Key Points
Wwox deletion in Vk∗MYC myeloma mice leads to aggressive plasmablastic tumors, reduced survival, and enhanced inflammatory signaling.
WWOX loss drives genomic instability in plasmablastic tumors, promoting mutations in cancer and DNA damage response genes.
Visual Abstract
Deletions and translocations affecting WWOX accompanied by loss of expression are frequently observed in B-cell neoplasms and are linked to poor prognosis. Our previous research showed that Wwox deletion early in B-cell development induces genomic instability, neoplastic transformation, and monoclonal gammopathies in mice. In this study, by crossing Cd19 Wwox knockout (KO) with Vk∗MYC myeloma model mice, we generated a model with concurrent Wwox deletion and MYC activation, reproducing 2 common oncogenic alterations in B- and plasma-cell cancers. We observed that Vk∗MYC:Wwox KO mice exhibited significantly reduced survival rates primarily due to the development of plasmablastic plasmacytomas and lymphomas. Transcriptome profiling from bone marrow derived CD138+ plasma cells and plasmablastic tumors revealed enrichment of biofunctions related to tumorigenic phenotype and inflammation activation upon Wwox deletion in Vk∗MYC mice. Wwox KO plasmablastic tumors displayed mutations affecting classical cancer genes, DNA damage response (DDR) genes, as well as overexpression of Aid/Apobec family members associated to hypermutation and DDR mutational signatures. These findings illustrate the significant pathobiological effects of B-cell–specific Wwox deletion and support a relevant role for WWOX loss of function in B-cell neoplastic progression toward more aggressive phenotypes.
Introduction
The WWOX/FRA16D chromosomal fragile locus is a hot spot for genomic instability.1-3 Consequently, WWOX is one of the most frequently deleted genes in cancer4-7 and its expression loss is indicative of poor prognosis.2 Previous research has demonstrated increase incidence of B-cell lymphomas (BCLs) in hypomorphic Wwox mice8 and in mice heterozygous for a functional Wwox allele upon carcinogen exposure,9 collectively indicating an increased vulnerability of B cells to neoplastic transformation upon Wwox deficiency. In humans, genetic alterations and depletion of WWOX have been linked to the pathogenesis of B-cell malignancies such as certain BCLs and multiple myeloma (MM). WWOX is frequently deactivated by copy number loss and exhibits structural variations in BCLs.10-13 In MM, WWOX is frequently affected by deletions and breakpoints leading to translocation events commonly associated with poor prognosis, for example, t(14;16).14-23 Importantly, WWOX has recently been identified among a small number of cancer genes displaying complete loss of function in MM cases from the Multiple Myeloma Research Foundation (MMRF) CoMMpass study, further supporting a potential role for WWOX in MM.24 Additionally, promoter methylation affecting WWOX expression has been associated with pathogenesis and advancement of MM.25 All these findings underscore a potential relevant role for WWOX deficiency in the progression and prognosis of B- and plasma-cell cancers.
To understand the impact of WWOX loss of function in B cells, we previously developed a Cd19 Wwox knockout (KO) mouse model, where Wwox was conditionally deleted early in B-cell development. These mice exhibited significantly reduced survival and developed B-cell tumors including lymphomas, plasma-cell neoplasias, and monoclonal gammopathies.26Wwox ablation in B cells also caused spontaneous translocations during class switch recombination and a shift from classical nonhomologous end-joining to the microhomology-mediated alternative- nonhomologous end-joining pathway, which is associated with chromosome translocations and genomic instability.26 This work suggested a propensity for oncogenic transformation in B cells upon Wwox deficiency.
In this study, we aimed to further elucidate the role of WWOX in B-cell neoplastic progression by crossing Cd19 Wwox KO mice with Vk∗MYC transgenic mice, a known model of MYC-driven plasma-cell neoplasia.27 Deletion of Wwox in B cells from Vk∗MYC mice (Vk∗MYC:Wwox KO) significantly reduced survival, primarily due to the development of extramedullary plasmablastic plasmacytomas (PBPs) and plasmablastic lymphomas (PBLs) (collectively referred to as plasmablastic tumors [PBTs] throughout the article), and BCLs. These tumors displayed a high-mutation burden. Transcriptome profiling of bone marrow (BM)–derived CD138+ plasma cells (PCs) and PBTs further revealed an enrichment of biofunctions associated with tumorigenesis and inflammation activation as consequence of Wwox deletion. Together, our findings demonstrate that B-cell–specific Wwox deletion in a myeloma mouse model leads to the development of aggressive PBTs characterized by hypermutation, overexpression of Aid/Apobec family members and proinflammatory signatures, further supporting the role of Wwox loss of function as a relevant event in malignant progression toward more aggressive phenotypes.
Materials and methods
Animals
The procedure to develop Vk∗MYC mice with Myc activation in germinal-center B cells was provided elsewhere,27 here we used a derivative strain described as Vk∗MYCΔloxP in which LoxP site flanking the transgenic 3′ kappa enhancer have been removed thus retaining expression in the presence of Cre recombination as recently described.28WwoxloxP/loxP and Cd19:Wwox KO mice have been previously described.26,29 We crossed Cd19:Wwox WT and Cd19:Wwox KO with Vk∗MYC mice to develop Vk∗MYC:Wwox WT (wildtype), Vk∗MYC:Wwox HET (heterozygous), and Vk∗MYC:Wwox KO (Knockout) mice. Mice of both genders were used in all experiments. Genotypes were confirmed as previously described.26,27
Histology and IHC
Tissues were processed by formalin fixation, paraffin embedding, and hematoxylin and eosin staining. Tumor samples were analyzed by histology and immunohistochemistry (IHC) using anti-CD138 (142502, BioLegend) and anti-CD19 (90176, Cell Signaling Technology) antibodies. Tumors were classified following guidelines of the Bethesda classification of lymphoid neoplasms in mice and a more recent classification of mouse plasmacytomas.30,31
SPEP
Blood samples were collected at various intervals throughout the lifespan of the mice. Samples were processed for serum protein electrophoresis (SPEP) as previously described.26
Purification of CD138+ PCs and CD19+ B cells from mouse BM
CD138+ PCs were purified from Vk∗MYC:Wwox WT (n = 5), Vk∗MYC:Wwox HET (n = 3), and Vk∗MYC:Wwox KO (n = 5) mice BM from tibia and femur bones using EasySep mouse CD138 positive selection kit (18957, Stemcell Technologies) using magnetic beads and following the manufacturer’s protocol. Magnetic beads with CD138+ BM cells were washed with cold PBS and the remaining supernatant was used to isolate CD19+ B cells using CD19 selection kit II (18954, Stemcell Technologies), to be used as a control or source of B cells in other experimental procedures.
