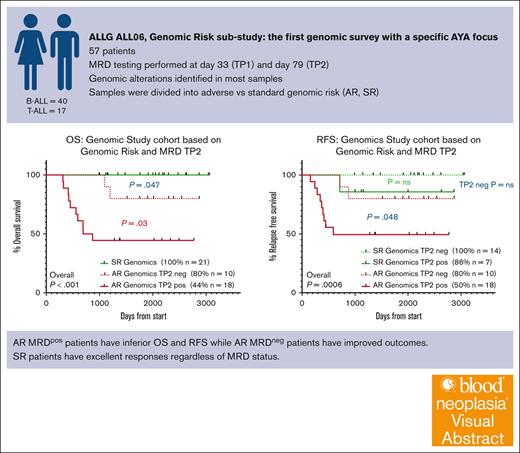
Key Points
One of the first AYA ALL cohorts (aged 15-39 years) treated on uniform intensive chemotherapy to report a detailed genomic description.
Adverse genomic risk lesions correlate with high end-of-induction MRD; both risk factors combined identify a group with high risk of relapse
Visual Abstract
Next-generation sequencing has enabled classification of multiple new recurrent genomic drivers of acute lymphoblastic leukemia (ALL). We aimed to describe the genomic drivers of ALL in an adolescent and young adult (AYA) cohort (ALL06; target age, 15-39 years; recruited age, 16.6-39 years), treated uniformly on a pediatric-inspired protocol (the Australasian Leukaemia and Lymphoma Group ALL06 study). ALL06 assessed the safety and efficacy of adapting a pediatric chemotherapy protocol in older patients. Genomic risk classification of enrolled B-ALL and T-ALL patients was based on multiple assays: messenger RNA sequencing, multiplex ligation-dependent probe amplification, immunophenotyping, and cytogenetics. Using this approach, 36 of 40 (90%) patients with B-ALL and 13 of 17 (76.5%) patients with T-ALL were classified according to genomic risk. A strong correlation existed between adverse genomic risk and minimal residual disease (MRD) at the end of consolidation, translating to inferior overall and relapse free survival. Patients with adverse risk genomics who achieved negative MRD status had improved responses compared with those with persistent MRD. Patients with standard-risk genomics had excellent responses regardless of MRD status. This is the first report of the impact of genomics in an individual cohort of AYA patients treated on a single protocol. These data argue strongly for incorporation of a genomic risk classification into future ALL treatment paradigms at the time of diagnosis, and also for the rigorous assessment of risk assignments in a group of patients who are not children and not older adults. This trial was registered at https://anzctr.org.au/ as #ACTRN12611000814976.
Introduction
Adolescent and young adult (AYA) patients with acute lymphoblastic leukemia (ALL) experience inferior survival and greater toxicity compared with young children, although recent reports highlight improving outcomes.1 The use of optimized chemotherapy schedules, particularly those based on pediatric regimens, has certainly contributed to this improvement. Improved risk stratification through minimal residual disease (MRD) detection to guide risk treatment and transplant decisions, better supportive care, and increased salvage options have also all contributed to improved outcomes for AYA patients with ALL.2-6 Notably, significant advances have been achieved in the diagnosis and subclassification of ALL based on genomic interrogation.7,8 A number of new genomic drivers have been identified by next-generation sequencing (NGS) diagnostics, which allows in-depth, unbiased interrogation of leukemic transcriptomes and genomes. These subtypes include recurrent alterations in the IKZF1 and PAX5 genes (including PAX5 p.P80R); rearrangements involving DUX4, MEF2D, and ZNF384; and the UBTF::ATXN7L3 gene fusion.7-14 In addition to these discrete pathogenic lesions, there are also molecular subtypes, based on global gene expression patterns identified using messenger RNA (mRNA) sequencing (mRNA-seq), such as the ETV6::RUNX1-like or Philadelphia chromosome (Ph)–like subtypes.15,16 In their entirety, these discoveries have provided a more in-depth characterization of disease biology and may facilitate the introduction of current targeted therapies and/or the development of new therapies. NGS also enables more refined disease risk stratification schemas to personalize existing therapies, minimizing the risk of relapse through treatment intensification for high-risk [HR] subtypes, and reducing toxicity in favorable-risk disease through de-escalation of therapy.17
