Key Points
Enitociclib synergizes with MI-463 to effectively decrease HOXA9 protein levels in infant KMT2A-r leukemia cells.
Enitociclib potentiates the cytotoxicity of venetoclax in relatively venetoclax-resistant KMT2A-r leukemic cells.
Visual Abstract
The KMT2A-rearranged (KMT2A-r) leukemia is one of the most challenging cancers to treat in children, owing to the higher relapse rates and chemoresistance frequently observed in this patient population. At the molecular level, chromosomal translocation in the KMT2A gene leads to a deregulated epigenetic landscape resulting in the upregulation of transcription factors like HOXA9, consequently contributing to leukemogenesis. One crucial component of the oncogenic KMT2A-r complex is the involvement of positive transcription elongation factor b, which is composed of cyclin T and cyclin-dependent kinase 9 (CDK9), which leads to the dysregulation of transcriptional elongation. This study investigated the function of enitociclib, a small molecule CDK9 inhibitor in clinical development that has shown effective activity in other tumor types. Enitociclib showed growth inhibition and an on-target effect in KMT2A-r leukemic cells with a significant decrease in MYC and MCL-1 protein levels. Moreover, enitociclib was found to reduce the growth advantage provided to leukemic cells by the bone marrow microenvironment. In addition, it demonstrates the ability to synergize with menin inhibitors, leading to an effective decrease in HOXA9 protein levels in KMT2A-r infant leukemia cells. Enitociclib also potentiates the cytotoxicity of venetoclax in relatively venetoclax-resistant KMT2A-r leukemic cells. Overall, enitociclib has shown measurable in vitro antitumor activity in KMT2A-r infant leukemia and is a rational therapeutic option to explore in future clinical trials.
Introduction
The overall outcome in children diagnosed with leukemia has improved in the last few decades with a survival rate of ∼95% for acute lymphoblastic leukemia (ALL) and 70% for acute myeloid leukemia (AML).1,2 However, the prognosis of high-risk disease phenotypes continues to be poor. Among the high-risk subgroups, infants diagnosed with leukemia that carry 11q23 translocations are characterized by poor survival rates and higher relapse rates within the first year of treatment.3 Approximately 70% of infants with ALL and 35% to 50% of infants with AML carry translocations in their leukemic cells in KMT2A, a histone methyl transferase gene.3 Translocations in KMT2A generate fusion proteins that deregulate the epigenetic landscape and lead to the upregulation of transcription factors, consequently contributing to leukemogenesis.3 Presently, the overall survival rate of these patients remains poor and therefore, novel therapies are needed for this population of infant leukemia patients. The KMT2A protein has H3K4 methyltransferase activity and associates with menin, the protein product of the MEN1 gene, to regulate the expression of multiple genes during normal hematopoiesis.3 One important downstream target of KMT2A is the transcription factor HOXA9 whose expression is tightly regulated during hematopoiesis.4 High expression of HOXA9 during the early stages of hematopoiesis is associated with self-renewal and increased proliferative capacity of the immature progenitor cells. Its expression diminishes as early hematopoietic cells become more differentiated into mature cell types. However, KMT2A-rearranged (KMT2A-r) leukemic cells have a constitutive expression of HOXA9, which has the potential function to drive leukemogenesis.4
In KMT2A-r leukemic cells, the 11q chromosomal translocation results in an abnormally expressed KMT2A fusion protein, in which truncated KMT2A protein is generally fused with members of the superelongation complex.3 The KMT2A fusion protein is part of a large multiprotein complex, components of which further mediate the transforming functions observed in KMT2A-r leukemia.3 One important protein in this complex is positive transcription elongation factor b (P-TEFb), which is composed of cyclin-dependent kinase 9 (CDK9) and cyclin T. The P-TEFb belongs to the class of transcriptional kinases and acts as a key regulator of the elongation phase of transcription.5 Generally, the main regulatory step at which the expression of most genes is controlled is transcriptional elongation as opposed to transcriptional initiation.6 The promoter regions of most genes contain stalled RNA polymerase II (RNAP II), resulting from the coordinated activity of negative elongation factor and DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole) sensitivity–inducing factor which keeps the RNAP Ⅱ in a transcriptionally paused state by blocking elongation.7 The P-TEFb factor, which contains a complex of CDK9 and cyclin T1, is recruited to the stalled RNAP II to begin the elongation phase of transcription. The CDK9 component within the P-TEFb complex phosphorylates the serine residue at position 2 in the heptapeptide sequence found in the carboxy-terminal domain (CTD) of Rpb1, a subunit of RNAPⅡ.5 It also phosphorylates negative elongation factor, resulting in its dissociation from RNAPⅡ. Additionally, CDK9 converts DRB sensitivity–inducing factor to a positive elongation factor for productive transcriptional elongation.7 In addition, P-TEFb also interacts with the components of superelongation complex in KMT2A-r leukemia.7 Consequently, once recruited to the appropriate regions on the chromatin, P-TEFb stimulates the transcriptional elongation as well as regulates the expression of short-lived messenger RNA transcripts like MYC and MCL-1.8
This study investigated the therapeutic implications of targeting CDK9/P-TEFb in infant KMT2A-r leukemias using enitociclib (Vincerx Pharma, Inc, Palo Alto, CA), a small molecule inhibitor that selectively targets CDK9.9 Enitociclib is tolerable and has shown effective antitumor activity in double-hit diffuse large B-cell lymphoma.10 The data obtained in this study show antitumor activity of enitociclib in KMT2A-r leukemic cells. In addition, enitociclib treatment decreases the survival support provided by the bone marrow (BM) stromal cells in coculture experiments. Enitociclib treatment also enhances the cytotoxic response to prednisolone in KMT2A-r cells. Furthermore, our findings demonstrate the synergistic drug interaction between enitociclib and the menin-KMT2A inhibitor MI-463 with increased loss of HOXA9 protein levels in KMT2A-r cells. Because enitociclib targets MCL-1, these findings also show the ability of enitociclib to enhance the cytotoxicity of venetoclax in KMT2A-r cell line models that are relatively resistant to venetoclax. Overall, the data presented provide important preclinical data to incorporate enitociclib in novel treatment protocols for KMT2A-r infant leukemia.