RNA-seq and data analysis
RNA sequencing (RNA-seq) and data analysis was performed as previously described.32,33 Briefly, total RNA was extracted from Vk∗MYC:Wwox WT-BM (n = 3), Vk∗MYC:Wwox KO-BM (n = 3), and Vk∗MYC:Wwox KO-PBT (n = 4). Differentially expressed genes between Vk∗MYC:Wwox WT-BM vs Vk∗MYC:Wwox KO-BM and Vk∗MYC:Wwox KO-BM vs Vk∗MYC:Wwox KO-PBT were identified using DESeq2 based on normalized log2 count per million values.34 Data integration and visualization of differentially expressed transcripts (false discovery rate [FDR] < 0.05) were done with MultiExperiment Viewer Software. Functional enrichment analysis of dysregulated transcripts was performed using Ingenuity Pathway Analysis (IPA; Qiagen Inc.)
Whole-exome sequencing and data analysis
DNA from Vk∗MYC:Wwox WT-BM (n = 5), Vk∗MYC:Wwox HET (n = 3), Vk∗MYC:Wwox KO-BM (n = 5), Vk∗MYC:Wwox KO-PBT (n = 4), and normal C57Bl6/J mice liver samples was purified and processed for Whole-exome sequencing (WES) as previously described.35 Sequenced 100 nt paired-end reads (∼200× coverage) were aligned against mm10 mouse genome using Burrows-Wheeler Aligner (BWA) v0.7.17 and Picard v2.27.4. MuTect2 (GATK v4.4) was employed to identify single-nucleotide variants (SNVs) and insertions/deletions. Following described methods,36 variants were filtered and compared against the Mouse Genome Project database of known germ line variants.37 Filtered variants were annotated using the Variant Effect Predictor38 and further filtered by functional consequence. Copy number variations (CNVs) were identified using CNVkit (version 0.9.9).39 Mutational signatures and mutational burden were computed using the SigProfiler Assignment resource40 according COSMIC signature database (https://cancer.sanger.ac.uk/signatures/) based on nonsynonyms somatic variants. Data integration and visualization of mutational signatures contribution across tumors were done with the MultiExperiment Viewer software (MeV v4.9).
qRT-PCR
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed on RNA extracted from CD19+ B cells using primers as previously described.33 Relative gene expression level was determined in triplicate using the SYBR Green–based method after normalization to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RNA expression, the average fold change was calculated using the 2ˆ-(ΔΔCt) method described elsewhere.41
Statistical analysis for mouse survival and tumor incidence
Overall survival and M-spike incidence analyses were performed using SPSS statistical software. Log-rank test was applied to determine statistical significance. Χ2 test was performed to compare tumor incidence and Student t test was performed to compare gene expression using GraphPad Prism V 10; P values <.05 were considered significant.
All animal research was conducted in facilities accredited by the AAALAC International at MD Anderson Cancer Center and approved by the Institutional Animal Care and Use Committee.
Results
Wwox ablation in B cells promotes PBT development in Vk∗MYC mice
We assessed the clinical relevance of WWOX in MM by analyzing 4 publicly available gene expression data sets (GSE4204, GSE9782, GSE24080, and GSE57317) using the Kaplan-Meier Plotter resource (https://kmplot.com/analysis/). A total of 1416 MM patients were stratified into 2 groups, high and low WWOX expression, based on the median expression value of the Affymetrix 219077_s_at probe. Lower WWOX expression was significantly associated with poorer outcomes, including shorter overall survival (hazard ratio [HR] = 0.46; P = 2.9×10-15) and event-free survival (HR = 0.34; P < 1×10-16) (supplemental Figure 1A), providing additional support for our in vivo investigation.
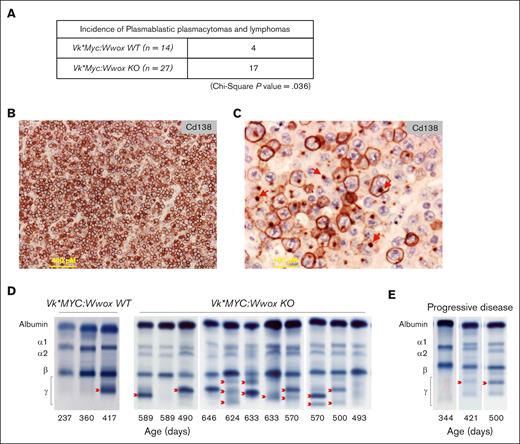
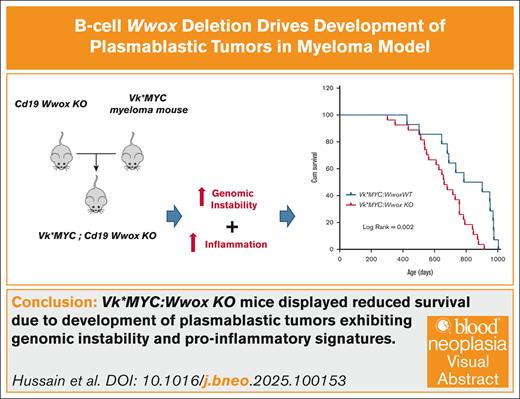
To experimentally investigate role of WWOX loss in B-cell malignancies, we generated a mouse model with targeted Wwox deletion early in B-cell development and MYC activation by crossing Cd19:Wwox KO26 and a derivative strain of Vk∗MYC mice.27,28Wwox deletion in CD19+ B cells purified from BM of Vk∗MYC:Wwox KO mice was confirmed by qRT-PCR (P < .05) (supplemental Figure 1B). The survival of Vk∗MYC:Wwox KO mice was compared with control littermates wild-type for Wwox expression. Overall survival was plotted using the Kaplan-Meier method and statistically analyzed by the log-rank test. Although most Vk∗MYC:Wwox KO mice displayed an indolent disease course, they exhibited a significantly shorter mean survival of 660 days vs 800 days observed in Vk∗MYC:Wwox WT mice (log-rank P = .002) (Figure 1). We noted that 63% (17/27) of Vk∗MYC:Wwox KO mice developed intra-abdominal tumors with the characteristics of PBTs and BCLs, particularly affecting mesenteric and peripancreatic lymph nodes (Figure 2A). This incidence was significantly higher than that observed in the Vk∗MYC:Wwox WT group, where only 28.5% (4/14) displayed intra-abdominal tumors (P = .036) (Figure 2A). Based on histopathology evaluation and IHC staining these tumors were broadly classified into, PBPs (n = 9), PBLs (n = 5) and BCLs (n = 3) (Figure 2B; supplemental Table 1). Interestingly, IHC revealed that samples from several Vk∗MYC:Wwox KO extramedullary plasmacytomas (ie, PBPs) displayed large areas characterized by the presence of CD138+ PCs with staining to cell membrane but also to small single polarized foci (Figure 2C). This striking intracellular pattern of CD138 staining has been previously described as resembling “uropods” shown to facilitate myeloma cells aggregation and adhesion to the tumor microenvironment. Furthermore, myeloma cells with high proportion of cells with CD138 staining uropods were shown to display high migratory potential, implying that they represent more aggressive tumor forms,42 which agrees with the histology observed. Abundant number of aberrant mitotic figures, enlarged nuclei, and multiple prominent nucleoli were also characteristic of all PBPs (Figure 2C).
