Abstract
Kaposi sarcoma–associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8) is causally associated with Kaposi sarcoma (KS). The absence of a cell culture system that effectively reproduces the composite mechanisms governing initiation and maintenance of HHV-8 infection (lytic and latent) in KS endothelial cells, however, has left important questions unanswered. Here, we report a culture system in which the earliest events that accompany HHV-8 infection could be surveyed in primary endothelial cells. Binding of HHV-8 to microvascular dermal endothelial cells (MVDECs) was directly compared with other primary target cells implicated in HHV-8–associated diseases. Virus attachment, fusion, internalization and transport within MVDECs was monitored by electron microscopy. Studies of genome configuration revealed that rapid circularization of the viral DNA occurred on entry, though by 72 hours after infection linear DNAs accumulated and early as well as late lytic RNAs (T1.1, K8.1) could be detected. The latency transcripts (LT1/LT2) were first detected on day 8, demonstrating that both lytic and latent infection were initiated. Although most lytic transcripts accrued until passage, open-reading frame–74 RNAs fluctuated with a fixed periodicity, suggesting that early replication after infection of MVDECs was synchronous.
Introduction
Kaposi sarcoma–associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8) is the first human γ-herpesvirus identified since the 1964 discovery of Epstein-Barr virus (EBV).1,2 HHV-8 has been causally implicated in the pathogenesis of 3 proliferative disorders that occur with increased frequency in the immunocompromised host: Kaposi sarcoma (KS), primary effusion lymphoma (PEL), and the plasmablastic variant of multicentric Castleman disease (MCD).1-4 Although KS is uncommon, it is the most common malignancy associated with human immunodeficiency virus infection. KS lesions may spontaneously resolve following immune reconstitution or may wax and wane with an individual's state of T-cell competence.5 6 This remarkable feature, shared by EBV-associated B-cell lymphoproliferative disease, suggests that a latent reservoir of HHV-8 infection in vivo gives rise to KS.
Many HHV-8–encoded proteins, which contribute to the pathogenesis of KS, PEL, and MCD, are homologs of cellular genes involved in inflammation, cell cycle regulation, and angiogenesis.7Two such proteins are those encoded by open-reading frame (ORF)–74 and ORF72. The respective proteins are expressed in distinct phases of the virus life cycle.7 ORF74 encodes a lytic viral G protein–coupled receptor (vGPCR), which is homologous to human interleukin-8 receptors.7,8 When injected into mice, vGPCR-transfected rodent fibroblasts cause spindle cell tumors with prominent vasculature.7,9 ORF72 encodes a latent viral cyclin that can activate kinases that inactivate retinoblastoma protein (pRB), a checkpoint protein that inhibits entry into S phase.7 10
In the complex lesions of KS, viral gene products are expressed in endothelial cells, in spindle cells of probable endothelial origin, in monocytes, and possibly in some infiltrating lymphocytes. The endothelial cells primarily harbor virus as circular episomes and synthesize latency-associated gene products, whereas the smaller monocyte subpopulation is productively infected.11 12
Normal primary endothelial cells can be infected with HHV-8 in vitro. However, the efficiency and outcome of infections have been variable despite diverse efforts to optimize in vitro culture.13-17In contrast, herpesvirus infection of relevant primary cells, whether productive or causing immortalization, is usually robust. Thus, the recent studies of endothelial cell infection raise several questions concerning whether a more physiologic target cell remains to be identified, whether variable transmission in vitro is caused by technical difficulties, whether in vitro infection is blocked at a particular phase of the virus life cycle, or whether indeed HHV-8 infection is inefficient when compared with infection by other members of the human herpesvirus family. To gain a better understanding of the viral and cellular events that occur upon initiation of infection, early in vitro transmission of HHV-8 to primary vascular endothelial cell populations was investigated.
Materials and methods
Cells
BCBL-1 cells18 obtained from the AIDS Research and Reference Reagent Program (ARRRP) (Rockville, MD) were routinely maintained in RPMI containing l-glutamine (Mediatech, Herndon, VA) to which had been added 2 mM Hepes buffer (Biowhittaker, Walkersville, MD), 50 μM beta-mercaptoethanol (Sigma, St Louis, MO), 100 U/mL penicillin G sodium (Biowhittaker), 100 μg/mL streptomycin sulfate (Biowhittaker), and 10% heat-inactivated (hi) bovine calf serum (BCS) (Atlanta Biologicals, Norcross, GA) in a 5% CO2 incubator at 37°C. B95-8 cells,19obtained from the American Type Culture Collection (ATCC) (Rockville, MD), were maintained as described for BCBL-1 cells except that beta-mercaptoethanol was omitted. Human neonatal foreskin microvascular dermal endothelial cells (fMVDECs) cryopreserved upon third or fourth passage, adult breast dermal microvascular endothelial cells (bMVDECs), human uterine–myometrial microvascular endothelial cells (uterine or mMVECs), and human umbilical vein endothelial cells (HUVECs) were purchased from Clonetics (San Diego, CA). Endothelial cells were cultured in EGM-2-MV medium with BulletKit supplements (Clonetics) in a 5% CO2 incubator at 37°C. We obtained 293 epithelial cells20 from the ATCC and 293L cells21 from the ARRRP. These lines were cultured in Dulbecco modified Eagle medium containingl-glutamine, 100 U/mL penicillin G sodium, 100 μg/mL streptomycin sulfate, and 10% hi BCS in a 10% CO2incubator at 37°C. Discarded peripheral blood mononuclear cells (PBMCs) were obtained from the blood of healthy, prescreened donors (Blood Component Laboratory, Dana-Farber Cancer Institute, Boston, MA). PBMCs were separated from contaminating cells, dead cells, and debris by Ficoll-Hypaque density gradient centrifugation by means of Lymphocyte Separation Medium, 1.077 g/mL Ficoll (Biowhittaker).
Virus preparation and labeling
HHV-8 was prepared and concentrated from the supernatant of BCBL-1 cells that had been maintained in culture for less than 3 months. The cells were grown to a density of 1 × 106/mL, either in spinner flasks (Bellco Glass, Vineland, NJ) (20 L capacity, filled to 8 L) or roller bottles (Falcon, Lincoln Park, NJ) (2 L, filled to 600 mL). Control virus, EBV, was identically prepared and concentrated from the supernatant of B95-8 cells. Virus production was induced by adding sodium butyrate (Sigma) to a final concentration of 3 mM for 72 hours. At the end of the incubation period, cells were allowed to settle for approximately 12 hours. No freeze-thaw step was performed.22 The supernatant was carefully removed and passed through a 0.8-μm fine porosity filter (Nalgene, Rochester, NY) to exclude residual cells and debris. Virus was centrifuged at 17 700g in a JA-10 rotor (Beckman, Palo Alto, CA) for 2 hours. The butyrate-containing supernatant was discarded, and the pellet was resuspended in RPMI to produce a 200-fold final concentration. In some experiments, resuspended virus was further purified on a continuous or a discontinuous Dextran gradient as previously described.23 Resuspended virus was dispersed into 1.8-mL CryoTube vials (Nunc, Naperville, IL) and frozen in a −80°C freezer until use. Gradient-purified HHV-8 was fluorescein isothiocyanate (FITC)–conjugated, as previously described for EBV,24 and designated FITC–HHV-8.