Materials and methods
Cell lines and cell culture
The leukemia cell line panel consisted of the KMT2A-r cell lines (MV4-11, THP-1, INL2,11 and KOPN-8) and the KMT2A wild-type (KMT2A-wt) cell lines (SUP-B15 and AML-193). All cell lines were cultured in RPMI 1640 (Sigma) with 10% fetal bovine serum at 37°C in a humidified CO2 incubator. The protocol for the isolation of peripheral blood mononuclear cells (PBMCs) from healthy donors and patient samples has been described previously.12 Leftover BM CD34+ from healthy donors were used. CD34+ cells were cultured in StemSpan SFEM II with StemSpan CD34+ Expansion Supplement (STEMCELL Technologies, Vancouver, Canada).
Small molecule inhibitors and chemotherapeutic agents
The CDK9 inhibitor enitociclib (formerly named BAY 1251152/VIP-152), venetoclax, and menin-KMT2A inhibitor MI-463 were purchased from Selleckchem. The stocks of all the single agents were prepared in 100% dimethyl sulfoxide (DMSO) at 10 mM concentration and stored at −20°C in aliquots. Chemotherapeutic agents were received from the Alberta Children’s Hospital oncology pharmacy (Alberta, Canada).
Cytotoxicity assays
Leukemic cells and PBMCs from healthy volunteers (1 × 104) were cultured in 200 μL of complete media (RPMI 1640 + 10% fetal bovine serum) and treated with various concentrations of enitociclib for 72 hours. DMSO was used as the vehicle control. Changes in cell viability were measured by Alamar blue assay (Thermo Fisher Scientific, MA) as per the manufacturer’s instructions and percent cell viability was calculated after normalization to DMSO control.
Apoptosis induction by Annexin/PI staining
Activation of cell death by apoptosis was measured by annexin V–fluorescein isothiocyanate and propidium iodide staining (Abcam, United Kingdom) and analysis was performed by flow cytometry. Briefly, 1 × 106 leukemic cells were plated in 6-well plates in 1.5 mL of complete culture media and incubated with various concentrations of enitociclib for 24 hours. Untreated and DMSO treated cells were used as controls. The experiment was performed according to the manufacturer’s instructions manual. Analysis was done by flow cytometry using BD FACSDiva software (NJ).
Immunoblotting
Immunoblotting was performed as described previously.11 List of primary antibodies used is given in supplemental Table 2. Densitometry analysis was done by the ImageJ program (version 1.54).
Leukemia and BM coculture experiments
Continuously growing BM-derived stromal cells (BMSCs) from a patient with leukemia were prepared as described previously.13 These cells were plated in 96 well plates (100 cells in 100 μL of medium) and allowed to adhere for 24 hours. Leukemia cells (1 × 104) in 100 μL of medium with enitociclib were added in each well to maintain a ratio of 1:100 stromal cells to leukemia cells. Wells with either the leukemia cells or the stromal cells were used as controls. Alamar blue assay was performed after 72-hour treatment with enitociclib.
Preparation of BM-CM and growth inhibition experiments
The preparation of BM-derived conditioned media (BM-CM) has been described previously.14 To investigate the effect of enitociclib on BM-CM–induced leukemic growth, 1 × 104 leukemia cells in 100 μL of complete medium were plated in a 96-well plate format. About 100 μL of enitociclib in varying concentrations was added to each well with or without 10% BM-CM. Additional control wells were also set up for leukemia cells in culture medium only and leukemia cells with 10% BM-CM. After a 72-hour incubation, Alamar blue assay was used to assess the differences in cell viability.
Drug combinations and synergy quantification
Leukemic cells were plated in 96-well plates and incubated with varying concentrations of chemotherapeutic agents or MI-463, with and without enitociclib (25% inhibitory concentration [IC25]) for 72 hours. To test the combination of venetoclax and enitociclib, leukemic cells were incubated with a constant ratio of the 2 drugs for 24 hours. The synergistic interaction between enitociclib and MI-463 was calculated based on the method of Chou and Talalay by using CompuSyn software (version 1.0).15 Combination index (CI) values of <1 indicates synergy, CI value of 1 indicates additivity, and CI values of >1 indicates antagonism between the 2 drugs.