Wwox ablation in B cells reduces overall survival in Vk∗MYC mice. Kaplan-Meier survival analysis comparing Vk∗MYC:Wwox KO (red line, n = 27) and Vk∗MYC:Wwox WT (blue line, n = 14) mice. Vk∗MYC:Wwox KO mice showed significantly reduced survival compared to the WT group (log-rank [Mantel-Cox] P = .002). The mean survival was 660 days for the Wwox KO group, compared to 800 days for the Wwox WT group. The x-axis indicates survival time in days, and the y-axis represents cumulative survival.
Wwox ablation in B cells reduces overall survival in Vk∗MYC mice. Kaplan-Meier survival analysis comparing Vk∗MYC:Wwox KO (red line, n = 27) and Vk∗MYC:Wwox WT (blue line, n = 14) mice. Vk∗MYC:Wwox KO mice showed significantly reduced survival compared to the WT group (log-rank [Mantel-Cox] P = .002). The mean survival was 660 days for the Wwox KO group, compared to 800 days for the Wwox WT group. The x-axis indicates survival time in days, and the y-axis represents cumulative survival.
Increased tumor incidence in Vk∗MYC:Wwox KO mice with high CD138+ expression and uropod formation in PBPs. (A) A significantly higher number of Vk∗MYC:Wwox KO mice (17/27) developed PBTs and BCLs compared to Vk∗MYC:Wwox WT mice (4/14), as determined by χ2 analysis (P = .036). (B) Low-magnification photomicrograph of extramedullary PBP histology section from a Vk∗MYC:Wwox KO mouse immunostained with anti-CD138 antibody. Scale bar on lower left, 400 μm in length. (C) High-magnification photomicrograph of histology section from another Vk∗MYC:Wwox KO PBP displaying areas of cells with immunostained uropod-like structures, that is, polarized intracellular CD138 staining to small membrane protrusions (red arrows). Scale bar on lower left, 100 μm in length. (D) Representative SPEP of serum samples from Vk∗MYC:Wwox WT and KO mice. Red arrows indicate representative M-spikes. (E) Representative longitudinal follow-up of serum samples analyzed by SPEP collected from the same mouse through time, displaying progressive disease with a marked increase in M-spike intensity over time.
Increased tumor incidence in Vk∗MYC:Wwox KO mice with high CD138+ expression and uropod formation in PBPs. (A) A significantly higher number of Vk∗MYC:Wwox KO mice (17/27) developed PBTs and BCLs compared to Vk∗MYC:Wwox WT mice (4/14), as determined by χ2 analysis (P = .036). (B) Low-magnification photomicrograph of extramedullary PBP histology section from a Vk∗MYC:Wwox KO mouse immunostained with anti-CD138 antibody. Scale bar on lower left, 400 μm in length. (C) High-magnification photomicrograph of histology section from another Vk∗MYC:Wwox KO PBP displaying areas of cells with immunostained uropod-like structures, that is, polarized intracellular CD138 staining to small membrane protrusions (red arrows). Scale bar on lower left, 100 μm in length. (D) Representative SPEP of serum samples from Vk∗MYC:Wwox WT and KO mice. Red arrows indicate representative M-spikes. (E) Representative longitudinal follow-up of serum samples analyzed by SPEP collected from the same mouse through time, displaying progressive disease with a marked increase in M-spike intensity over time.
A characteristic of monoclonal gammopathies is the secretion of immunoglobulin, which manifests as immunoglobulin monoclonal bands (M-spikes) in SPEP. We performed a longitudinal follow-up with SPEP in 24 of 27 Vk∗MYC:Wwox KO mice, as 3 mice died before the SPEP follow-up could be completed. All 24 Wwox KO mice (100%) showed detectable M-spikes, accompanied by a progressive increase in the intensity of secreted gamma globulin as mice aged, indicating disease progression (Figure 2D-E; supplemental Figure 2). M-spikes were also observed in all Vk∗MYC:Wwox WT mice, with 14 of 14 (100%) showing secreted gamma globulin. Overall, there was no difference in the age of M-spikes detection when comparing Vk∗MYC:Wwox KO vs WT counterparts (supplemental Figure 2).
Proinflammatory and tumorigenic transcriptomic signatures in PCs with Wwox deletion
To gain insight into transcriptome changes associated with B-cell specific Wwox deletion in Vk∗MYC mice, we performed RNA-seq analysis of BM derived CD138+ PCs from Vk∗MYC:Wwox KO and Vk∗MYC:Wwox WT mice. The differential gene expression (DGE) data set identified 87 dysregulated genes (FDR < 0.05) in Vk∗MYC:Wwox KO vs Vk∗MYC:Wwox WT CD138+ BM cells (supplemental File 1). Unsupervised hierarchical clustering of DGE profiles robustly segregated Wwox WT from Wwox KO BM samples (Figure 3A). Various immune regulatory and protumorigenic genes, such as Foxq1, Vegfa, Jun, and Tgfb2, were significantly upregulated in the Wwox KO group (supplemental File 1).
Vk∗MYC:Wwox KO CD138+ PCs display proinflammatory and tumorigenic transcriptomic signatures. (A) Unsupervised clustering and heat map of 87 differentially expressed genes (FDR < 0.05) comparing Vk∗MYC:Wwox WT and Vk∗MYC:Wwox KO CD138+ PCs (n = 3 mice per group). Red or blue colors indicate differentially upregulated or downregulated genes, respectively. Mean signals were background corrected and log2 transformed. (B-C) Bar graph depicting enrichment of biofunctions associated with tumorigenic phenotypes (B) and disease processes (C) (highlighted by red arrows), driven by dysregulated gene expression in Vk∗MYC:Wwox KO PCs (z score cutoff ±2; P < .05). (D) Bar graph showing top-activated and inhibited upstream regulators as per IPA (z score cutoff ±2; P < .05) based on DGE profiles in Vk∗MYC:Wwox KO compared to the control group.
