position 1559 of intron 9 of normal DNA (T1559TGTTGTTGTTGTTGTA1575), there was a mutation in 17 consecutive base pairs in Fitzgerald DNA (G1559GTGGTGGTGGTGGTGG1575). Also at nucleotide position 1578, a GT sequence in normal DNA was changed to TG in Fitzgerald's intron 9. Last, 3 single base pair polymorphisms were found in Fitzgerald intron 9 at nucleotide positions 119 (C to T), 1586 (T to G), and 1736 (A to G), which were not present in the DNA of the 4 healthy individuals.
Discussion
HK deficiency is extremely rare. It is of note that the HK deficiency in the patient and his family only was recognized after a traumatic injury that was associated with thrombosis. Kininogens have been believed to contribute to the constitutive anticoagulant nature of the intravascular compartment by their ability to inhibit thrombin activation of platelets, allow for kinetically favorable single chain urokinase formation, and stimulate nitric oxide, prostacyclin, and tissue plasminogen activator liberation.1,5,7,9,10,14,19 The absence of HK and the presence of a posttraumatic arterial lesion may have summated into the episode of thrombosis in this individual. It is of interest that this patient also had a slightly reduced factor XII coagulant activity. The reason for this abnormality is not known, but it too may have contributed to the summation of risk factors that resulted in clinical thrombosis in this patient. However, until such time as appropriate animal models are developed to examine the hypothesis that kininogens are anticoagulant, this interpretation should be considered as conjecture.
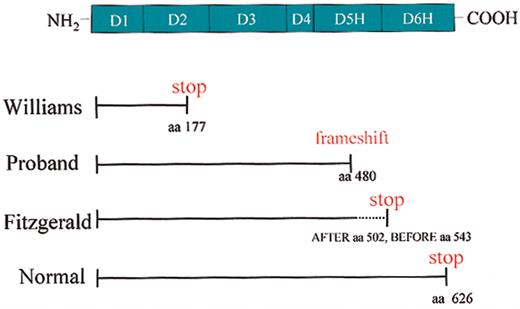
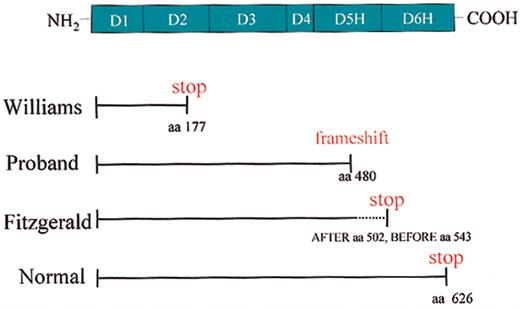
These investigations provide an opportunity to better understand the molecular basis of HK deficiency. Williams trait is characterized by a premature stop codon at positions 586 to 588 due to a C to T transition2 resulting in a shortened mRNA encoding the signal peptide (18 amino acid residues), domain 1 (112 residues), and a portion of domain 2 (65 residues) (Figure 8). Presently, it is not known whether the truncated mRNA is actually translated into a protein of 177 residues (after removal of the signal peptide) or whether this truncated form of Williams kininogen is stored and/or rapidly degraded in hepatocytes of the liver (ie, the major source of human kininogen). Clearly, this truncated form is not secreted since no kininogen antigen is detected in Williams plasma.
Characterization of molecular defects of HK-deficient patients. The domain structure of HK is shown. Solid lines indicate the size of the plasma proteins. The dotted lines indicate the size of the DNA that has the potential to make the protein. Williams trait (Williams) has a premature stop codon at amino acid (aa) 177 of the mature protein. The propositus (Proband) has no plasma HK as result of a single base pair deletion at amino acid 480 of the mature protein and the degeneration into a stop codon downstream. Fitzgerald trait (Fitzgerald) has identified protein antigen through amino acid 502 of the mature protein. Normal full-length HK is 626 amino acids in the mature, secreted protein.
Characterization of molecular defects of HK-deficient patients. The domain structure of HK is shown. Solid lines indicate the size of the plasma proteins. The dotted lines indicate the size of the DNA that has the potential to make the protein. Williams trait (Williams) has a premature stop codon at amino acid (aa) 177 of the mature protein. The propositus (Proband) has no plasma HK as result of a single base pair deletion at amino acid 480 of the mature protein and the degeneration into a stop codon downstream. Fitzgerald trait (Fitzgerald) has identified protein antigen through amino acid 502 of the mature protein. Normal full-length HK is 626 amino acids in the mature, secreted protein.
The propositus presented in this study has a single nucleotide deletion at position 1492 (nucleotide sequence) causing a frame-shift from position 480 on (amino acid sequence of mature HK) and premature termination of HK biosynthesis after position 532 (stop codon at positions 1597 to 1599 of the nucleotide sequence) (Figure 8). Accordingly, the aberrant mRNA encodes a shortened form of HK that comprises 479 residues of the authentic HK sequence covering domains 1 to 4 and most of domain 5, followed by 53 residues of an unrelated sequence starting with a Ser residue such that a mature protein of 532 residues would result after removal of the signal peptide. Because no liver biopsy of the propositus is available to us, questions could not be addressed whether this truncated form of HK is synthesized and properly processed, but accumulated and degraded in hepatocytes. Future studies using the mutated HK cDNA for recombinant expression in mammalian cells should be helpful to answer these questions and to address the possibility that the newly added C terminus of 53 residues may expose “retention” signals preventing the secretion of the truncated HK form or inducing a rapid intracellular degradation without significant secretion of the aberrant HK protein.