Statistical analysis
Data were plotted as means ± standard error of the mean using GraphPad Prism 10 software. A P value <.05 was considered statistically significant (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001).
This study was conducted in compliance with the ethical guidelines laid down in the Declaration of Helsinki and appropriate institutional review board approval was obtained -Conjoint Health Research Ethics Board (CHREB), University of Calgary (Approval no: HREBA.CC-16-0286).
Results
Enitociclib decreases cell viability and induces apoptosis in KMT2A-r leukemia cell lines
In the first set of experiments, the sensitivity of KMT2A-r leukemic cells to CDK9 inhibition was evaluated. Cytotoxicity of 3 CDK9 inhibitors were assessed in 2 KMT2A-r cell lines, MV4-11, and KOPN-8 (Figure 1A). KMT2A-r cells were found to be sensitive to all 3 CDK9 inhibitors. However, dinaciclib and alvocidib are classified as pan-CDK inhibitors as they are known to target other members of CDK family.16 Consequently, these off target specificities leads to severe toxicities limiting the clinical use of these agents. Prior studies have demonstrated the enhanced potency of enitociclib over other CDK9 inhibitors.17 Therefore, enitociclib was used as the CDK9 inhibitor for the rest of this study. To further investigate the sensitivity of leukemia cells with KMT2A-r to enitociclib, a panel of leukemia cell lines with and without KMT2A-r were tested (supplemental Table 1). PBMCs and BM-derived CD34+ cells from healthy volunteers were used as controls. Treatment of leukemic cell lines with enitociclib decreased cell viability in a dose-dependent manner in all cell lines and controls (Figure 1B). However, by comparing the IC50 values (supplemental Table 1), cells carrying KMT2A-r were found to be relatively more sensitive to enitociclib treatment than KMT2A-wt cells. Furthermore, the effect of enitociclib on apoptotic cell death was measured by annexin V/propidium iodide flow cytometry assay in representative KMT2A-r (KOPN-8) cell line and PBMCs from healthy donors as controls (Figure 1C). Dose-dependent treatment of cells with enitociclib for 24 hours, increased the percentage of apoptotic cells with ∼48% of cells in the apoptotic stage after 0.1 μM treatment with enitociclib in KOPN-8. However, relative to KOPN-8, minimal difference was observed in healthy PBMCs after 24-hour treatment with enitociclib. Taken together, these results demonstrate the ability of enitociclib to effectively decrease cell viability and induce cell death by apoptosis in KMT2A-r leukemia cells. The antitumor effect of enitociclib was further demonstrated in MV4-11 rat model (Figure 1D). MV4-11 cells were suspended in 0.2 mL of 100% matrigel and were implanted into the flank of female nude rats. Randomization occurred after a tumor size of 35 to 65 mm2 was achieved (14-18 days after inoculation). About 4.5 mg/kg IV of enitociclib was administered once per week for 3 weeks. After 3 weekly administrations, enitociclib achieved complete remission in 10 of 12 animals, lasting until the end of study (day 96).
Enitociclib demonstrates cytotoxic activity in KMT2A-r pediatric leukemia cells. (A) Cytotoxicity of 3 CDK9 inhibitors was assessed in MV4-11 and KOPN-8 cells using Alamar blue assay. (B) Cell viability of various leukemia cell lines, patient samples, and healthy controls after 72-hour treatment with enitociclib were measured by Alamar blue assay. Data represented as percent viability normalized to respective DMSO controls. (C) Enitociclib treatment increased apoptosis in a dose-dependent manner in KOPN-8 cells, as measured by annexin V/PI staining. A minimal effect was observed on healthy PBMCs. (D) Enitociclib demonstrates antileukemic activity in an MV4-11 rat model. PI, propidium iodide; QW, once weekly; SD, standard deviation.
Enitociclib demonstrates cytotoxic activity in KMT2A-r pediatric leukemia cells. (A) Cytotoxicity of 3 CDK9 inhibitors was assessed in MV4-11 and KOPN-8 cells using Alamar blue assay. (B) Cell viability of various leukemia cell lines, patient samples, and healthy controls after 72-hour treatment with enitociclib were measured by Alamar blue assay. Data represented as percent viability normalized to respective DMSO controls. (C) Enitociclib treatment increased apoptosis in a dose-dependent manner in KOPN-8 cells, as measured by annexin V/PI staining. A minimal effect was observed on healthy PBMCs. (D) Enitociclib demonstrates antileukemic activity in an MV4-11 rat model. PI, propidium iodide; QW, once weekly; SD, standard deviation.