Virus infection
Endothelial cells were seeded at 2 × 105 per 100 mm2 tissue-culture dish (Falcon, Franklin Lakes, NJ) and cultured until 40% to 50% confluent. The cells were washed once with phosphate-buffered saline (PBS), overlaid with 2 mL virus suspension that was diluted 1:1 with endothelial cell tissue-culture media, and incubated for 3 hours at 37°C. At the end of the incubation, the cells were washed 2 or 3 times with PBS and reincubated as described above. Cells were harvested at specified time points and processed for preparation of total RNA, DNA, or protein as described below. Cells were split 1:2 or 1:3 on day 10.
Antibodies and reagents for immunofluorescence
Phycoerythrin (PE)–conjugated monoclonal antibodies (mAbs) used for direct staining and cytometric analysis were as follows: Anti-CD19, anti–CD-3, anti-CD14, and anti–MOPC-21 (control) were from Sigma. Anti-CD16 was from Becton Dickinson (San Jose, CA). Streptavidin-fluorescein conjugate, designated FITC-avidin and used as a control for FITC–HHV-8 staining, was purchased from Biosource (Camarillo, CA). The 2L10, a murine mAb directed to gp350/220, was a gift of Dr Gary Pearson. UPC10, an isotype-matched irrelevant control mAb (immunoglobulin G2a [IgG2a]), was purchased from ICN/Cappel (Aurora, OH). FITC-labeled goat F(ab′)2antimouse IgG was purchased from Biosource.
Fluorescence cytometry
HHV-8.
Purified PBMCs were washed 3 times in RPMI, 2% hi FCS, and 0.1% sodium azide and were incubated with either FITC–HHV-8 or FITC-avidin for 30 minutes on ice. To remove unbound ligand, 2 washes in ice-cold medium containing 2% hi FCS were performed. The cells were reincubated under identical conditions with one of the respective PE-conjugated mAbs: anti-CD3 directed to T cells, anti-CD19 directed to B cells, anti-CD14 directed to monocyte/macrophages, anti-CD16 directed primarily to natural killer (NK) cells, or with a PE-conjugated control antibody (MOPC-21) that has no known specificity for human cells. Following the second incubation, cells were washed in media 3 additional times, fixed with 2% paraformaldehyde in media, and analyzed by cytometry on a FACScan (Becton Dickinson) (FITC, FL1 channel; and PE, FL2 channel). Cytometric analyses of the purified PBMCs with the respective PE-conjugated antibodies and the PE-conjugated control were performed prior to definitive experiments to optimize the setting of gates. Adherent cells (fMVDECs, HUVECs, and 293 cells) were removed from tissue-culture dishes by incubation in media containing 2 mM EDTA at 37°C for 10 minutes. The cells were pipetted gently 3 times to assure complete detachment and then washed 3 times in media containing 2% hi FCS and 0.1% sodium azide prior to staining with FITC–HHV-8 or FITC-avidin as described above.
EBV.
Suspended cells (fMVDECs, HUVECs, 293 cells, and JY-B-cell line) were incubated with purified EBV for 30 minutes on ice, washed twice, and reincubated with either mAb 2L10 directed to EBV membrane glycoprotein 350 or mAb UPC10 (control IgG2a antibody) for another 30 minutes on ice. They were then rewashed twice and reincubated with FITC-goat F(ab′)2 anti-mouse IgG for 30 minutes on ice, washed 2 additional times, fixed, and analyzed as described above.
Electron microscopy
The fMVDECs were infected with HHV-8 or were mock-infected for 1 hour at 37°C. The supernatant was discarded, and the cells were rinsed 3 times in PBS to remove unbound virus. The cell monolayer was covered with 2% glutaraldehyde in 0.1 M cacodylate buffer and stored at 4°C. The monolayer was subsequently imbedded in situ in Epon resin, postfixed in 1% osmium tetroxide, dehydrated in a graded series of solutions of ethanol and propylene oxide, cut into thin sections, and reimbedded in Epon resin. The thin sections were stained with uranyl acetate and lead citrate and were examined on a JEOL 100cx transmission electron microscope (JEOL USA, Peabody, MA).
Polymerase chain reaction–based in situ lysis hybridization
BCBL-1 cells (uninduced or induced with 3 mM sodium butyrate for 72 hours) and fMVDECs (uninfected or infected with HHV-8 for various times) were harvested, lysed in wells, and electrophoresed through in situ lysis gels as described.25 A grid was created and gel slices were prepared. DNA was extracted from the individual slices, resuspended, and aliquoted as described.26 DNA polymerase chain reaction (PCR) was performed with upstream and downstream oligonucleotide primers (described below) that amplified a 394–base pair (bp) fragment from the genomic region encoding T1.1.27,28 The conditions were 94°C for 3 minutes, 35 cycles of 94°C for 1 minute, 62°C for 1 minute, 72°C for 1 minute, and a final extension step at 72°C for 7 minutes. The entire amplified product from each of the slices was electrophoresed through a 1% agarose gel (left to right on the agarose gel correlated with top to bottom slices of the in situ lysis gel), transferred to nitrocellulose, and probed with a32P-labeled T1.1 complementary DNA (cDNA) (described below) generously provided by Dr Don Ganem (University of California–San Francisco, CA).28 The relative positions of migration of covalently closed circular versus linear HHV-8 genomes derived from BCBL-1 were determined in all experiments as a positive control. Uninfected fMVDECs were included in all experiments as a negative control.
Probes and RNA blot hybridization
Probes used for detection of total RNA or DNA by blot hybridization were as follows: for detection of T1.1, also known as polyadenylated nuclear RNA (PAN) or nut-1, ORFK7, a 1.1-kilobase (kb) cDNA EcoRI-XhoI fragment excised from pBS-T1.1 generously provided by Don Ganem was used. For detection of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a cDNA clone, pHcGAP,29 was obtained from the ATCC. To detect K8.1 (gp35/37), a 750-bp cDNA fragment isolated by reverse-transcription PCR (RT-PCR), as described below, was used. For detection of latent transcript 1 (LT1) and LT2,30 a 795-bp cDNA fragment encoding ORF72, the viral cyclin (vCYC) was synthesized by RT-PCR as described below. A 1045-bp cDNA probe was synthesized for detection of ORF74, the vGPCR, as described below. The BCBL-1 cell line was used as the source for total RNA in these syntheses. All probes were random prime-labeled as described previously.31 Total RNA was isolated, and RNA blot hybridization was performed as previously described.31
RT-PCR analysis
Total RNA (0.5 μg), isolated at various times from relevant primary cells and cell lines, was first treated with RQ1 DNAse (Promega, Madison, WI) as recommended by the manufacturer. The RNA was then reverse transcribed and sequentially amplified by means of the GeneAmp RNA PCR kit (Perkin-Elmer, Foster City, CA) according to directions provided by the manufacturer. Standard amplification conditions were 94°C for 3 minutes, 35 cycles of 94°C for 1 minute, annealing temperature for 1 minute, and 72°C for 1 minute, followed by 7 minutes' final extension at 72°C. Primers for KS330,1 ORF29,32 K8.1,33 and GAPDH34 are described in the respective references cited here. The remaining primers were as follows. ORF72 (vCYC): sense, 5′-GGAATTCCTATATGGCAACTGCCAAT-3′; antisense, 5′TCACAAGCTTAATAGCTGTCCAGAAT-3′. ORF74 (GPCR): sense, 5′-CCCAAGCTTATGGCGGCCGAGGATTTCCTA-3′; antisense, 5′-GGAATTCCTACGTGGTGGCGCCGGACAT-3′. T1.1 (PAN, nut1, ORFK7): sense, 5′-TTGGCTGCCGCTTCACCTAT-3′; antisense, 5′-CACCAGTGGGCGCTGCTTTC-3′.