Fitzgerald trait, the first HK deficiency ever reported,4 differs from other reported HK deficiencies in that a smaller form of HK of 92 kDa (as compared with 120 kDa of normal kininogen) is synthesized, processed, and secreted to be present in its plasma (Figure 8). Antibody mapping studies indicate that the 92-kDa form lacks major portions of domain 6 and that a premature stop may occur between residues 502 and 543, though the precise site has not been determined (Figure 8). The fact that the missing portion contains at least 4 O-glycosylation sites (positions 553, 559, 575, and 610 of the mature protein) explains the difference in the predicted molecular weights (∼ 28 kDa) between the mutated form (92 kDa) and native HK (120 kDa). The antigen mapping studies indicate that a shortened form of HK comprising domains 1 through 5 and a short stretch of the N-terminal portion of domain 6 is sufficient for biosynthesis, processing, and secretion of HK. Importantly, the 92-kDa form of Fitzgerald HK lacks the overlapping binding sites for factor XI (residues 556 to 613)12 and prekallikrein (residues 569 to 595),31,32 and thus is functionally defective because it cannot support contact activation.32 The “dropping” of the factor XI/prekallikrein binding sites also removes a cysteine residue that forms a disulfide bridge with the extreme N-terminal portion of domain 1 (position 10), thereby forcing the HK molecule into a “tensed” conformation that is only relieved by cutting out the kinin sequence. Accordingly, one may expect that the 92-kDa form may have a “free” cysteine residue in domain 1 unless Cys10 is engaged in a dimer or forms an unusual bridge to an “extra” Cys residue present in the C-terminal extension caused by a frameshift. These findings also solve the conundrum that Fitzgerald plasma contains normal amounts of total releasable kinin5 because the 92-kDa form contains the complete kinin domain D4, and is therefore accessible for kininogenases such as plasma and tissue kallikreins.
Further efforts identified a possible gene defect(s) in Fitzgerald trait DNA that may give rise to a 92-kDa HK protein form in plasma. DNA sequencing of the PCR product of Fitzgerald plasma DNA does not show any mutation in exon 10. The reliability of this result was confirmed by determining the exon 5 defect in plasma DNA from the Williams trait. Using deductive PCR sequencing of Fitzgerald DNA, the Fitzgerald intron 9 was found to contain a 17-bp mutation as well as one double and 3 single nucleotide polymorphisms when compared with the DNA sequence of intron 9 from 4 healthy individuals. How these intron defects lead to the absence of the synthesis of domain 6 of Fitzgerald HK is not known. HK protein is produced by an alternative splicing mechanism in exon 10 of the single human kininogen gene.33,34 Each of the intron 9 mutations alone or combined could affect the alternative usage of termination sites and/or change the alternative slicing of the primary transcript of Fitzgerald DNA generating a modified mRNA to produce an altered HK that is eventually targeted to the plasma through the secretory pathways of hepatocytes.33,34 This latter possibility is reminiscent of the finding of a Japanese patient lacking HK antigen in the plasma where a partial deletion in intron 7 was proposed as a cause of this defect.35
Last, it is of interest that the plasma PK levels in the propositus and his family did not correct to normal upon addition of purified HK in their plasmas. This finding is different from that previously reported in HK-deficient plasmas.28 It is possible that the plasma PK was damaged as result of lyophilization, shipment, and resuspension. However, a polymorphism in the Apple domain 2 resulting in a replacement of an asparagine with a serine at amino acid position 124 in the heavy chain of PK was detected in the propositus and his family after sequencing the HK binding regions of Apple domains 1, 2, and 4 on PK.29,30 This polymorphism is previously recognized to exist in 30% of the population in the United States.27 It is presently unknown whether this polymorphism influences the plasma level of PK. A single base pair polymorphism was also noted at amino acid 563 of domain 6 of the mature HK, which is in the prekallikrein binding region of HK.13 In 2 laboratories it has been shown that amino acids 569 to 595 on domain 6 are the essential ones to bind PK.31,36 The influence of this latter polymorphism to plasma PK levels, if any, is not known.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-11-3329.
Supported by National Institutes of Health grants HL52779, HL57346, HL61981, and HL65194.
Y.K. and V.P. contributed equally to this report.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Professor Marc Tardieu for assistance in the clinical management of the recent patient. We appreciate the generosity of Dr Robert Waldmann of Clinton, MI, who provided us with the stained skin window studies performed on Fitzgerald granulocytes. We also appreciate the generosity of Dr Oscar Carretero of Detroit, MI, who provided us with additional Fitzgerald trait plasma.