Given that molecular subtypes of ALL vary depending on the age of the cohort studied, the goal of incorporating genomics into risk stratification requires an appreciation of the more common molecular subtypes and correlated outcomes within each specific age cohort. The associated biological behavior and subsequent clinical implications for similar lesions, may also differ between the age groups and treatment used. For AYA ALL, further refinement of stratification schemas along these lines are limited by the current paucity of information on molecular subtypes of ALL and their outcomes in AYA cohorts.18 Currently available data sets come mainly from either pediatric or adult populations, or from cohorts in which AYAs represent a small subset of patients, treated using a mix of protocols. Here, we report, to our knowledge, the first comprehensive genomic examination of a cohort of AYA patients with ALL treated on a single MRD-stratified platform, the Australasian Leukaemia and Lymphoma Group (ALLG) ALL06 study.19
Methods
ALLG-ALL06 was a prospective, single-arm phase 2 study that enrolled 82 patients aged 15 to 39 years, with newly diagnosed Philadelphia-negative ALL. All patients were treated using a standardized combination chemotherapy regimen based on the Australian and New Zealand Children’s Haematology/Oncology Group Study 8 protocol,20 which was modeled on the BFM2000 regimen.21 The ALLG sponsored and registered the trial (ACTRN12611000814976). Details of the protocol assessments, treatment, risk group stratification, and outcomes have been published elsewhere.19 Briefly, MRD testing was performed at day 33 (time point 1 [TP1]) and day 79 (TP2) using real-time quantitative polymerase chain reaction to measure immunoglobulin (IG) or T-cell receptor (TCR) gene rearrangements according to EuroMRD guidelines, as previously described.22 MRD negativity was defined as no amplification with a minimum test sensitivity of 1 × 10−4. Patients were given a trial-specific classification of standard, medium, medium-high, high, and very-high risk based on clinical factors such as presenting white cell count, day-8 prednisolone response, morphological remission status at TP1, and MRD status. This classification then determined chemotherapy intensity and guided the decision to offer an allogeneic stem cell transplant (SCT).19
We now report outcomes correlated with a genomic risk algorithm, independent of the clinical trial risk classification. We assigned patients into standard risk (SR) vs adverse risk (AR) groups based on genomic subtyping at diagnosis, using information from flow cytometry, cytogenetics, mRNA-seq, and multiplex ligation-dependent probe amplification (genomic analyses). This classification was a per-protocol retrospective analysis that had no effect on treatment offered. For this genomic study, 57 patient samples of sufficient quantity and quality were available for analysis. Transcriptomes were generated using TruSeq library preparation or Universal Plus mRNA-seq (NuQuant) and an Illumina NextSeq sequencer, using a poly-A–selected mRNA 75–base-pair paired-end read protocol to a median depth of 69 million reads. Sequencing data were analyzed for detection of gene fusions, single-nucleotide variants, and insertion-deletion mutations, as previously described.23 Briefly, sequencing reads were aligned to the human reference genome (GRCh37) using STAR aligner (version 2.5.3a),24 variant calling was performed by GATK HaplotypeCaller25 and annotated with Annovar.26 Only variants with >10 reads and variant allele frequency of >0.1 were reported; detected variants with an allele frequency of 0.1 to 0.2 were verified within Integrative Genomics Viewer.27 Gene fusions were detected using SOAPfuse version 1.26,28 JAFFA version 1.09,29 and FusionCatcher (fusioncatcher.py0.99.7cbeta).30 Only fusions detected by multiple callers were considered, except fusions involving IGH, which had a lower calling threshold. DUX4r was confirmed by hierarchical clustering using published gene lists,31,32 as previously described.33 Bioinformatic tools developed in-house, RaScALL and ALLSpice,23,34 in addition to manual curation, were used to derive a list of pathologic genomic events. Global transcriptomic patterns were also assessed using T-distributed stochastic neighbor embedding visualizations.35 Multiplex ligation-dependent probe amplification (P202, P335, P383, P327; MRC Holland) was used to confirm focal gene deletions for which a DNA sample was available, according to the manufacturer’s instructions.