Amplified cDNAs were separated on a 1% agarose gel, transferred to nitrocellulose membranes by means of a Turboblotter downward transfer system (Schleicher and Schuell, Keene, NH), and detected with32P-labeled probes made by the random priming method.31
Results
Differential attachment of HHV-8 to primary microvascular endothelial cells, PBMCs, and the 293 epithelial cell line
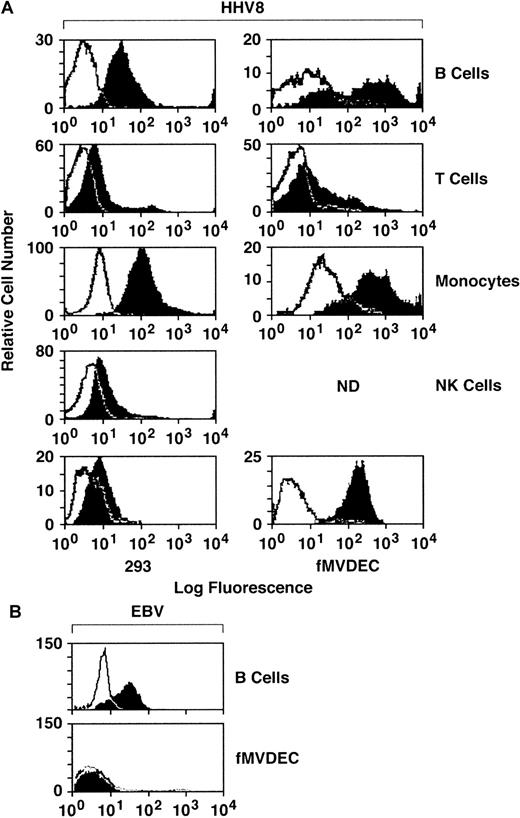
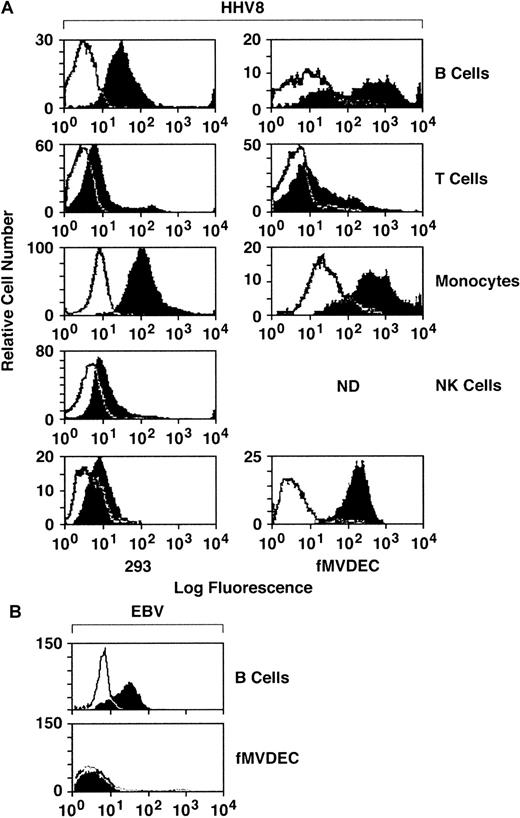
To explore the relationship of cellular attachment to HHV-8 entry, filtered and concentrated virus from the supernatant of the BCBL-1 cell line was labeled with FITC and was column purified. Binding of FITC–HHV-8 to primary PBMCs, fMVDECs, and the 293 cell line was assessed by flow cytometry. PE-conjugated mAbs were used to gate on PBMC subpopulations, permitting independent identification of B-cell (anti-CD19), T-cell (anti-CD3), monocyte (anti-CD14), and NK-cell (anti-CD16) subpopulations in identical assays. As shown in Figure 1, FITC–HHV-8 bound to the majority of peripheral blood B cells and monocytes, demonstrated by the relative number of virus-bearing cells (block) located to the right and outside of the negative control (FITC-avidin) curve (line). Comparative attachment of FITC-virus to T cells and NK cells was significantly less. Of note, FITC–HHV-8 bound to virtually all fMVDECs, exceeding virus attachment to either B cells or monocytes (Figure 1). In addition, the change in peak fluorescence of fMVDECs bearing FITC-virus compared with FITC-avidin (autofluorescence control) was greater than the corresponding change in peak fluorescence of the other cell populations studied. These results not only indicated that a greater number of fMVDECs bound virus, but also showed that fMVDECs contained a larger number of virus attachment sites per cell. Although the related γ-herpesvirus EBV readily bound to the surface of B-lymphoblastoid cells, EBV did not bind to fMVDECs (Figure 1, bottom). Binding of HHV-8 and EBV to HUVECs was similar to binding to fMVDECs (not shown). The 293 cells are susceptible to HHV-8 infection; however, in a representative experiment (Figure 1, bottom left), attachment of FITC-virus to these cells was comparatively low. Virus attachment to 293L cells (a clonal derivative of 293 epithelial cells reported to be more susceptible to HHV-8 infection than the parent line21) was similar. Unlabeled virus blocked binding of FITC–HHV-8 to all cell populations in a concentration-dependent manner, indicating that attachment based on competition-binding assay was specific (not shown).
Comparative attachment of FITC–HHV-8 to PBMC subpopulations (B, T, monocyte, and NK), primary MVDECs, and the 293 epithelial cell line.
(A) HHV-8 histograms. Binding of FITC HHV-8 to PBMCs isolated from leukopaks of 2 distinct donors (right and left histograms) was assessed by flow cytometry. The top 4 histograms compare binding of FITC-virus (block) with FITC-avidin (control) (line) on normal B-cell, T-cell, monocyte/macrophage, and NK-cell populations identified by prior staining of the PBMCs with PE-conjugated monospecific antibodies as described in the text. The bottom left panel demonstrates staining of 293 cells performed and analyzed at the same time as the leukopak on the left, and the bottom right panel shows staining of fMVDECs at the same time as the leukopak on the right. (B) EBV histograms. Binding of purified EBV to a B-lymphoblastoid cell line (JY) compared with fMVDEC displays a pattern distinct from HHV-8. Virus was detected by indirect immunofluorescence (“Materials and methods”) and analyzed by flow cytometry.
Comparative attachment of FITC–HHV-8 to PBMC subpopulations (B, T, monocyte, and NK), primary MVDECs, and the 293 epithelial cell line.
(A) HHV-8 histograms. Binding of FITC HHV-8 to PBMCs isolated from leukopaks of 2 distinct donors (right and left histograms) was assessed by flow cytometry. The top 4 histograms compare binding of FITC-virus (block) with FITC-avidin (control) (line) on normal B-cell, T-cell, monocyte/macrophage, and NK-cell populations identified by prior staining of the PBMCs with PE-conjugated monospecific antibodies as described in the text. The bottom left panel demonstrates staining of 293 cells performed and analyzed at the same time as the leukopak on the left, and the bottom right panel shows staining of fMVDECs at the same time as the leukopak on the right. (B) EBV histograms. Binding of purified EBV to a B-lymphoblastoid cell line (JY) compared with fMVDEC displays a pattern distinct from HHV-8. Virus was detected by indirect immunofluorescence (“Materials and methods”) and analyzed by flow cytometry.