Enitociclib disrupts transcriptional elongation and leads to decreased c-MYC and MCL-1 protein levels
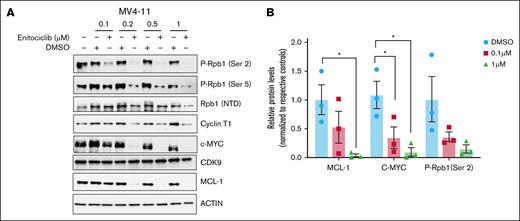
Enitociclib has been shown to be highly selective for CDK9,9 consequently leading to the transcriptional downregulation of an oncodriver c-MYC and antiapoptotic protein MCL-1 in other tumor models.17 Reduction in transcriptional elongation by P-TEFb/CDK9 inhibition is mediated by the loss of phosphorylation of serine residue on position 2 on the CTD of Rpb1, a subunit of RNAP II.5 To confirm the on-target activity of enitociclib in KMT2A-r leukemic cells, MV4-11 cells were treated with varying concentrations of enitociclib for 6 hours. At 0.2 μM, a measurable decrease in phosphorylation at Ser 2 was observed with ∼87% reduction following treatment for 6 hours (Figure 2A-B). Consequently, enitociclib treated cells also exhibited 93% and 95% reduction in protein levels of MCL-1 and c-MYC, respectively, at 0.2 μM after 6-hour treatment. No significant changes were observed in protein levels of CDK9. Together, these results demonstrate the effective on-target activity of enitociclib in KMT2A-r leukemia cells.
Enitociclib has effective on-target activity in KMT2A-r leukemia cells. (A) MV4-11 (KMT2A-r) cells were treated with varying concentrations of enitociclib and respective DMSO control for 6 hours. Western blot was performed to measure changes in Ser 2 phosphorylation of Rpb1, as well as changes in protein levels of MCL-1 and c-MYC after enitociclib treatment. Western blots are representative images of 3 independent biological repeats. (B) Densitometry analysis shows changes in protein levels of MCL-1 and c-MYC as well as changes in phosphorylation levels at Ser 2 of Rpb1 from 3 independent biological experiments. For group comparisons, 1-way analysis of variance test was used. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. NTD, Amino-Terminal Domain; P-Rpb1, Phospho-RNAP II subunit B1.
Enitociclib has effective on-target activity in KMT2A-r leukemia cells. (A) MV4-11 (KMT2A-r) cells were treated with varying concentrations of enitociclib and respective DMSO control for 6 hours. Western blot was performed to measure changes in Ser 2 phosphorylation of Rpb1, as well as changes in protein levels of MCL-1 and c-MYC after enitociclib treatment. Western blots are representative images of 3 independent biological repeats. (B) Densitometry analysis shows changes in protein levels of MCL-1 and c-MYC as well as changes in phosphorylation levels at Ser 2 of Rpb1 from 3 independent biological experiments. For group comparisons, 1-way analysis of variance test was used. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. NTD, Amino-Terminal Domain; P-Rpb1, Phospho-RNAP II subunit B1.
Enitociclib decreases the BM stroma–mediated survival advantage in KMT2A-r leukemic cells
Evidence from multiple studies has shown the ability of BM to support the survival of leukemic cells, consequently contributing to relapse and chemoresistance.18-20 Therefore, this study investigated the potential of enitociclib to reduce the stromal-mediated survival benefit to leukemic cells. Initially, the cytotoxic effects of enitociclib on cell viability were measured when the leukemic cells were cocultured directly in physical contact with BMSCs. Leukemic cells with KMT2A-r were treated with enitociclib for 72 hours in BMSCs cocultures. All KMT2A-r cell lines showed increased viability in a coculture experimental condition. The highest viability increase was observed in MV4-11 cells where stroma increased the cell viability of leukemic cells by ∼120% compared to leukemic cells alone after 72 hours of coculture. Treatment with 125 nM of enitociclib for 72 hours decreased the cell viability of leukemic cells cocultured with stroma by 76% compared to control leukemic cells in coculture (Figure 3A). Similarly, enitociclib was able to reduce BM stromal–mediated growth support compared to controls in all KMT2A-r cell lines tested. BMSCs interact with leukemic blast cells by both physical contact and by secretion of soluble factors.21 Therefore, the next set of experiments tested the cytotoxic effects of enitociclib when leukemic cells were cultured with and without BM stromal cells–derived conditioned media. Previously, different percentages of conditioned media were tested to choose the optimal percentage of BM-CM (10%) that increases the viability of leukemic cells.14 Four KMT2A-r leukemic cells were cultured with and without 10% BM-CM and the cytotoxic effect of enitociclib was monitored. Treatment with 125nM of enitociclib in MV4-11 cells decreased the cell viability of cells cultured in 10% BM-CM by 63% compared to control leukemic cells in condition media (Figure 3B). Collectively, these results show the ability of enitociclib to reduce the survival benefit provided to leukemic cells by the BM microenvironment.