Vk∗MYC:Wwox KO CD138+ PCs display proinflammatory and tumorigenic transcriptomic signatures. (A) Unsupervised clustering and heat map of 87 differentially expressed genes (FDR < 0.05) comparing Vk∗MYC:Wwox WT and Vk∗MYC:Wwox KO CD138+ PCs (n = 3 mice per group). Red or blue colors indicate differentially upregulated or downregulated genes, respectively. Mean signals were background corrected and log2 transformed. (B-C) Bar graph depicting enrichment of biofunctions associated with tumorigenic phenotypes (B) and disease processes (C) (highlighted by red arrows), driven by dysregulated gene expression in Vk∗MYC:Wwox KO PCs (z score cutoff ±2; P < .05). (D) Bar graph showing top-activated and inhibited upstream regulators as per IPA (z score cutoff ±2; P < .05) based on DGE profiles in Vk∗MYC:Wwox KO compared to the control group.
IPA-based functional annotation of dysregulated genes in Vk∗MYC:Wwox KO PCs revealed enrichment of biofunctions related to tumorigenic phenotype such as cellular growth and proliferation, cellular movement, cell death and survival, and cell-to-cell signaling and interaction (Figure 3B). Additionally, enriched disease processes included, inflammatory response, hematological disease, inflammatory disease, and cancer (Figure 3C). IPA analysis also identified putative top-activated and inhibited upstream regulators (z score cutoff ±2; P < .05). Tumor necrosis factor (TNF; activation z score 2.47) was identified as the topmost activated regulator along with other inflammatory pathway factors like the nuclear factor κB (NF-κB) complex and interleukin 6 (IL-6). Furthermore, TNF-based regulation of several differentially expressed genes in our data set pointed to proliferation of tumor cells as a significant outcome effect in Vk∗MYC:Wwox KO CD138+ BM cells (supplemental Figure 3). In addition, protumorigenic signaling pathway molecules such as MAPK and transforming growth factor-β1, along with transcription factor Stat5a/b were among other significantly activated upstream regulators, whereas Runx1, Erg, and Gata1 were found as inhibited regulators (Figure 3D). These observations strongly link transcription factors, signaling pathways, and inflammatory changes that contribute to a tumorigenic phenotype with the specific deletion of Wwox in B cells from Vk∗MYC mice.
PBTs displayed highly enriched inflammatory gene expression signatures
We further compared the transcriptome profiles of CD138+ BM PCs and PBTs from Vk∗MYC:Wwox KO mice. This analysis identified 2355 differentially regulated genes (FDR < 0.05) in Vk∗MYC:Wwox KO-PBT vs Vk∗MYC:Wwox KO-BM cells (supplemental File 1). Hierarchical clustering of the DGE data set clearly separated BM PCs from PBTs (Figure 4A). In Wwox KO PBTs, a noteworthy observation was the pronounced upregulation of genes implicated in cancer cell migration, invasion, and metastasis such as Ccl21a, Cilp, Mmp3, and Pdpn, exhibiting over a ninefold difference in expression (log2 FC range, 9.09-12.53) (supplemental File 1). Interestingly, the Aicda gene (activation induced cytidine deaminase, also known as Aid), which encodes a key enzyme expressed in germinal centers involved in somatic hypermutation, gene conversion and class switch recombination of Ig genes, was found to be overexpressed 11 log2 fold higher on average in PBTs vs BM cells (supplemental Figure 4). Likewise, Utf1, a gene linked with stem cell properties and cancer stemness, was significantly upregulated in Wwox KO-PBT samples (log2 FC = 9.32) (supplemental File 1). Similar to observations in the Wwox KO vs WT BM comparison, transcriptome profiling of Wwox KO-PBT vs Wwox KO-BM samples, also showed enrichment of cell death and survival, cellular movement, cellular growth and proliferation, and cell-to-cell signaling and interaction tumorigenic biofunctions and cancer, hematological disease, inflammatory disease, and inflammatory response as significantly enriched disease conditions (Figure 4B-C). However, the number of deregulated genes and magnitude of statistical significance for all enriched biofunctions and disease processes were far more pronounce in Vk∗MYC:Wwox KO-PBT (P value range, 4.28×10-40 to 1.69×10-19) in comparison to Vk∗MYC:Wwox KO-BM (P value range, 2.06×10-7 to 1.19×10-3) (Figure 4D-E). Interestingly, one of the significant regulator effects identified in Wwox KO-PBT is based on the upstream regulator CCL2 (also known as Mcp10), a potent chemoattractant cytokine with an activation z score of 4.64 and capable of upregulating multiple target genes in the DGE data set, ultimately leading to increased homing of cells and inflammatory response (supplemental Figure 5). The enrichment of the same biofunctions and diseases suggests consistency between Vk∗MYC KO-BM cells and PBTs. However, markedly higher number of differentially expressed genes and magnitude of statistical significance in the enrichment analysis strongly indicate a more pronounced tumorigenic and inflammatory phenotype in PBTs, likely driving the plasmablastic/anaplastic progression of malignant PCs.
Highly enriched inflammatory gene expression signatures in Vk∗MYC:Wwox KO PBTs. (A) Unsupervised clustering and heat map of 2355 differentially expressed genes (FDR < 0.05) comparing Vk∗MYC:Wwox KO-BM (n = 3) and Vk∗MYC:Wwox KO-PBT (n = 4) groups. Red or blue colors indicate differentially upregulated or downregulated genes, respectively. Mean signals were background corrected and log2 transformed. (B-C) Bar graph depicting enrichment of biofunctions associated with tumorigenic phenotypes (B) and disease processes (C) (highlighted by red arrows), driven by dysregulated gene expression in Vk∗MYC:Wwox KO PBTs (z score cutoff ±2; P < .05). (D) Bar graph showing the number of deregulated genes and the magnitude of statistical significance for enriched biofunctions and disease processes in Vk∗MYC:Wwox KO-PBT vs Vk∗MYC:Wwox KO-BM and Vk∗MYC:Wwox KO-BM vs Vk∗MYC:Wwox WT-BM groups. The enrichment was more pronounced in Vk∗MYC Wwox KO-PBT vs Vk∗MYC Wwox KO-BM (P value range, 4.28×10-40 to 1.69×10-19) compared to Vk∗MYC:Wwox KO-BM vs Vk∗MYC:Wwox WT-BM (P value range, 2.06×10-7 to 1.19E×10-3). Numbers in parentheses beside each bar indicate the number of differentially expressed genes in each category.