Electron-microscopic analyses of attachment and internalization of HHV-8
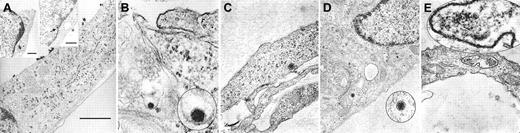
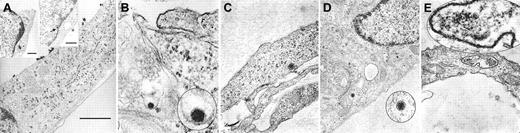
To visually confirm the attachment of HHV-8 to fMVDECs and to delineate the cellular events associated with virus entry, fMVDECs were incubated with HHV-8 at 37°C for 1 hour. Although precise knowledge of herpesvirus entry is limited, it is believed that herpesviruses enter cells by fusion of the viral envelope with the plasma membrane, resulting in the release of tegument proteins and the nucleocapsid into the cytoplasm, or alternatively by endocytosis followed by nucleocapsid release through the endosomal membrane.35-37 Of note, in the case of herpes simplex virus–1 (HSV-1), it has been suggested that entry by endocytosis may not result in productive infection.37 It was predicted that at the 1-hour time point, diverse events related to virus entry and uncoating might be concurrently observed,38 documenting that transport within fMVDECs followed attachment. Figure2 confirms that virions at various stages of intracellular transport within distinct fMVDECs could be readily observed by EM at 1 hour after infection. Figure 2A depicts a single cell harboring multiple virions at different stages along the entry pathway. The large micrograph in Figure 2A and the right inset show surface attachment of electron-dense virions measuring, on average, approximately 110 nm, in agreement with the reported size range of enveloped herpesvirions (∼80-250 nM).39-44 Size bars are indicated, as the fine structure of these virions cannot be appreciated. The virus particles, in nearly linear array, are bound to the exposed surface of the fMVDECs, but not to the cell surface that is attached to the culture plate. A single virion (inset) as well as clusters of virions could be observed, whereas no similar structures were detected in uninfected cells (not shown). Virions enclosed within a thin-walled cytoplasmic vacuole could be seen between the plasma membrane and nucleus in Figure 2A, left inset. A structure suggestive of a disintegrating nucleocapsid at the nuclear-cytoplasmic border was also visible (Figure 2A, arrow, right inset).37,41 In Figure 2B, a virion appears to be fusing with a cellular membrane, probably the plasma membrane,35,37,42 as invagination from the cell surface could be observed in a larger micrograph (not shown). Figure 2C and 2D each show a de-enveloped nucleocapsid that has penetrated into the cytosol. Decondensation of the tightly packaged genome, a typical feature that distinguishes virions entering the cell from newly synthesized exiting virions, was observed.37,42Figure 2D further demonstrates the nucleocapsid in close proximity to microtubular systems, the likely transport mechanism, as has been observed for other herpesviruses.37,38 Although free nucleocapsids were present in the cytosol, some virions were clearly contained within cytoplasmic vesicles, probably endosomes, as shown in Figure 2A, left inset, and in Figure 2E. Disintegrating nucleocapsids and empty capsids could also be identified at the nuclear membrane at 1 hour (not shown).41,43 44
Entry and uncoating of HHV-8: electron-microscopic (EM) analyses of fMVDEC 1 hour after incubation with HHV-8.
Please note that in panels B, D, and E, an enlargement of the virion is circled and superimposed on the base figure. (A) An overview. The main panel shows multiple electron-dense herpeslike virions (average diameter, approximately 110 nm) attached to a cytoplasmic extension on the superior surface of an fMVDEC (× 47 000). The virions are aligned and give the appearance of centripetal movement; bar, 1000 nM. Insets demonstrate virions on the cell surface and in the cytoplasm in the perinuclear region of the same fMVDEC. The left inset (× 25 000) shows electron-dense virions enclosed within a thin-walled vacuole, consistent with recent endocytosis; bar, 500 nM. The right inset (× 31 000) shows an enveloped virion attached to the cell surface with membranelike material in contact with the cell, while a dense, symmetric nucleocapsid-type structure is present at the periphery of the nucleus (arrow); bar 500 nM. (B) The viral envelope fusing with a cellular membrane, most likely the plasma membrane (× 125 000). (C) A nucleocapsid devoid of its envelope that has been released in the cytosol (× 75 000). (D) An intracytoplasmic nucleocapsid distant from the nucleus and in close proximity to a microtubularlike structure (× 55 000). (E) A nucleocapsid can be clearly visualized within a cytoplasmic vesicle; it is probably an endosome or lysosome (× 95 000).
Entry and uncoating of HHV-8: electron-microscopic (EM) analyses of fMVDEC 1 hour after incubation with HHV-8.
Please note that in panels B, D, and E, an enlargement of the virion is circled and superimposed on the base figure. (A) An overview. The main panel shows multiple electron-dense herpeslike virions (average diameter, approximately 110 nm) attached to a cytoplasmic extension on the superior surface of an fMVDEC (× 47 000). The virions are aligned and give the appearance of centripetal movement; bar, 1000 nM. Insets demonstrate virions on the cell surface and in the cytoplasm in the perinuclear region of the same fMVDEC. The left inset (× 25 000) shows electron-dense virions enclosed within a thin-walled vacuole, consistent with recent endocytosis; bar, 500 nM. The right inset (× 31 000) shows an enveloped virion attached to the cell surface with membranelike material in contact with the cell, while a dense, symmetric nucleocapsid-type structure is present at the periphery of the nucleus (arrow); bar 500 nM. (B) The viral envelope fusing with a cellular membrane, most likely the plasma membrane (× 125 000). (C) A nucleocapsid devoid of its envelope that has been released in the cytosol (× 75 000). (D) An intracytoplasmic nucleocapsid distant from the nucleus and in close proximity to a microtubularlike structure (× 55 000). (E) A nucleocapsid can be clearly visualized within a cytoplasmic vesicle; it is probably an endosome or lysosome (× 95 000).