Enitociclib diminishes the BM-derived stromal–mediated survival advantage in KMT2A-r leukemic cells. (A) Four KMT2A-r leukemic cell lines were cultured with and without BMSCs and treated with enitociclib for 72 hours. Data represented as means ± standard error of the mean (SEM) from 3 independent biological experimental replicates. (B) Four KMT2A-r leukemic cell lines were cultured with and without 10% BM-CM and treated with enitociclib for 72 hours. Data represented as means ± SEM from 3 independent biological experimental replicates. For 2 group comparisons, a Student t test was used. A P value <.05 was considered statistically significant. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Enitociclib diminishes the BM-derived stromal–mediated survival advantage in KMT2A-r leukemic cells. (A) Four KMT2A-r leukemic cell lines were cultured with and without BMSCs and treated with enitociclib for 72 hours. Data represented as means ± standard error of the mean (SEM) from 3 independent biological experimental replicates. (B) Four KMT2A-r leukemic cell lines were cultured with and without 10% BM-CM and treated with enitociclib for 72 hours. Data represented as means ± SEM from 3 independent biological experimental replicates. For 2 group comparisons, a Student t test was used. A P value <.05 was considered statistically significant. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Enitociclib enhances the cytotoxic effect of prednisolone in KMT2A-r leukemia cells
The conventional chemotherapy backbone for the treatment of pediatric leukemia consists of various classes of drugs including glucocorticoids, antimetabolites, mitotic inhibitors, and anthracyclines. Therefore, this experiment investigated the effective agents within the chemotherapy backbone that can be combined with enitociclib to achieve enhanced antileukemic effects in patients that carry 11q23 translocations in their leukemic blast cells. The cytotoxic effect of combining low dose (IC25 value) enitociclib in combination with frequently used chemotherapeutic agents for treating childhood leukemia was measured. These agents included cytarabine (antimetabolite), methotrexate (antimetabolite), vincristine (vinca alkaloid), prednisolone (glucocorticoid), and doxorubicin (anthracycline). In MV4-11 cells, enitociclib demonstrated an additive effect with most chemotherapeutic agents but exhibited a synergistic effect with doxorubicin and prednisolone with an overall CI value of 0.9 and 0.5, respectively (Figure 4A-B). Strong synergy is noted with higher doses of prednisolone and doxorubicin. The differences in IC50 values of chemotherapeutic agents alone and in combination with enitociclib are given in Table 1. The most effective synergy was observed between prednisolone and enitociclib, which correlates well with the differences observed in the IC50 values between prednisolone alone (25 μM) and a combination of prednisolone with enitociclib (0.1 μM).
Enitociclib demonstrated effective synergistic activity with doxorubicin and prednisolone in KMT2A-r pediatric leukemia cells. (A) MV4-11 cells were treated with various concentrations of different chemotherapeutic agents, either alone or in combination with low dose (IC25 value) of enitociclib, for 72 hours. Percent cell viability is calculated by normalizing to appropriate controls, and results are presented from 3 independent biological replicates. Data are shown as mean ± SEM. (B) Fa-CI plots were derived by Chou-Talalay method using CompuSyn software, and overall CI values are given for each combination. For 2 group comparisons, a Student t test was used. A P value <.05 was considered statistically significant. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Fa, fraction affected.
Enitociclib demonstrated effective synergistic activity with doxorubicin and prednisolone in KMT2A-r pediatric leukemia cells. (A) MV4-11 cells were treated with various concentrations of different chemotherapeutic agents, either alone or in combination with low dose (IC25 value) of enitociclib, for 72 hours. Percent cell viability is calculated by normalizing to appropriate controls, and results are presented from 3 independent biological replicates. Data are shown as mean ± SEM. (B) Fa-CI plots were derived by Chou-Talalay method using CompuSyn software, and overall CI values are given for each combination. For 2 group comparisons, a Student t test was used. A P value <.05 was considered statistically significant. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Fa, fraction affected.
IC50 values for MV4-11 cells treated with single chemotherapeutic agent alone or in combination with enitociclib
| . | Doxorubicin (μM) . | Prednisolone (μM) . | Methotrexate (μM) . | Cytarabine (μM) . | Vincristine (μM) . |
|---|---|---|---|---|---|
| Single agent | 0.01 | 25 | 0.005 | 0.006 | 0.0004 |
| Single agent + enitociclib | 0.002 | 0.1 | 0.005 | 0.008 | 0.0004 |
| . | Doxorubicin (μM) . | Prednisolone (μM) . | Methotrexate (μM) . | Cytarabine (μM) . | Vincristine (μM) . |
|---|---|---|---|---|---|
| Single agent | 0.01 | 25 | 0.005 | 0.006 | 0.0004 |
| Single agent + enitociclib | 0.002 | 0.1 | 0.005 | 0.008 | 0.0004 |
The drug combination between prednisolone and enitociclib was further evaluated in steroid-sensitive (KOPN-8) and relatively steroid-resistant (INL2) infant KMT2A-r cell lines (supplemental Figure 1). In KOPN-8, the IC50 value of 0.17 μM for prednisolone was reduced to 0.03 μM by a combination of prednisolone with enitociclib. In steroid-resistant INL2 cells, after treatment with 100 μM prednisolone for 72 hours, ∼86% of cells were found to be viable; therefore, IC50 and IC25 values could not be defined for prednisolone as a single agent. However, the IC25 value for the combination of prednisolone and enitociclib was found to decrease to 0.5 μM in INL2 cells. These results suggest the potential of enitociclib to be an effective antileukemic agent to be included in treatment regimens with prednisolone in the chemotherapy backbone for KMT2A-r leukemia.