ALL06 samples subjected to comprehensive genomic analyses were then given a subclassification, most closely aligned with any identified recurrent drivers of disease (Table 1). These subtypes were based largely on the 2016 World Health Organization (WHO) classification of ALL (contemporaneous at the time of the study), and still closely align with the most recent WHO and International Consensus Criteria for classification of hematological malignancies, since published in 2022.36-38 Each subtype was assigned as either SR or AR, depending on prognostic significance suggested by relevant literature. Cases in which there was >1 driver were deemed AR if any of the drivers were identified as such. Our classification algorithm, including an expanded analysis of DUX4-rearranged cases, is detailed in supplemental Methods. Briefly, adverse-risk cases include rearrangements of ABL1/2, CRLF2, KMT2A, DUX4, MEF2D, MLLT10, NUP214, and NUP98; the UBTF::ATXN7L3 fusion; intrachromosomal amplification of chromosome 21; Early T-cell Precursor (ETP)-ALL immunophenotype; PAX5 alteration; deletions in IKZF1, CDKN2A/B, and PAX5; and hypodiploidy.
Distribution of genomic lesions
| B-ALL AR . | n = 25 . | T-ALL AR . | n = 10 . |
|---|---|---|---|
| CRLF2r∗ | 2 | SET:NUP214 (ETP†) | 2 |
| DUX4r | 7 | NUP214::ABL1 | 1 |
| Hyperdiploid with chr9p del | 1 | NUP98r | 2 |
| Hypodiploid | 1 | KMT2Ar | 1 |
| KMT2Ar | 7 | PICALM::MLLT10 | 1 |
| MEF2Dr | 2 | ETP-ALL | 2 |
| NUP214::ABL1∗ | 1 | CDKN2A/B del | 1 |
| PAX5alt | 1 | ||
| PAX5::JAK2∗ | 1 | ||
| UBTF::ATXN7L3 | 2 | ||
| B-ALL SR | n = 15 | T-ALL SR | n = 7 |
| ETV6::RUNX1 | 1 | Hyperdiploid | 2 |
| Hyperdiploid | 3 | TAL1r | 1 |
| PAX5 p.P80R | 3 | Not classified (NOS)‡ | 4 |
| ZNF384r | 4 | ||
| NOS‡ | 4 | ||
| TOTAL | N = 40 | N = 17 |
| B-ALL AR . | n = 25 . | T-ALL AR . | n = 10 . |
|---|---|---|---|
| CRLF2r∗ | 2 | SET:NUP214 (ETP†) | 2 |
| DUX4r | 7 | NUP214::ABL1 | 1 |
| Hyperdiploid with chr9p del | 1 | NUP98r | 2 |
| Hypodiploid | 1 | KMT2Ar | 1 |
| KMT2Ar | 7 | PICALM::MLLT10 | 1 |
| MEF2Dr | 2 | ETP-ALL | 2 |
| NUP214::ABL1∗ | 1 | CDKN2A/B del | 1 |
| PAX5alt | 1 | ||
| PAX5::JAK2∗ | 1 | ||
| UBTF::ATXN7L3 | 2 | ||
| B-ALL SR | n = 15 | T-ALL SR | n = 7 |
| ETV6::RUNX1 | 1 | Hyperdiploid | 2 |
| Hyperdiploid | 3 | TAL1r | 1 |
| PAX5 p.P80R | 3 | Not classified (NOS)‡ | 4 |
| ZNF384r | 4 | ||
| NOS‡ | 4 | ||
| TOTAL | N = 40 | N = 17 |
See supplemental Table 1 for individual patient listings and detailed genomic alterations.
ETP, Early T-cell Precursor ALL; Ph-like, Philadelphia-like; NOS, not-otherwise-specified.
Ph-like genomic features.
1 patient has ETP-ALL immunophenotype.
NOS genomic classification.
Statistical tests were performed using GraphPad Prism version 9.4.0 (GraphPad Prism Inc). Kaplan-Meier survival analyses were performed, with log rank used to determine statistical differences. The χ2 (Mantel-Cox) and Fisher exact tests were used for categorical variables. Differences were considered statistically significant when the P value was <.05. The ALLG ALL06 treatment protocol was approved by the ethics review board of each of the 15 participating institutions. All patients provided written informed consent for the correlative studies presented here.