The state of the viral genome during initial infection of fMVDECs
In cells latently infected with HHV-8, viral DNA persists in the form of covalently closed circular episomes (CCCs), whereas in productively infected cells, linear DNA accumulates as virus replicates by a rolling circle mechanism. The configuration of the virus genome in infected cells can be distinguished by electrophoresis of extrachromosomal DNA through in situ lysis gels and can be verified by DNA blot hybridization. Application of PCR technology to DNA extracted from consecutive gel slices has enhanced the sensitivity of this procedure.26
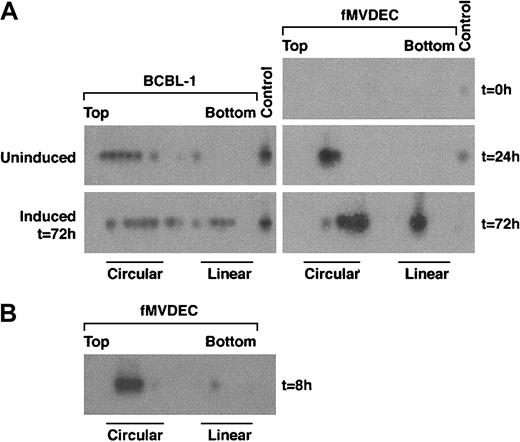
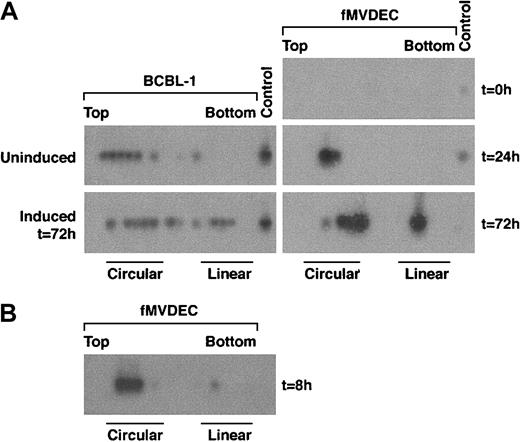
To examine the genome configuration of HHV-8 after infection of fMVDECs, cell lysates were prepared before and at various times after virus infection, electrophoresed through in situ lysis gels, and analyzed by DNA PCR of sequential slices. A nonrepetitive genomic subfragment of 394 bp from T1.1 was amplified in these experiments (“Materials and methods”). The relative migration of circular versus linear HHV-8 genomes was analyzed by comparing lysates from latently infected BCBL-1 cells with BCBL-1 cells induced to produce HHV-8. During latency, nearly all genomes were present as CCCs (Figure3A, left, top) whereas, following induction of lytic replication, linear DNAs accumulated (Figure 3A, left, bottom). Because only a subset of BCBL-1 cells were sensitive to induction,45 both circular and linear forms of HHV-8 persisted after treatment with butyrate (Figure 3A, left, bottom). When fMVDEC lysates were analyzed prior to HHV-8 infection, no PCR product could be detected (Figure 3A, right, top panel), demonstrating that the cells were free of endogenous virus and that the PCR reaction was specific. PCR-based in situ lysis gel analysis of infected fMVDECs performed 24 hours later (ie, for 21 hours after a 3-hour virus incubation) revealed HHV-8 genomes almost exclusively in the form of CCCs (Figure 3A, right, middle panel). In fact, circular genomes were readily detected as early as 8 hours after entry (Figure 3B), as shown in a separate experiment. These results demonstrated that linear virion DNAs introduced at the time of entry into fMVDECs rapidly circularized. However, by 72 hours after infection, the genome configuration of HHV-8 in infected fMVDECs had clearly shifted. A mixed pattern consisting of both circular and linear genomes, similar to that observed after BCBL-1 induction, was observed (Figure 3A, right, bottom). This rapid evolution of the configuration of genomic DNAs following transmission of HHV-8 to fMVDECs demonstrated that primary infection was complex and dynamic. Concurrent detection of CCCs and linear DNAs indicated that both lytic and latent infections were initiated shortly after exposure to virus (Figure 3B).
PCR-based in situ lysis gel analysis of HHV-8 genomes during initial infection.
Cells were lysed in the wells of an agarose gel at the indicated times, and extrachromosomal DNA that had been separated by electrophoresis was extracted from sequential slices of the gel and analyzed by PCR amplification of a 394-bp fragment from the HHV-8 T1.1 gene. (A) At left is the configuration of HHV-8 genomes in latently infected BCBL-1 cells before and after lytic cycle induction with 3 mM sodium butyrate for 72 hours. At right is the configuration of HHV-8 before (top), at 24 hours after (middle), and at 72 hours after (bottom) primary infection in fMVDECs with HHV-8. (B) The configuration of HHV-8 genomes 8 hours after infection with HHV-8 in fMVDECs in an independent experiment. The positions of sequential PCR-amplified DNAs relative to the top and bottom of the original in situ lysis gel are indicated from left to right. The relative positions of circular and linear forms are also indicated.
PCR-based in situ lysis gel analysis of HHV-8 genomes during initial infection.
Cells were lysed in the wells of an agarose gel at the indicated times, and extrachromosomal DNA that had been separated by electrophoresis was extracted from sequential slices of the gel and analyzed by PCR amplification of a 394-bp fragment from the HHV-8 T1.1 gene. (A) At left is the configuration of HHV-8 genomes in latently infected BCBL-1 cells before and after lytic cycle induction with 3 mM sodium butyrate for 72 hours. At right is the configuration of HHV-8 before (top), at 24 hours after (middle), and at 72 hours after (bottom) primary infection in fMVDECs with HHV-8. (B) The configuration of HHV-8 genomes 8 hours after infection with HHV-8 in fMVDECs in an independent experiment. The positions of sequential PCR-amplified DNAs relative to the top and bottom of the original in situ lysis gel are indicated from left to right. The relative positions of circular and linear forms are also indicated.
Viral transcripts associated with lytic replication after HHV-8 infection of fMVDECs
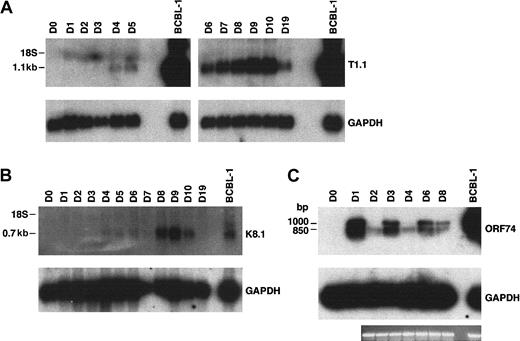
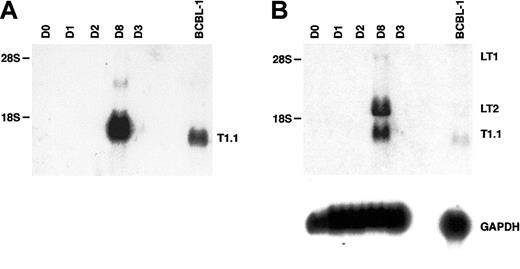
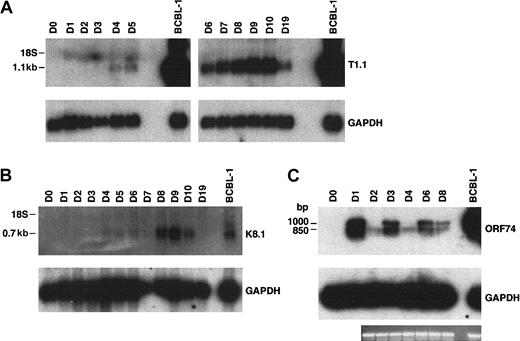
Total RNA prepared from fMVDECs was analyzed just prior to virus infection, on sequential days after infection, and 9 days after the first passage (“Materials and methods”) (Figure4). T1.1 encodes a polyadenylated nuclear RNA of unknown function, which is highly transcribed during the HHV-8 lytic cycle.27,28,45 Figure 4A, top, shows that T1.1 could be readily detected by Northern blot hybridization, indicating that lytic RNAs were more rapidly and effectively transcribed following HHV-8 infection of fMVDECs than had been demonstrated. Steady-state levels of T1.1 RNAs increased throughout the 10 days of primary infection (Figure 4A), suggesting that infection spread through the cell population. RNAs specific for late antigens, such as the virion envelope glycoprotein gp35/37 encoded by genomic fragment K8.1,33,46,47 also increased during this period, as evidenced by appearance of a major 0.7-kb RNA (Figure 4B). Additional lytic transcripts, ie, ORF26 (early), KS Bam330 (late), and ORF29 (late),7 which were analyzed by RT-PCR, could be readily detected (not shown).
Analysis of HHV-8 lytic cycle RNAs (T1.1, K8.1, and ORF74) on sequential days after primary infection.
(A) At top is the detection of T1.1 by RNA blot hybridization of 10 μg total RNA from fMVDECs harvested before infection, on days 1 through day 10 after primary infection, and on day 19 (9 days after passage). At bottom, the blot was reprobed to detect cellular GAPDH. The position of 18S RNA (2.37 kb) is indicated on the left. (B) At top is the detection of K8.1 RNA before infection and on days 1 through 10 and on day 19 after infection by the same methods described in panel A. At bottom, the blot was reprobed to detect cellular GAPDH RNA. The position 18S RNA is indicated on the left. (C) At top, ORF74 transcripts were detected in 0.5 μg total RNA from uninfected (day 0) and infected (days 1-8) fMVDECs or from BCBL-1 cells (control, right lane) by RT-PCR. Amplification products were detected by DNA blot hybridization. Positions of cDNAs are indicated in base pairs on the left. GAPDH transcripts were detected under identical conditions in the same experiment. An ethidium bromide stain of the gel (GAPDH) is also displayed.