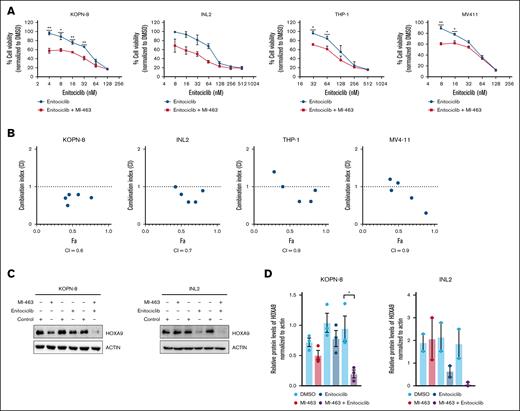
Enitociclib in combination with MI-463 decreases HOXA9 levels in KMT2A-r leukemia cells
Menin, a small protein that binds to the KMT2A protein, tethers the KMT2A complex to the target regions on the DNA.22 Menin inhibitors are designed primarily to target KMT2A-r leukemia cells by disrupting the critical protein interaction between menin, and KMT2A that drives leukemogenesis, primarily by overexpression of HOXA9.22 The results described have demonstrated effective targeting of KMT2A-r leukemic cells by enitociclib. In this set of experiments, the combined effect of simultaneously targeting P-TEFb and menin-KMT2A interaction in KMT2A-r leukemia cells was studied. Supplemental Figure 2 shows the growth inhibition curve for MV4-11, KOPN-8, and THP-1 after treatment with MI-463. The viability response of INL2 with MI-463 treatment has been published previously.11 KMT2A-r cell lines were treated with varying concentrations of enitociclib alone or in combination with a low dose (IC25 value) of MI-463 for 72 hours. Synergy was calculated with the Chou-Talalay method. Synergistic interaction between enitociclib and MI-463 was observed in all KMT2A-r cell lines (Figure 5A-B). Strong synergy was observed in INL2 and KOPN-8 across all drug concentrations. MV4-11 and THP-1 showed strongest synergy at higher drug concentrations. Furthermore, the changes in HOXA9 protein levels after combination treatment relative to individual treatment with the drugs were assessed (Figure 5C). In representative infant KMT2A-r cell lines, HOXA9 protein levels were strongly diminished with ∼75% to 85% decrease after 24-hour treatment with the drug combination relative to the DMSO control (Figure 5C-D). Together, these results suggest that simultaneously targeting the 2 critical interactions (KMT2A-menin and P-TEFb–KMT2A-r complex) in KMT2A-r complex will effectively decrease HOXA9 protein levels in KMT2A-r cells.
Enitociclib synergized with MI-463 in KMT2A-r leukemia cells. (A) KMT2A-r cell lines cells were treated with varying doses of enitociclib, either alone in combination with MI-463 (IC25 value), for 72 hours. Data represented as mean ± SEM from 3 biological repeats. (B) Fa-CI plots were derived by Chou-Talalay method using CompuSyn software, and overall CI values are given for each combination. (C) Western blot shows changes in HOXA9 protein levels in KOPN-8 and INL2 cells treated with enitociclib (IC50 value), MI-463 (IC50 value), or a combination of both the agents for 24 hours. (D) Densitometry analysis shows changes in HOXA9 protein levels from 2 to 3 independent biological experiments. For 2 group comparisons, a Student t test was used. A P value <.05 was considered statistically significant. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Enitociclib synergized with MI-463 in KMT2A-r leukemia cells. (A) KMT2A-r cell lines cells were treated with varying doses of enitociclib, either alone in combination with MI-463 (IC25 value), for 72 hours. Data represented as mean ± SEM from 3 biological repeats. (B) Fa-CI plots were derived by Chou-Talalay method using CompuSyn software, and overall CI values are given for each combination. (C) Western blot shows changes in HOXA9 protein levels in KOPN-8 and INL2 cells treated with enitociclib (IC50 value), MI-463 (IC50 value), or a combination of both the agents for 24 hours. (D) Densitometry analysis shows changes in HOXA9 protein levels from 2 to 3 independent biological experiments. For 2 group comparisons, a Student t test was used. A P value <.05 was considered statistically significant. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Enitociclib potentiates the cytotoxic effect of venetoclax in KMT2A-r leukemia cells that are relatively resistant to venetoclax
Various leukemia subtypes including KMT2A-r leukemic cells have overexpressed B-cell lymphoma 2 (BCL-2).23,24 Venetoclax, a small molecule BCL-2 inhibitor is currently being investigated for the treatment of various BCL-2 overexpressing pediatric hematological malignancies.25 One potential mechanism of venetoclax resistance is the overexpression of MCL-1.25 Because MCL-1 is one of the targets of enitociclib, we investigated if combining enitociclib with venetoclax can reduce the observed relative resistance in venetoclax-resistant KMT2A-r cell line model. We studied the cytotoxic effect of combining enitociclib with venetoclax in both venetoclax-sensitive (MV4-11 and KOPN-8) and relatively resistant (THP-1 and INL2) KMT2A-r leukemia cells in constant ratios for 24 hours (Figure 6A). Increased levels of MCL-1 produced by THP-1 and INL2 support the observed relative resistance to venetoclax (Figure 6B). All 4 KMT2A-r cell lines exhibited enhanced cytotoxicity with the 2-drug combination (Figure 6A). In addition, drug combination led to a marked increase in cell death as observed by increased poly (ADP-ribose) polymerase (PARP) cleavage after 6 and 24 hours of treatment in representative venetoclax-sensitive (MV4-11) and venetoclax-resistant (INL2) cell lines (Figure 6C). A marked decrease in BCL-2 protein levels was also observed in INL2 cell line after treatment with drug combination for 6 hours (Figure 6D). Collectively, these findings demonstrate the ability of enitociclib to potentiate the cytotoxic effect of venetoclax in KMT2A-r leukemia cells as observed by enhanced cell death of both venetoclax-sensitive and -resistant cell line upon combination treatment.