Results
Overall outcomes of the genomic study cohort
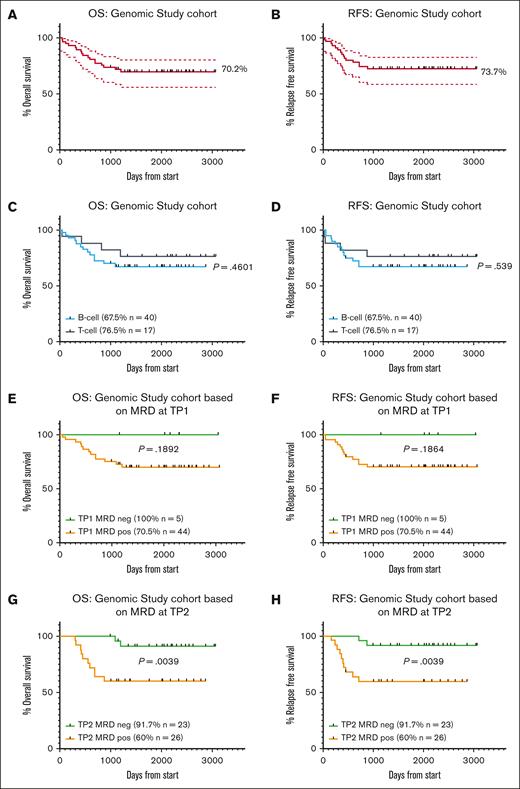
Results of the ALL06 study were previously described.19 Briefly, 82 patients were treated between 2012 and 2018 (complete cohort). At the last report, with a median follow-up of 44 months (1-96 months), the estimated 3-year relapse-free survival (RFS) was 72.8% (95% confidence interval [CI], 62.8-82.7), and the estimated 3-year overall survival (OS) was 74.9% (95% CI, 65.3-84.5). For the genomic study cohort, patients were grouped according to AR vs SR genomic lesions, then by MRD status at TP2 (day 79) and transplant status (Figure 1). The OS and RFS for this genomic study cohort were 70.2% and 73.7%, respectively (95% CI, 59.6-83.7, and 95% CI, 62.8-86.5, respectively; Figure 2A-B) and were not significantly different to results from the complete cohort. Within the genomic study cohort there were 40 patients with B-cell ALL (B-ALL) and 17 patients with T-cell ALL (T-ALL; Table 2; supplemental Table 1). As observed in the complete cohort, patients with T-ALL had a better OS, but this did not reach statistical significance (T-ALL 76.5% vs B-ALL 67.5%; OS, P = .4601; and RFS, P = .539; Figure 2C-D).
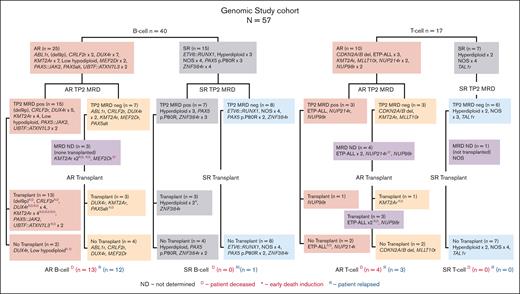
Genomic study cohort CONSORT diagram. The flow diagram shows the genomic lesions identified in patients with B-ALL (n = 40) and those with T-ALL (n = 17) comprising the genomic study cohort. Patients are grouped according to AR vs SR genomic lesions, then by MRD status at TP2 (day 79) and transplant status. Note that 1 patient with T-ALL experienced toxicity and died early in the treatment course (NUP214rD∗) and does not appear in the posttransplant section of the flow diagram. neg, negative; pos, positive.
Genomic study cohort CONSORT diagram. The flow diagram shows the genomic lesions identified in patients with B-ALL (n = 40) and those with T-ALL (n = 17) comprising the genomic study cohort. Patients are grouped according to AR vs SR genomic lesions, then by MRD status at TP2 (day 79) and transplant status. Note that 1 patient with T-ALL experienced toxicity and died early in the treatment course (NUP214rD∗) and does not appear in the posttransplant section of the flow diagram. neg, negative; pos, positive.
Patients who are MRDneg demonstrated a survival advantage compared with patients who are MRDpos in the ALL06 genomic study cohort. Kaplan-Meier curves assessed OS and RFS for (A-B) genomic study cohort (57 patients), (C-D) patients with T-ALL (17 patients) vs B-ALL (40 patients), (E-F) patients who were MRDneg vs MRDpos at TP1 (day 33), (G-H) patients who were MRDneg vs MRDpos at TP2 day 79). Bold lines represent percentage survival, and dotted lines represent 95% CI. Statistical significance was determined by the log-rank survival (Mantel-Cox) test.
Patients who are MRDneg demonstrated a survival advantage compared with patients who are MRDpos in the ALL06 genomic study cohort. Kaplan-Meier curves assessed OS and RFS for (A-B) genomic study cohort (57 patients), (C-D) patients with T-ALL (17 patients) vs B-ALL (40 patients), (E-F) patients who were MRDneg vs MRDpos at TP1 (day 33), (G-H) patients who were MRDneg vs MRDpos at TP2 day 79). Bold lines represent percentage survival, and dotted lines represent 95% CI. Statistical significance was determined by the log-rank survival (Mantel-Cox) test.