Analysis of HHV-8 lytic cycle RNAs (T1.1, K8.1, and ORF74) on sequential days after primary infection.
(A) At top is the detection of T1.1 by RNA blot hybridization of 10 μg total RNA from fMVDECs harvested before infection, on days 1 through day 10 after primary infection, and on day 19 (9 days after passage). At bottom, the blot was reprobed to detect cellular GAPDH. The position of 18S RNA (2.37 kb) is indicated on the left. (B) At top is the detection of K8.1 RNA before infection and on days 1 through 10 and on day 19 after infection by the same methods described in panel A. At bottom, the blot was reprobed to detect cellular GAPDH RNA. The position 18S RNA is indicated on the left. (C) At top, ORF74 transcripts were detected in 0.5 μg total RNA from uninfected (day 0) and infected (days 1-8) fMVDECs or from BCBL-1 cells (control, right lane) by RT-PCR. Amplification products were detected by DNA blot hybridization. Positions of cDNAs are indicated in base pairs on the left. GAPDH transcripts were detected under identical conditions in the same experiment. An ethidium bromide stain of the gel (GAPDH) is also displayed.
Interestingly, RT-PCR analysis of RNAs encoded by ORF74, the lytic cycle vGPCR,8,9,48 displayed cyclic variation in the relative abundance of these RNAs on sequential days (Figure 4C, top) when compared with a GAPDH control in the same experiment (Figure 4C, bottom). The level of steady-state transcripts of this gene fluctuated with a periodicity of approximately 48 to 72 hours, consistent with the duration of a typical γ-herpesvirus lytic cycle. Approximately 3 cycles of alternating ORF74 expression were observed during 9 days (D0-D8) in primary culture (Figure 4C), suggesting that primary lytic HHV-8 infection of fMVDECs at first proceeds synchronously. A similar temporal variation in the level of steady-state transcripts of ORF74 was observed during a 48-hour period when these were monitored after lytic induction of BCBL-1 cells (a PEL line).49
The oligonucleotide primers synthesized to reverse-transcribe the ORF74 coding sequence 129372-130400,50 generated 2 PCR fragments, the expected 1045-bp fragment as well as an approximately 850-bp fragment (Figure 4C). Splicing of a potential exon between 130116 and 130336, inclusive, could generate an additional 857-bp fragment,48 49 the size of the second fragment observed. Taken together, these results demonstrate that abundant transcription of lytic cycle genes accompanies initial infection of fMVDECs with HHV-8.
Detection of HHV-8 transcripts associated with latency by RNA blot hybridization
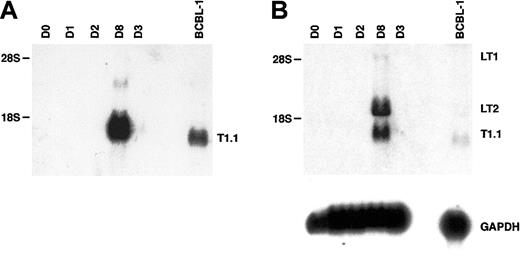
The transcriptional program associated with HHV-8 persistence or latency is complex and differs in distinct cell types.50-53 Moreover, latent herpesviral RNAs are often less abundantly transcribed than lytic transcripts.7,27The HHV-8 cyclin (vCYC) encoded by ORF72 constitutes a predominantly latent-cycle product in both immortalized B lymphocytes (PEL lines) in vitro53,54 and spindle cells (KS biopsies) in vivo.55 In PEL lines, 2 major latent transcripts of 6 kb and 2 kb, known as LT1 and LT2 respectively, encode vCYC. As seen in Figure 5A (and Figure 4), expression of the abundant lytic transcript T1.1 could be detected by RNA blot hybridization beginning at day 3 to 4 after HHV-8 infection of fMVDECs and was more strongly detected after 8 days. When a cDNA probe encoding the entire ORF72 reading frame was used to reprobe the identical blot, LT1 and LT2 transcripts of characteristic size, although undetectable at 3 days, were apparent by day 8 (Figure 5B, top) (a minor 1-kb fragment additionally detected in some PEL lines upon hybridization with an ORF72 probe was not assessed).56 Thus, both lytic and latent transcripts could be detected after the end of the first week in culture.
Analysis of HHV-8 latent-cycle ORF72 (vCYC) RNA (encoded by LT1 and LT2) and comparison with lytic T1.1 RNA on days 3 and 8 after primary infection of fMVDECs.
(A) Detection of T1.1 RNA by Northern blot hybridization as described in Figure 4, before infection and on days 1 through 3 and on day 8 after infection. BCBL-1 RNA was analyzed as a control. Positions of 28S (6.33 kb) and 18S RNA (2.37 kb) are indicated on the left. (B) At top, the blot was reprobed with a cDNA encoding full-length ORF72. The positions of LT1 and LT2 transcripts and of residual T1.1 are indicated on the right. Positions of 28S and 18S RNAs are indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA.
Analysis of HHV-8 latent-cycle ORF72 (vCYC) RNA (encoded by LT1 and LT2) and comparison with lytic T1.1 RNA on days 3 and 8 after primary infection of fMVDECs.
(A) Detection of T1.1 RNA by Northern blot hybridization as described in Figure 4, before infection and on days 1 through 3 and on day 8 after infection. BCBL-1 RNA was analyzed as a control. Positions of 28S (6.33 kb) and 18S RNA (2.37 kb) are indicated on the left. (B) At top, the blot was reprobed with a cDNA encoding full-length ORF72. The positions of LT1 and LT2 transcripts and of residual T1.1 are indicated on the right. Positions of 28S and 18S RNAs are indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA.
MVDECs from distinct tissues and HUVECs support efficient HHV-8 infection, whereas 293 cells do not
To determine whether other primary vascular endothelial cell populations were susceptible to HHV-8 infection and to assess whether significant differences in susceptibility to infection existed among these cell populations, adult bMVDECs, mMVECs, HUVECs, as well as 293L cells (a clonal derivative of 293 epithelial cells reported to be more susceptible to HHV-8 infection than the parent line21) were analyzed by RNA blot hybridization following HHV-8 infection. As shown in Figure 6A-B, bMVDECs and mMVECs as well as HUVECs could be efficiently infected, as demonstrated by the cumulative detection of steady-state T1.1 RNAs from days 3 and 4 to day 8 after infection. Major differences in viral gene transcription were not detected between the different sources. In contrast, no RNA could be detected in 293L cells by RNA blot hybridization (Figure 6A) although, as previously reported,21 transcripts were detectable by RT-PCR. Figure6A, bottom inset, shows detection of the 394-bp T1.1 fragment during the first 4 days after infection; although the fragment did not persist to day 7, its presence is consistent with the prior suggestion that virus titer was not the limiting factor in efficient infection of 293 cells.32 Comparison of different lots of MVDECs (Figure 6C, left and right) further revealed that there was variation in the infectivity of cells from the distinct lots controlled for the same passage. As shown in Figure 6C, infection under identical conditions with the same virus preparation (either VP2 or VP3) at 2 different dilutions resulted in increased production of T1.1 RNA in the fMVDEC lot displayed on the right as compared with that on the left. The current results demonstrate that vascular endothelial cells derived from diverse tissues can support initial infection and lytic transcription of HHV-8 and that no major differences were detected among the cell populations studied.