Highly enriched inflammatory gene expression signatures in Vk∗MYC:Wwox KO PBTs. (A) Unsupervised clustering and heat map of 2355 differentially expressed genes (FDR < 0.05) comparing Vk∗MYC:Wwox KO-BM (n = 3) and Vk∗MYC:Wwox KO-PBT (n = 4) groups. Red or blue colors indicate differentially upregulated or downregulated genes, respectively. Mean signals were background corrected and log2 transformed. (B-C) Bar graph depicting enrichment of biofunctions associated with tumorigenic phenotypes (B) and disease processes (C) (highlighted by red arrows), driven by dysregulated gene expression in Vk∗MYC:Wwox KO PBTs (z score cutoff ±2; P < .05). (D) Bar graph showing the number of deregulated genes and the magnitude of statistical significance for enriched biofunctions and disease processes in Vk∗MYC:Wwox KO-PBT vs Vk∗MYC:Wwox KO-BM and Vk∗MYC:Wwox KO-BM vs Vk∗MYC:Wwox WT-BM groups. The enrichment was more pronounced in Vk∗MYC Wwox KO-PBT vs Vk∗MYC Wwox KO-BM (P value range, 4.28×10-40 to 1.69×10-19) compared to Vk∗MYC:Wwox KO-BM vs Vk∗MYC:Wwox WT-BM (P value range, 2.06×10-7 to 1.19E×10-3). Numbers in parentheses beside each bar indicate the number of differentially expressed genes in each category.
Vk∗MYC:Wwox KO PBTs display hypermutation profiles and multiple cancer driver mutations
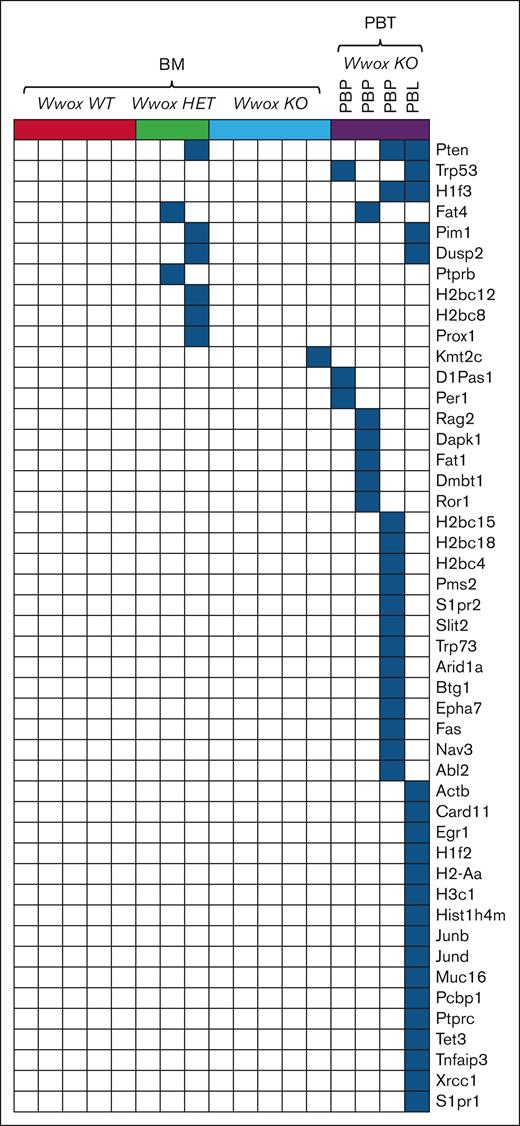
By means of whole-exome sequencing, we analyzed the mutational profile of CD138+ BM cells from Vk∗MYC:Wwox KO, HET, and WT mice, and from Vk∗MYC:Wwox KO PBT tissue samples. Among the 4 PBT samples analyzed, 3 were classified as PBPs, whereas 1 was identified as a PBL. Nonsynonymous SNVs and insertions/deletions were analyzed, and variant effect predictor and Sorting Intolerant From Tolerant (SIFT) impact scores were employed to identify relevant protein mutations affecting cancer driver genes. No cancer driver mutations were detected in any of the Vk∗MYC:Wwox WT-BM samples, whereas 2 of the Wwox HET-BM and 1 Wwox KO-BM displayed deleterious mutations affecting some cancer genes (Figure 5). The scarcity of cancer driver mutations in most of the BM samples regardless of genotype agrees with previous observations; however, it must be noted that our study limited to WES analyses is unable to capture structural variants also known to be major cancer drivers as shown previously.28 On the other hand, all Vk∗MYC:Wwox KO PBT samples exhibited multiple mutations affecting various cancer genes commonly associated with both MM and BCLs. Notably, the highest number of mutations was observed in the Wwox KO PBL sample, which harbored 21 distinct cancer driver mutations.
Somatic, nonsynonymous SNVs in cancer driver genes in Vk∗MYC:Wwox KO PBTs. Oncoplot displaying nonsynonymous SNVs in cancer driver genes identified through exome sequencing of CD138+ BM PCs and Vk∗MYC:Wwox KO PBTs. Each column represents an individual mouse, grouped as indicated at the top of the plot, and each row corresponds to mutations detected in a specific gene for these mice. In the Vk∗MYC:Wwox KO-PBT group, the first 3 columns represent PBPs, whereas the last column represents a PBL. Blue squares indicate the presence of mutations in the corresponding gene for each sample.
Somatic, nonsynonymous SNVs in cancer driver genes in Vk∗MYC:Wwox KO PBTs. Oncoplot displaying nonsynonymous SNVs in cancer driver genes identified through exome sequencing of CD138+ BM PCs and Vk∗MYC:Wwox KO PBTs. Each column represents an individual mouse, grouped as indicated at the top of the plot, and each row corresponds to mutations detected in a specific gene for these mice. In the Vk∗MYC:Wwox KO-PBT group, the first 3 columns represent PBPs, whereas the last column represents a PBL. Blue squares indicate the presence of mutations in the corresponding gene for each sample.