Enitociclib potentiates the cytotoxicity of venetoclax in KMT2A-r leukemia cells. (A) Four KMT2A-r leukemia cell lines were treated with enitociclib, venetoclax, or a combination of both (constant ratios) for 24 hours. (B) Endogenous levels of BCL-2 and MCL-1 are shown in 4 KMT2A-r cell lines. (C) Venetoclax-sensitive (MV4-11) and venetoclax-resistant (INL2) KMT2A-r cell lines were treated with enitociclib (50 nM for MV4-11 and 100 nM for INL2), venetoclax (15 nM for MV4-11 and 5 μM for INL2), or a combination of the 2 drugs for 6 and 24 hours. Levels of cleaved PARP were measured by western blotting. (D) Venetoclax-resistant (INL2) KMT2A-r cell line was treated with enitociclib (100 nM), venetoclax (5 μM), or a combination of the 2 drugs for 6 hours; and protein levels of MCL-1 and BCL-2 were measured by western blotting.
Enitociclib potentiates the cytotoxicity of venetoclax in KMT2A-r leukemia cells. (A) Four KMT2A-r leukemia cell lines were treated with enitociclib, venetoclax, or a combination of both (constant ratios) for 24 hours. (B) Endogenous levels of BCL-2 and MCL-1 are shown in 4 KMT2A-r cell lines. (C) Venetoclax-sensitive (MV4-11) and venetoclax-resistant (INL2) KMT2A-r cell lines were treated with enitociclib (50 nM for MV4-11 and 100 nM for INL2), venetoclax (15 nM for MV4-11 and 5 μM for INL2), or a combination of the 2 drugs for 6 and 24 hours. Levels of cleaved PARP were measured by western blotting. (D) Venetoclax-resistant (INL2) KMT2A-r cell line was treated with enitociclib (100 nM), venetoclax (5 μM), or a combination of the 2 drugs for 6 hours; and protein levels of MCL-1 and BCL-2 were measured by western blotting.
Discussion
KMT2A-r leukemia is characterized by the fusion of the truncated KMT2A protein with various binding partners. These translocations lead to the upregulation of an important transcription factor HOXA9, a negative prognostic factor that affects the overall survival of patients diagnosed with this malignancy.3 Several attempts have been made to design single agent or combination therapies that target the KMT2A-r leukemic cells to downregulate HOXA9. Currently, multiple combination therapies with novel small molecule inhibitors are being evaluated in clinical trials for mostly adult patients diagnosed with KMT2A-r leukemia. However, KMT2A-r leukemia is frequently observed in the infant patient population and treatment of these patients has proven challenging, in part, due to the development of chemoresistance and high relapse rates.3
This study investigated the use of enitociclib, a CDK9 inhibitor, for the treatment of infants diagnosed with KMT2A-r leukemias. It has been shown that KMT2A-r leukemias have deregulated transcriptional elongation.26 Therefore, we hypothesized that KMT2A-r leukemic cells are relatively more sensitive to CDK9 inhibition than control cells. The efficacy of enitociclib as an antitumor agent has been previously studied in multiple hematologic malignancies.10,17,27 The findings from the present study report the preclinical evidence of enitociclib as a potential therapeutic agent for the treatment of KMT2A-r infant leukemia.
In this present study, the first set of experiments tested the sensitivity of KMT2A-r and KMT2A-wt leukemic cells to enitociclib. Enitociclib demonstrated growth inhibition and cell death by apoptosis in KMT2A-r cells. The IC50 values for KMT2A-r cell lines were found to be as low as 30 nM compared to 883 nM for normal PBMCs. A direct comparison of the IC50 values showed that KMT2A-r cells were relatively more sensitive to enitociclib than KMT2A-wt cells as well as PBMCs from healthy donors. As previously demonstrated, CDK9 inhibition by enitociclib promotes cell death by disrupting the assembly of transcriptional machinery and decreasing the level of Ser 2 phosphorylation on the CTD of RNAPⅡ.17 To examine the on-target effect of enitociclib on KMT2A-r leukemic cells, MV4-11 cells were treated with varying concentrations of enitociclib for 6 hours. The results showed that enitociclib has an effective on-target activity in KMT2A-r cells by reducing Ser 2 phosphorylation levels within 6 hours of treatment. Subsequently, a significant reduction in levels of oncogene MYC and antiapoptotic protein MCL-1 was also observed in MV4-11 cells. Additionally, no significant reduction in the protein levels of CDK9 was observed after treatment.