Comparative infectivity of primary vascular endothelial cells and 293 cells as determined by RNA blot hybridization.
(A) At top, detection of T1.1 RNA by Northern blot hybridization of 15 μg total RNA from adult MVDECs (bMVDECs), HUVECs, and 293 cells infected at the same time and under identical conditions. Transcripts were analyzed on day 0 before infection and on days 4 and 8 after infection. BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA (2.37 kb) is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA. In bottom inset, T1.1 transcripts were detected by RT-PCR in 0.5 μg total RNA from 293L cells and fMVDECs that were infected with HHV-8 at the same time and under identical conditions. RNAs were analyzed on day 0 (D0) prior to infection and variably on days 1 through 7 (D1-D7) after infection as separately indicated for the various cells. BCBL-1 RNA was analyzed as a control (right lane). Primers are described in “Materials and methods.” Amplification products were detected by DNA blot hybridization with the use of probes as described in “Materials and methods.” The position of the 394-bp T1.1 amplification product is indicated on the right. (B) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from fMVDECs and mMVECs that were infected at the same time and under the identical conditions described in panel A. RNA loading was not equal (fMVDECs = 1.5 μg; mMVECs = 15 μg). However, comparison with GAPDH permits assessment of the relative amount of T1.1 RNA in the various cells. (C) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from 2 independent lots of fMVDECs (shown on the left and right with a lane between the 2 sets of experiments) that were infected after the same passage and under identical conditions with GAPDH purified HHV-8 (virus preparation 2 [VP2] and VP3) undiluted and diluted 1:5. RNA loading was not equal (left lot = 10 μg; right lot = 5 μg). BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA.
Comparative infectivity of primary vascular endothelial cells and 293 cells as determined by RNA blot hybridization.
(A) At top, detection of T1.1 RNA by Northern blot hybridization of 15 μg total RNA from adult MVDECs (bMVDECs), HUVECs, and 293 cells infected at the same time and under identical conditions. Transcripts were analyzed on day 0 before infection and on days 4 and 8 after infection. BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA (2.37 kb) is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA. In bottom inset, T1.1 transcripts were detected by RT-PCR in 0.5 μg total RNA from 293L cells and fMVDECs that were infected with HHV-8 at the same time and under identical conditions. RNAs were analyzed on day 0 (D0) prior to infection and variably on days 1 through 7 (D1-D7) after infection as separately indicated for the various cells. BCBL-1 RNA was analyzed as a control (right lane). Primers are described in “Materials and methods.” Amplification products were detected by DNA blot hybridization with the use of probes as described in “Materials and methods.” The position of the 394-bp T1.1 amplification product is indicated on the right. (B) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from fMVDECs and mMVECs that were infected at the same time and under the identical conditions described in panel A. RNA loading was not equal (fMVDECs = 1.5 μg; mMVECs = 15 μg). However, comparison with GAPDH permits assessment of the relative amount of T1.1 RNA in the various cells. (C) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from 2 independent lots of fMVDECs (shown on the left and right with a lane between the 2 sets of experiments) that were infected after the same passage and under identical conditions with GAPDH purified HHV-8 (virus preparation 2 [VP2] and VP3) undiluted and diluted 1:5. RNA loading was not equal (left lot = 10 μg; right lot = 5 μg). BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA.
Discussion
Since its discovery in 1994, substantial information has accumulated about the biology of HHV-8 in KS on the basis of analyses of biopsy specimens and investigations of isolated viral genes. The absence of an appropriate culture system that effectively duplicates the composite mechanisms believed to govern the initiation and maintenance of HHV-8 infection (lytic and latent) in KS endothelial cells, however, has left many questions about pathogenesis unresolved. Although infections of several cell types have been carefully documented, they have proved relatively inefficient and often self-abort. In this report, we provide evidence that the initial events that occur upon HHV-8 infection of normal vascular endothelial cells in vitro are efficient. In addition, we examine the cell biology of the virus-cell interactions that occur at these earliest time points. HHV-8 attachment, internalization, genome configuration, and patterns of viral gene expression during initial infection were surveyed in normal primary fMVDECs and compared with those of other susceptible endothelial populations. The results of this study demonstrate the usefulness of primary vascular endothelial cell culture for studying virion entry and for investigating the cellular and viral events regulating initiation of HHV-8 transcription.
Although many cell types susceptible to HHV-8 infection have been identified, little is known about their relative ability to bind virus. Attachment of HHV-8 to the cell populations targeted by KSHV, which subsequently become involved in virus-associated diseases such as PEL and MCD, has not been characterized. Although we observed that FITC-virus attached to all of the cell populations examined, we identified a highly reproducible rank order for binding: greatest for fMVDECs, intermediate for B cells or monocytes, and significantly less for T/NK cells. Interestingly, this rank order reflects the known cellular tropism of HHV-8 in pathologic specimens from patients with HHV-8–associated disease (reviewed in Moore and Chang7). These observations suggest that receptor density may be a salient determinant of susceptibility to HHV-8 infection and are consistent with recent studies suggesting that low-affinity interactions between several viral membrane proteins and widely expressed cell surface glycosaminoglycans, such as heparan sulfate, constitute the initial HHV-8–binding event.17,57 A high density of low-affinity cellular receptors could compensate for the absence of a single high-affinity attachment receptor such as that found for the related γ-herpesvirus, EBV. Coreceptors that mediate subsequent steps in attachment, fusion, and transfer of virus into the cytoplasm are also likely to be important determinants of entry—as has been observed for other members of the herpesvirus family.58-60
EM analysis with HHV-8 confirmed that virions attached to multiple sites on the exposed endothelial cell surface, providing visual evidence that these virions rapidly fused with cellular membranes and became internalized into the cytosol or cytoplasmic vesicles, probably endosomes. Because γ-herpesvirus replication is less robust than that of α-herpesviruses61 and virion preparation is tedious, few virus-entry studies of γ-herpesviruses are available. The reproducibility and efficiency of these early events (attachment, fusion, internalization), however, established that this culture system could be used for real-time evaluation of γ-herpesvirus entry. Interestingly although naked nucleocapsids were clearly present in the cytosol, many of the internalized virions were also localized to cytoplasmic vesicles, endosomes, and possibly lysosomes (Figure 2). Although the significance of nucleocapsid routing to the endosome is not established for HHV-8, such routing for HSV-1 has been shown to lead to nonproductive infection with minimal replication.37,62 Thus, it is possible that the differences among various HHV-8–endothelial cell infection systems13-17 may result not just from delivery of high-titer virus to the endothelial surface, but also from differential routing of the virion in the endothelial cells as a result of the distinct cell culture conditions employed.
Although complex and potentially novel patterns of HHV-8 gene transcription were observed after virus entry, a temporal analysis that coordinated viral genome structure with the appearance of specific RNAs provided clear evidence that both lytic and latent infection were initiated. The stable lytic cycle RNAs T1.1 (early) and K8.1 (late) could be detected by RNA blot hybridization at 72 hours and accrued until passage, whereas ORF74 RNAs (early lytic) fluctuated on sequential days. Assessment of a primary infected culture during a 9-day time interval revealed alternating cycles of ORF74 RNA detection with a periodicity typical of herpesvirus replicative cycles (48 to 72 hours). This suggested that the initial rounds of lytic replication were synchronous. ORF74 transcripts have been shown to be early lytic RNAs, detectable only between 20 and 30 hours after lytic cycle induction of a PEL cell line.63 Their alternating pattern of expression in newly infected fMVDECs was highly suggestive that replication was synchronous, as this pattern could not otherwise have been identified. ORF74 encodes a vGPCR that, when expressed independently of virus signals constitutively as well as in response to certain chemokines, up-regulates vascular endothelial growth factor and, in animal models, induces angiogenesis of endothelial cells.9 64-68 In KS lesions, vGPCR is hypothesized to drive proliferation of neighboring cells; however, vGPCR is expressed only in lytically infected cells, which are the minority, and for a limited window of time. The role of ORF74 in KS pathogenesis and specifically in facilitating lytic replication remains unknown.