Among all the identified mutations, the tumor suppressor Pten emerged as the most frequently mutated gene, detected in 1 Wwox HET-BM sample and in 2 Wwox KO PBTs (1 PBP and 1 PBL). Other mutated genes, common to both BM and PBT groups, included the tumor suppressor Fat4, identified in 1 Wwox HET-BM and 1 Wwox KO-PBP sample. Additionally, mutations in the oncogene Pim1 and Dusp2 were both found in 1 Wwox HET-BM sample and the Wwox KO-PBL. Notably, all these genes have been previously reported as MM drivers43-45 and in the Vk∗MYC model28 (Figure 5). Among other recurrent mutations in Vk∗Myc:Wwox KO-PBTs were the tumor suppressor gene Trp53 and the histone gene H1f3, each present in 2 of 4 samples, including both PBP and PBL. Several other histone genes, H2bc12, H2bc8, H2bc15, H2bc18, H2bc4, H1f2, H2-Aa, H3c1, and Hist1h4m, were also found mutated, underscoring a potential disruption of chromatin dynamics and gene expression in these tumors. Mutations in other tumor suppressors including Fat1, Dapk1, and the Ror1 oncogene were also observed. Both PBPs and PBL also showed mutations in DDR genes (Arid1a, Pms2, Xrcc1) and genes involved in proliferation and cell cycle regulation (Junb, Jund, Per1, Egr1, Btg1, Tet3), further highlighting genomic instability. Moreover, several mutations were detected in cell-signaling genes, suggesting broad perturbations across oncogenic networks (Figure 5). Mutations in the described cancer drivers have been reported not only in MM43-45 but also in BCLs.46
We also performed single based substitution (SBS) signature analyses to characterize the most represented signatures and uncover underlying mutagenic processes. All samples exhibited signatures SBS1, associated with spontaneous 5-methylcytosine deamination, and SBS5, known for its clock-like mutation profile (Figure 6A; supplemental Figure 6). Both signatures are associated with age-related mutational processes, with SBS5 further linked to DNA replication stress. The consistent presence of these signatures indicates a background of aging-related mutations and ongoing replication stress across all samples. The most significant finding from our analysis was the detection of DNA repair related mutational activity, marked by the presence of SBS26 and SBS30 (Figure 6A; supplemental Figure 6). SBS26, associated with defective DNA mismatch repair, was observed in samples from all BM groups including 2 samples from the HET and 1 from the KO groups (Figure 6A; supplemental Figure 6). Additionally, SBS30 linked to defective base excision repair, was observed only in Vk∗MYC:Wwox KO-PBPs (2/3 samples) (Figure 6A; supplemental Figure 6). Additionally, we identified SBS32 and SBS87, both associated with thiopurine family drugs exposure. These drugs are known to be mutagenic via the integration of the final metabolite 6-thioguanine into DNA. Importantly, one route of escape of thiopurines cytotoxicity is by inactivation of mismatch repair mechanisms.47 Perhaps and like the described SBS26 and SBS30 associated with defective base excision repair and mismatch repair, the finding of SBS32 and SBS87 signatures also reflects associations with DNA mismatch repair mechanisms. Additionally, we detected the SBS85 signature, which reflects the indirect effects of Aid activity in lymphoid cells. Other signatures, including SBS12, SBS37, and SBS40a, of unknown origin, were also detected (Figure 6A).
Distinct SBS signatures and the occurrence of kataegis-like clustered hypermutation profiles in Vk∗MYC:Wwox KO BM and PBTs. (A) Heat map displaying the most frequent SBS mutational signatures identified in BM PCs and Vk∗MYC:Wwox KO PBPs. The y-axis represents the distinct SBS signatures detected in these samples, whereas the x-axis denotes the individual samples. Each column represents an individual mouse, grouped as indicated at the top of the heat map. In the Vk∗MYC:Wwox KO-PBT group, the first column represents PBL, whereas the remaining 3 columns represent PBPs. Color intensity on the heat map indicates the strength of association with each SBS signature. (B) Rainfall plots of all the mutations across the genome of representative samples from Vk∗MYC:Wwox KO BM and PBTs demonstrating a widespread distribution of clustered hypermutations resembling kataegis. The y-axis represents the distance of each mutation from the preceding mutation. Different colors represent the different types of the mutation substitutions. Clustering of mutations closer to the x-axis indicates a smaller distance between mutations, reflecting hypermutation. (C) Bar graphs illustrating the average normalized counts of Aicda and Apobec2 messenger RNA expression in Vk∗MYC:Wwox WT (n = 5), HET (n = 3), KO (n = 5) BM and Vk∗MYC:Wwox KO PBT (n = 4) samples. Each data point represents the expression level for an individual mouse, with error bars indicating the mean ± standard error of the mean. The green data point indicates Aicda and Apobec2 expression in the Vk∗MYC:Wwox KO1-PBT (PBP), whereas the red data point indicates expression in the Vk∗MYC:Wwox KO4-PBT (PBL). A marked difference in expression was observed; statistical analysis using 1-way analysis of variance did not reach significance, likely due to intersample variability (P = .2646 for Aicda; P = .2680 for Apobec2).
Distinct SBS signatures and the occurrence of kataegis-like clustered hypermutation profiles in Vk∗MYC:Wwox KO BM and PBTs. (A) Heat map displaying the most frequent SBS mutational signatures identified in BM PCs and Vk∗MYC:Wwox KO PBPs. The y-axis represents the distinct SBS signatures detected in these samples, whereas the x-axis denotes the individual samples. Each column represents an individual mouse, grouped as indicated at the top of the heat map. In the Vk∗MYC:Wwox KO-PBT group, the first column represents PBL, whereas the remaining 3 columns represent PBPs. Color intensity on the heat map indicates the strength of association with each SBS signature. (B) Rainfall plots of all the mutations across the genome of representative samples from Vk∗MYC:Wwox KO BM and PBTs demonstrating a widespread distribution of clustered hypermutations resembling kataegis. The y-axis represents the distance of each mutation from the preceding mutation. Different colors represent the different types of the mutation substitutions. Clustering of mutations closer to the x-axis indicates a smaller distance between mutations, reflecting hypermutation. (C) Bar graphs illustrating the average normalized counts of Aicda and Apobec2 messenger RNA expression in Vk∗MYC:Wwox WT (n = 5), HET (n = 3), KO (n = 5) BM and Vk∗MYC:Wwox KO PBT (n = 4) samples. Each data point represents the expression level for an individual mouse, with error bars indicating the mean ± standard error of the mean. The green data point indicates Aicda and Apobec2 expression in the Vk∗MYC:Wwox KO1-PBT (PBP), whereas the red data point indicates expression in the Vk∗MYC:Wwox KO4-PBT (PBL). A marked difference in expression was observed; statistical analysis using 1-way analysis of variance did not reach significance, likely due to intersample variability (P = .2646 for Aicda; P = .2680 for Apobec2).