Furthermore, enitociclib also demonstrates the ability to reduce the cell survival support to leukemic cells provided by the BM stromal cells. This was observed when leukemic cells were cultured in both direct contact BM stromal coculture experiments as well as in BM-derived conditioned media supplemented culture conditions. The BM microenvironment can increase survival of leukemia cells by upregulation of signaling pathways and oncogenes like MYC and MCL-1 in leukemia cells.28,29 Additionally, coculturing stromal cells with leukemia cells have demonstrated to increase the total RNA synthesis in the cells.30 Because MCL-1, MYC, and P-Ser2 are potential targets of enitociclib, it could potentially counteract the stroma-mediated survival advantage provided to leukemic cells. The potential of enitociclib to decrease the increased advantage conferred to leukemia cells by the BM microenvironment merits further in-depth investigation.
Additionally, steroid resistance is frequently observed, in ∼30% of infants diagnosed with KMT2A-r ALL.31,32 Among the chemotherapeutic agents tested, enitociclib demonstrated an effective synergistic combination with prednisolone, a constituent in the common chemotherapy backbone. Enitociclib was further tested in combination with prednisolone in both steroid-sensitive and steroid-sensitive KMT2A-r cell lines. In both cases, enitociclib enhances the cytotoxicity in combination with prednisolone. One potential explanation for the drug synergy could be the ability of enitociclib to decrease MCL-1 protein levels, which is highly expressed in steroid-resistant cells.33 Future studies with the potential role of enitociclib in overcoming steroid resistance can uncover the mechanisms and pathways involved in chemoresistance in KMT2A-r cells.
In addition to the combination of enitociclib with chemotherapeutic agents, this study also explored small molecule inhibitors that could more effectively target KMT2A-r leukemic cells. Simultaneous targeting of various critical interactions in the KMT2A-r complex can also be utilized to achieve enhanced and more targeted cytotoxic effects on KMT2A-r leukemic cells.34 The menin-KMT2A inhibitor, MI-463, was studied in combination with enitociclib. Reduction in HOXA9 protein level by MI-463, combined with the ability of enitociclib to downregulate c-MYC and MCL-1, is a feasible mechanism behind the synergistic response seen in KMT2A-r infant leukemic cells under this treatment condition. To further expand the therapeutic potential of enitociclib for the treatment of KMT2A-r leukemia, the potential of combining venetoclax with enitociclib was investigated. It is based on the rationale that the combined effect of enitociclib and venetoclax would lead to enhanced cytotoxicity by targeting both antiapoptotic proteins MCL-1 and BCL-2. Enhanced cytotoxicity was observed in KMT2A-r leukemia cell lines after drug combination treatment. In addition, a decrease in BCL-2 protein levels and enhanced cell death, as measured by PARP activation, were observed after combination treatment observed, especially in venetoclax-resistant cell line (INL2), supporting the combination of enitociclib with venetoclax for the treatment of KMT2A-r leukemia.
Overall, the data presented demonstrated the ability of enitociclib to induce growth inhibition, cell death by apoptosis and exert an on-target effect on the transcription elongation process in KMT2A-r cells. Additionally, the findings showed a synergistic interaction of enitociclib with a menin inhibitor to target KMT2A-r leukemic cells more specifically and effectively. These preclinical findings justify future experiments in xenograft models to assess the efficacy and tolerability of this drug combination. Collectively, our findings support further evaluation of enitociclib in future clinical studies for the treatment of KMT2A-r infant leukemia.
Acknowledgments
This research study was funded in part by the Kids Cancer Care Foundation, Alberta Children’s Hospital Foundation, and a preclinical study research grant from Vincerx Pharma Inc. A.N. laboratory received a pediatric cancer research award from Vincerx Pharma, Inc.
Visual abstract was created with BioRender.com.
Authorship
Contribution: R. Sharma planned and conducted the experiments, analyzed data, interpreted the results, and wrote the manuscript; C.Z. provided the technical expertise in generating data; A.N. contributed to experimental planning, interpretation of data, and reviewed the manuscript; and R. Shah., M.M.F., A.J.J., D.S.M., L.D.M., N.J.L., and A.N. contributed to conceptualization of the overall approach, development, and formulation of the studies.
Conflict-of-interest disclosure: M.M.F. and A.J.J. report employment with Vincerx Pharma, Inc. The remaining authors declare no competing financial interests.
Correspondence: Aru Narendran, Department of Oncology, University of Calgary, 3330 Hospital Dr NW, HMRB 326, Calgary, AB, Canada, T2N 4N1; email: a.narendran@ucalgary.ca.
References
Author notes
Data are available on request from the corresponding author, Aru Narendran (a.narendran@ucalgary.ca).
The full-text version of this article contains a data supplement.