The baseline characteristics and genomic risk characterization of the genomic study cohort of the ALL06 study
| . | Patient characteristics . | Protocol risk stratification . |
|---|---|---|
| Total | N = 57 | |
| Age, y; median [range] | 22 [16-38] | |
| Female; n (% of total) | 15 (26%) | |
| BMI (kg/m2); median [range] | 24.8 [15.5-50.6] | |
| ≥30 | n = 10 | |
| <30 | n = 47 | |
| MRD at TP1 | N = 49 | |
| Negative; n (%) | 5 (10%) | |
| Positive; n (%) | 44 (90%) | |
| MRD at TP2 | N = 49 | |
| Negative; n (%) | 24 (49%) | |
| Positive; n (%) | 25 (51%) | |
| B-ALL | N = 40 | |
| Adverse genomic risk; n (%) | 25 (63%) | 16/23 high or very high, 3 medium-high, and 4 standard/medium risk∗ |
| Standard genomic risk; n (%) | 15 (38%) | 2/15 high or very-high, 4 medium-high, and 8 standard/medium risk |
| T-ALL | N = 17 | |
| Adverse genomic risk; n (%) | 10 (59%) | 6/9 high or very-high, 1 medium-high, 2 standard/medium risk† |
| Standard genomic risk; n (%) | 7 (41%) | 6/6 standard or medium risk† |
| . | Patient characteristics . | Protocol risk stratification . |
|---|---|---|
| Total | N = 57 | |
| Age, y; median [range] | 22 [16-38] | |
| Female; n (% of total) | 15 (26%) | |
| BMI (kg/m2); median [range] | 24.8 [15.5-50.6] | |
| ≥30 | n = 10 | |
| <30 | n = 47 | |
| MRD at TP1 | N = 49 | |
| Negative; n (%) | 5 (10%) | |
| Positive; n (%) | 44 (90%) | |
| MRD at TP2 | N = 49 | |
| Negative; n (%) | 24 (49%) | |
| Positive; n (%) | 25 (51%) | |
| B-ALL | N = 40 | |
| Adverse genomic risk; n (%) | 25 (63%) | 16/23 high or very high, 3 medium-high, and 4 standard/medium risk∗ |
| Standard genomic risk; n (%) | 15 (38%) | 2/15 high or very-high, 4 medium-high, and 8 standard/medium risk |
| T-ALL | N = 17 | |
| Adverse genomic risk; n (%) | 10 (59%) | 6/9 high or very-high, 1 medium-high, 2 standard/medium risk† |
| Standard genomic risk; n (%) | 7 (41%) | 6/6 standard or medium risk† |
n = 2 not assigned.
n = 1 not assigned.
Outcomes of the genomic study cohort based on MRD
As previously published for the complete cohort, MRD status at TP1 was associated with survival, but was not statistically significant. In the genomics cohort at TP1, only 5 patients were MRD negative (MRDneg), with 44 patients MRD positive (MRDpos; Table 3). The OS was 100% vs 70.5% in these 2 groups, respectively (P = .1892) and RFS was 100% vs 70.5%, respectively (P = .1864; Figure 2E-F). At TP2, 23 patients were MRDneg, and 26 were MRDpos. The survival differences based on MRD at TP2 were more pronounced. OS (as well as RFS) were 91.7% for patients who were MRDneg vs 60% for those who were MRDpos (P = .0039; Figure 2G-H). Of note, there were 8 patients within the genomic study cohort for whom MRD was not available because of the absence of identifiable IG/TCR gene rearrangements. Of these patients, 5 were patients with T-ALL and, overall, the absence of identifiable IG/TCR gene rearrangements was associated with patients with adverse-risk genomic features (7/8 patients).