Interestingly, 2 of 5 Vk∗MYC:Wwox KO-BM samples and 2 of 4 Vk∗MYC:Wwox KO-PBTs, including 1 PBP and 1 PBL, displayed a widespread distribution of clustered mutations resembling kataegis, with a broader mutational pattern consistent with hypermutated genomic regions (Figure 6B; supplemental Figure 7). This profile was particularly prominent in the PBL, which exhibited an exceptionally pronounced hypermutated pattern (Figure 6B). Mutation clusters and hypermutation in cancer have been hypothesized to result from the activity of AID/APOBEC editing deaminases.43 Consistent with this hypothesis, expression analysis of Aid/Apobec family members in Vk∗MYC:Wwox KO-PBT samples showed marked overexpression of Aicda and Apobec2 in PBP and PBL tumors exhibiting hypermutation (Figure 6C). However, these differences did not reach statistical significance (P = .2646 for Aicda; P = .2680 for Apobec2), likely due to high intersample variability as commonly seen in MM patients (supplemental Figure 8). Overall, these findings and those of mutational signatures described earlier suggest that Wwox deletion affects DNA repair processes with Wwox KO samples exhibiting mutational profiles compatible with DNA repair deficiencies and hypermutation.
Extensive chromosomal copy number alterations are characteristic of Vk∗MYC:Wwox KO PBTs
We next analyzed CNVs by comparing the genomic profiles of BM and PBT samples. Similar to SNV analyses, copy number alterations were relatively low and comparable between all BM sample groups again pointing to a rather indolent disease status mainly driven by MYC overexpression (Figure 7A; supplemental Figure 9A-C; supplemental Table 2). In contrast, both copy number gains and losses were significantly elevated in Vk∗MYC:Wwox KO PBTs (Figure 7A-B; supplemental Table 2).
Vk∗MYC:Wwox KO PBTs display recurrent focal and large copy number alterations. (A) Heat map illustrating copy number abnormalities in BM PCs and Vk∗MYC:Wwox KO PBPs. The y-axis represents individual samples, whereas the x-axis displays individual chromosomes, separated by solid lines. Color coding indicates copy number losses or gains at various chromosomal loci, with blue representing copy number loss and red indicating copy number gain. The intensity of the colors reflects the magnitude of the copy number alterations. (B) Scatter plots from Vk∗MYC:Wwox KO-PBT samples illustrate widespread and pronounced copy number alterations across all samples. Focal and large copy number alterations are represented by red markers; those above zero indicate copy number gains, whereas markers below zero signify copy number losses. The names of the genes affected by specific copy number alterations are displayed near the corresponding alterations, with red indicating copy number gains and blue indicating copy number losses for the respective genes.
Vk∗MYC:Wwox KO PBTs display recurrent focal and large copy number alterations. (A) Heat map illustrating copy number abnormalities in BM PCs and Vk∗MYC:Wwox KO PBPs. The y-axis represents individual samples, whereas the x-axis displays individual chromosomes, separated by solid lines. Color coding indicates copy number losses or gains at various chromosomal loci, with blue representing copy number loss and red indicating copy number gain. The intensity of the colors reflects the magnitude of the copy number alterations. (B) Scatter plots from Vk∗MYC:Wwox KO-PBT samples illustrate widespread and pronounced copy number alterations across all samples. Focal and large copy number alterations are represented by red markers; those above zero indicate copy number gains, whereas markers below zero signify copy number losses. The names of the genes affected by specific copy number alterations are displayed near the corresponding alterations, with red indicating copy number gains and blue indicating copy number losses for the respective genes.
Monosomy or large deletions of chr.5 were observed across all groups, with 2 of 5 Vk∗MYC:Wwox WT-BM, 3 of 3 Vk∗MYC:Wwox HET-BM, and 3 of 5 Vk∗MYC:Wwox KO-BM (supplemental Figure 9A-C). Additionally, 1 Vk∗MYC:Wwox KO PBT case exhibited losses of chr.5, whereas 2 cases displayed small focal gains (Figure 7B). Notably, such extensive chr.5 deletions have previously been implicated as drivers of tumor progression in the Vk∗MYC myeloma model.28,48,49
Focal deep deletions affecting chr.12 were identified in 1 sample from each BM group and various PBTs, with 2 PBPs showing extensive chr.12 deletions (Figure 7B; supplemental Figure 9A-C). Focal deletions affecting chr.6 were also observed in a Wwox KO-BM sample and a PBT. These deletions primarily affected the Igh locus on chr.12 and the Igk locus on chr.6, both crucial for antibody diversity (supplemental Table 2). Interestingly, the chr.12 large deletions observed in 2 PBPs also affected Traf3, Max, and Cdc42bpb genes (Figure 7B), all described among the handful of targets in addition to Wwox displaying complete loss of function in the latest CoMMpass study.24Vk∗MYC:Wwox KO plasmacytomas also displayed large chr.14 losses in 2 samples affecting 2 critical genes, Dis3 and the tumor suppressor Rb1 (Figure 7B). Mouse chr.14 shows significant synteny with human chr.13q, known for being among the most common large chromosomal losses found in MM and other human lymphoid malignancies48,50 and involving DIS3 and RB1 genes, both recognized as key drivers in MM.24 Large deletions affecting chr.8, which includes the Cyld locus, were also observed in 2 of the Vk∗MYC:Wwox KO PBPs (Figure 7B).
Additionally, an amplicon was detected affecting chr.11 in PBT samples involving the Rel oncogenic NF-κB subunit, Xpo1, Bcl11a, Il9r, Tlx3, and Spdl1 all of which play critical roles in lymphoid development and oncogenesis51-55 (Figure 7B). A focal amplification including the Braf locus (chr.6) was identified in 1 BM and 1 PBP sample, gene also displaying gain of function in MM24 (Figure 7B; supplemental Figure 9C). Additional CNVs identified in individual PBTs were focal losses in Kdm4c, Ptprd, Trp53, chr.19 losses containing Pten and Fas as well as gains of chromosomes containing oncogenes such as Hras, Nras, Mcl1, and Pclo oncogenes (Figure 7B).
Discussion
We generated a Vk∗MYC:Wwox KO mouse cancer model, harboring 2 oncogenic alterations commonly found in B- and plasma-cell cancers: Myc activation and Wwox deletion. The Vk∗MYC:Wwox KO mice exhibited significantly reduced survival rates when compared with Vk∗MYC:Wwox WT counterparts, demonstrating that B cell-specific Wwox deficiency in mice with MYC activation leads to the development of more lethal cancer phenotypes. These mice displayed a significant high incidence of PBTs characterized by distinct histopathological features and immunohistochemical staining patterns compatible mostly with aggressive extramedullary PBTs and also PBLs.