Detection of the latent vCYC RNA by RNA blot hybridization later in the course of the same primary infection additionally suggested that a potentially immortalizing HHV-8 infection was concurrently or consecutively established. The detection of the vCYC encoded by the latent ORF72 is important in that vCYC inactivates pRB, the checkpoint protein that inhibits entry into S phase.10 69-74 By manipulating the host cell cycle, HHV-8 contributes to cell immortalization and tumorigenesis.
Although several groups have now reported HHV-8 infection of MVDECs, robust initiation and durable maintenance of immortalization in culture systems has remained elusive.13-17 MVDECs immortalized with the human papilloma virus–16 oncoproteins E6 and E7 have also been shown to support establishment and effective spread of HHV-8 latency; however, the modifications of cell cycle regulation and apoptosis by E6 and E7 potentially complicate the studies of the unique contributions by HHV-8.
The current studies demonstrate that HHV-8, concentrated and devoid of inducing agent, obtained from the PEL line BCBL-1, efficiently mediates viral entry and transcriptional initiation of a mixed lytic and latent infection in cultured vascular endothelial cells. The model presented in this study will facilitate detailed analysis of early pathogenic events relevant to the development of KS.
We thank Michael Gill for critical evaluation of the manuscript, Larry Coffin for computer assistance, Don Ganem for supplying pBS T1.1, and Dan Brown of the EM facility at Beth Israel Deaconess Medical Center.
Supported by National Institutes of Health grants P30AR42689 (Pilot Project), R01DE12186, and K24CA85083, and by an Established Investigator Award from the American Heart Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Joyce D. Fingeroth, Harvard Institutes of Medicine, 4 Blackfan Circle, Room 353, Boston, MA 02115; e-mail: jfingero@caregroup.harvard.edu.

![Fig. 6. Comparative infectivity of primary vascular endothelial cells and 293 cells as determined by RNA blot hybridization. / (A) At top, detection of T1.1 RNA by Northern blot hybridization of 15 μg total RNA from adult MVDECs (bMVDECs), HUVECs, and 293 cells infected at the same time and under identical conditions. Transcripts were analyzed on day 0 before infection and on days 4 and 8 after infection. BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA (2.37 kb) is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA. In bottom inset, T1.1 transcripts were detected by RT-PCR in 0.5 μg total RNA from 293L cells and fMVDECs that were infected with HHV-8 at the same time and under identical conditions. RNAs were analyzed on day 0 (D0) prior to infection and variably on days 1 through 7 (D1-D7) after infection as separately indicated for the various cells. BCBL-1 RNA was analyzed as a control (right lane). Primers are described in “Materials and methods.” Amplification products were detected by DNA blot hybridization with the use of probes as described in “Materials and methods.” The position of the 394-bp T1.1 amplification product is indicated on the right. (B) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from fMVDECs and mMVECs that were infected at the same time and under the identical conditions described in panel A. RNA loading was not equal (fMVDECs = 1.5 μg; mMVECs = 15 μg). However, comparison with GAPDH permits assessment of the relative amount of T1.1 RNA in the various cells. (C) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from 2 independent lots of fMVDECs (shown on the left and right with a lane between the 2 sets of experiments) that were infected after the same passage and under identical conditions with GAPDH purified HHV-8 (virus preparation 2 [VP2] and VP3) undiluted and diluted 1:5. RNA loading was not equal (left lot = 10 μg; right lot = 5 μg). BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/3/10.1182_blood.v100.3.888/5/m_h81522914006.jpeg?Expires=1767823895&Signature=Fjxh~637PK~Xbj6XUKQX0p4v-NbBEzXvoi0YNg8wir60rT3T7--eh2~Li9r1wjjq3JSBGpmU~xf5wp0VBy6-5UVYv5m9dbtTpHWAMIlyK56eaHyFcYK5lAbWmM15xfn78zlT5iG2jOBLLoTqRXyCQB6seG2opNKWNmn8NbVeSOd9U8krtlil8-6pabTz9nuj4cXELrrItd5k7hNkW4i~n8enBzmI1sieMybAf3WE3GGtPPZpi5g6T1XhzBFrzHF0pvnlSPZKEjDALbTAExnmXLv4UliHdvyiWXuBOhZ8TRaAXhFSDTsjesZsgSonc9LhI2LTv5ypxCu2iQ0hrrhv5Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 6. Comparative infectivity of primary vascular endothelial cells and 293 cells as determined by RNA blot hybridization. / (A) At top, detection of T1.1 RNA by Northern blot hybridization of 15 μg total RNA from adult MVDECs (bMVDECs), HUVECs, and 293 cells infected at the same time and under identical conditions. Transcripts were analyzed on day 0 before infection and on days 4 and 8 after infection. BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA (2.37 kb) is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA. In bottom inset, T1.1 transcripts were detected by RT-PCR in 0.5 μg total RNA from 293L cells and fMVDECs that were infected with HHV-8 at the same time and under identical conditions. RNAs were analyzed on day 0 (D0) prior to infection and variably on days 1 through 7 (D1-D7) after infection as separately indicated for the various cells. BCBL-1 RNA was analyzed as a control (right lane). Primers are described in “Materials and methods.” Amplification products were detected by DNA blot hybridization with the use of probes as described in “Materials and methods.” The position of the 394-bp T1.1 amplification product is indicated on the right. (B) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from fMVDECs and mMVECs that were infected at the same time and under the identical conditions described in panel A. RNA loading was not equal (fMVDECs = 1.5 μg; mMVECs = 15 μg). However, comparison with GAPDH permits assessment of the relative amount of T1.1 RNA in the various cells. (C) At top, detection of T1.1 RNA by Northern blot hybridization of total RNA from 2 independent lots of fMVDECs (shown on the left and right with a lane between the 2 sets of experiments) that were infected after the same passage and under identical conditions with GAPDH purified HHV-8 (virus preparation 2 [VP2] and VP3) undiluted and diluted 1:5. RNA loading was not equal (left lot = 10 μg; right lot = 5 μg). BCBL-1 RNA was analyzed as a control. The position of the major T1.1 transcript is indicated on the right. The position of 18S RNA is indicated on the left. At bottom, the blot was reprobed to detect cellular GAPDH RNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/3/10.1182_blood.v100.3.888/5/m_h81522914006.jpeg?Expires=1767897895&Signature=YwTM9pNBmMDWcC~E2F7~BRjUQ2dn3aYACEJorb~EAKRtHNYV4UcPm3rFrFU~-TorfGSOtaRckFPKqqPh92xbkz9ssAjhPVf7CPO7Sl~t-kYZvNkN4h9vIUiAYL5sG1I~bHvwb7ReQxok2MdJ5DMWsG1cyCNzan0TAa3dv9K8ChcGQIY-GT27Wyt7CE9c~Nhp5xz5Umks8HRRCrX1T7c7rlimF-xUo76AlT05eXRoub7OFWjhG2y90oSfTajO62dhXJuVjWTagukbDS0~nLgKqxrx-PIfG6AdogJZWrfniIJUK-lK79QQADl0JTVG31lXXLJ-Xou8tHNCXNKeDRSHjQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)