An interesting finding in plasmacytomas was the presence of clustered CD138 staining to polarized cellular foci previously described as resembling uropods in Vk∗MYC:Wwox KO plasmacytoma cells. This phenomenon is known to correlate with heightened migratory potential and aggressiveness in myeloma cells and thus highlighting the potential role of Wwox deletion in fostering a more aggressive tumor phenotype.42
Among the most important observations from these studies was the significant activation of proinflammatory pathways in Wwox KO BM cells and PBTs. Key transcriptome regulators such as TNF, NF-κB, IL-6, and CCL2 appear prominently involved, highlighting the central role of inflammation in driving tumor progression in Wwox deficient models. These findings are in agreement with previous observations across cancers, CNS disorders, and various inflammatory conditions indicating a significant role for WWOX in inflammation regulation due to direct associations with the NF-kB and IL-6/JAK2/STAT3 signaling pathways.32,33,56-63 Chronic inflammation is well known to be crucial in cancer development, fostering an immunosuppressive environment, promoting cell proliferation, survival, migration and also contributing to genomic instability.64,65 NF-κB is a known master regulator of inflammatory response transcriptionally controlling expression of proinflammatory cytokines as well as antiapoptotic genes.66 Furthermore, production of cytokines like TNF and IL-6 and growth factors that promote tumor progression, have been associated with more aggressive forms of BCLs and MM.65
Exome sequencing analysis identified a single oncogenic mutation and minimal copy number alterations in Vk∗MYC:Wwox KO-BM cells, indicating a rather indolent disease state in the BM regardless of genotype accompanied by the common copy number losses affecting chr.5. In contrast, Vk∗MYC:Wwox KO PBTs exhibited multiple mutations and significant number of CNVs affecting key cancer driver genes, indicating increased genomic instability and a clear shift toward a more aggressive disease state. Mutation analysis identified various classical myeloma cancer target genes, including Pten, Trp53, Pim1, Dusp2, Btg1, Egr1, as well as various histone targets such as H1f3, H1f2, and others that align with those recently reported in Vk∗MYC derived models,28 thereby underscoring their relevance in hematological malignancies, including BCLs and MM.67-69 Likewise, CNV findings aligned with the recent comprehensive MMRF CoMMpass study of newly diagnosed MM patients that led to the identification of only 12 target genes with recurrent and complete loss of function in >2% of the cohort that include and coincide with target losses in our study such as TRAF3, DIS3, RB1, CYLD, TP53, CDC42BPB, and indeed WWOX by study design in our case.24 Furthermore, the authors of this study described that although loss of 1 WWOX allele is expected in t(14;16), they frequently detected complete loss of function at similar frequency to total loss of classical targets such as RB and TP53, indeed supporting a possible role for WWOX in MM.24 Similarly, copy number gains including Braf and Ras family oncogenes, identified in our PBPs and also described in the CoMMpass study were also found.
Wwox KO PBTs also displayed mutations of various DDR genes, mutational signatures associated with defective DNA damage repair, and overexpression of Aid/Apobec gene family members, known for inducing somatic hypermutations through cytosine deamination all processes contributing to genomic instability, tumorigenesis, and described in MM.70,71 Notably, we and others have shown that loss of WWOX function significantly impairs DNA repair efficiency, resulting in the accumulation of DNA damage that triggers genomic instability, a hallmark of cancer progression.3,26,72-74
In conclusion, the described findings strongly support that the loss of WWOX function is a key driver for the development of aggressive plasmablastic phenotypes, primarily through 2 mechanisms: escalating genomic instability and fostering a protumorigenic inflammatory microenvironment.
Acknowledgments
The authors thank the University of Texas MD Anderson Cancer Center Research Animal Support Facility and Laboratory Animal Genetic Services for animal support, and Advanced Technology Genomics Core for sequencing facility (P30 NIH CA016672).
This work was supported by a grant (C.M.A.) from the Leukemia and Lymphoma Society Specialized Center of Research award number 7016-18 (project 2). M.C. is supported by grants from the National Cancer Institute (RO1 CA272426, CA186781).
Authorship
Contribution: T.H. performed experiments and wrote the manuscript; M.D.B. performed exome sequencing (exome-seq) data analysis; B.L. performed RNA-sequencing and exome-seq data analysis; M.C.A. performed exome-seq data analysis and provided intellectual input; M.C. provided Vk∗MYC mice and intellectual input; and C.M.A. conceptualized the study, performed experimental designing and data analysis, wrote the manuscript, and handled correspondence.
Conflict-of-interest disclosure: M.C. receives royalties from the licensing of Vk∗MYC mice and derivative lines. The remaining authors declare no competing financial interests.
Correspondence: C. Marcelo Aldaz, Department of Epigenetics and Molecular Carcinogenesis, The University of Texas MD Anderson Cancer Center, 1901 East Rd, Houston, TX 77054; email: maaldaz@mdanderson.org.
References
Author notes
RNA sequencing data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (available at: doi:10.1093/nar/30.1.207; accession number GSE288262).
Exome sequencing data are available on request from the corresponding author, C. Marcelo Aldaz (maaldaz@mdanderson.org).
The full-text version of this article contains a data supplement.

![Wwox ablation in B cells reduces overall survival in Vk∗MYC mice. Kaplan-Meier survival analysis comparing Vk∗MYC:Wwox KO (red line, n = 27) and Vk∗MYC:Wwox WT (blue line, n = 14) mice. Vk∗MYC:Wwox KO mice showed significantly reduced survival compared to the WT group (log-rank [Mantel-Cox] P = .002). The mean survival was 660 days for the Wwox KO group, compared to 800 days for the Wwox WT group. The x-axis indicates survival time in days, and the y-axis represents cumulative survival.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodneoplasia/2/4/10.1016_j.bneo.2025.100153/3/m_bneo_neo-2025-000654-gr1.jpeg?Expires=1768222827&Signature=Avzzwxk9ilkszbi4HCXDIKu3-L5s7mjrE2T1FbYoJ~5-LWDf8g7TgUsJFR9YyGFpq5XRwixCTdd3H95o5HjFMzN4ZRlSDvn2jaLQyooyK~B9fhtjfmMnZ28GSrw0-NMM-W1Y8bC10XTTGxeNs944sBXIcz33RkAbXPmv2QGJoMLcHb6giZpTvzcCAMZrOUyaV-~mHB2AhinV1Ws9sdFYW1KX3a-My46QOr7UWsqX1W-5Qmnn5OmVBpAfc5H9pMMIYGPZLj8HoiaxedOk9f1pzOMXt7nLUtrHsGPHiWmmvrKQ4d6JNXwnLNlynAB7gFxnAjc8M7ADic6vp2o3JOOGIA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)