MRD at TP1 and TP2, as stratified by genomic risk group
| B-ALL . | MRD at TP1 . | MRD at TP2 . | ||
|---|---|---|---|---|
| . | POS . | NEG . | POS . | NEG . |
| Adverse genomic risk | 22 (96%) | 1 (4%) | 15 (68%) | 7 (32%) |
| Standard genomic risk | 14 (93%) | 1 (7%) | 7 (47%) | 8 (53%) |
| T-ALL | MRD at TP1 | MRD at TP2 | ||
| POS | NEG | POS | NEG | |
| Adverse genomic risk | 5 (100%) | 0 (0%) | 3 (50%) | 3 (50%) |
| Standard genomic risk | 3 (50%) | 3 (50%) | 0 (0%) | 6 (100%) |
| Combined | MRD at TP1 | MRD at TP2 | ||
| POS | NEG | POS | NEG | |
| Adverse genomic risk | 27 (96%) | 1 (4%) | 18 (64%) | 10 (36%) |
| Standard genomic risk | 17 (81%) | 4 (19%) | 7 (33%) | 14 (67%) |
| P = .0002 | P = .045 | |||
| B-ALL . | MRD at TP1 . | MRD at TP2 . | ||
|---|---|---|---|---|
| . | POS . | NEG . | POS . | NEG . |
| Adverse genomic risk | 22 (96%) | 1 (4%) | 15 (68%) | 7 (32%) |
| Standard genomic risk | 14 (93%) | 1 (7%) | 7 (47%) | 8 (53%) |
| T-ALL | MRD at TP1 | MRD at TP2 | ||
| POS | NEG | POS | NEG | |
| Adverse genomic risk | 5 (100%) | 0 (0%) | 3 (50%) | 3 (50%) |
| Standard genomic risk | 3 (50%) | 3 (50%) | 0 (0%) | 6 (100%) |
| Combined | MRD at TP1 | MRD at TP2 | ||
| POS | NEG | POS | NEG | |
| Adverse genomic risk | 27 (96%) | 1 (4%) | 18 (64%) | 10 (36%) |
| Standard genomic risk | 17 (81%) | 4 (19%) | 7 (33%) | 14 (67%) |
| P = .0002 | P = .045 | |||
NEG, MRD negative; POS, MRD positive.
Outcomes of the genomic study cohort based on genomic analyses
Of 57 cases available for genomic analyses (the genomic study cohort), 40 and 17 were B-ALL and T-ALL, respectively. These cases were assigned to 14 different B-ALL, and 9 different T-ALL genomic subtypes (Figure 1; Table 1; supplemental Table 1). Only 8 patients (4 with B-ALL and 4 with T-ALL) had a not-otherwise-specified (NOS) genomic classification.
Of patients with B-ALL, 25 (62.5%) were considered to have AR genomic lesions. These included the recently described UBTF::ATXN7L3 fusion and CDX2 overexpressing subtype8 (n = 2); both of these patients underwent SCT in the first year after diagnosis and, whereas 1 patient remains in remission at last follow-up, the other relapsed and died in the second year. Compared with recent analyses of AYA B-ALL, our cohort was most strikingly characterized by an increased proportion of patients with DUX4r (7/40, 17.5% of B-ALL), KMT2Ar (7/40, 17.5% of B ALL and 1/17, 5.8% T-ALL), and ZNF384r (4/10, 10% of B-ALL). In contrast, we had a reduced proportion of Ph-like ALL, 10% compared with previous reports of up to 27% (Table 1; supplemental Table 1).1
Seven cases were driven by DUX4 rearrangements, which are cytogenetically cryptic.33 Although we acknowledge that some recent reports classify these lesions as having excellent prognosis, we considered them as AR lesions (see supplemental Methods for detailed justification). Of these 7 patients, 2 had SCT, relapsed, and died; the other 5 patients remain alive at last follow-up, including an additional 3 who received transplantation in the first year. Furthermore, 5 of 7 cases were MRD positive at TP2. There were 2 cases that involved CRLF2 rearrangements, 1 of whom died of relapsed disease after SCT. Both patients had features of Ph-like ALL: P2RY8::CRLF2 in the context of intrachromosomal amplification of chromosome 21; and CRLF2r with a concomitant activating CRLF2 p.F232C mutation and IKZF1 deletion.
NUP214::ABL1 was noted in 1 patient with T-ALL who remains alive at last follow-up. Four patients had PAX5-related driver lesions. For the 3 patients with PAX5 p.P80R who, in line with recent evidence, were classified as SR,11,13 all remain alive at last follow-up. One patient with a PAX5alt transcriptome relapsed and died after SCT, and 1 with PAX5::JAK2 remains alive after SCT at last follow-up (both classified as AR). The latter demonstrated an IKZF1 deletion (exons 2-7).
Seven patients with B-ALL had KMT2A rearrangements, of which 6 were KMT2A::AFF1; the remaining patient harbored a KMT2A::MLLT1 gene fusion. All were classified as AR. Of 6 patients with KMT2A::AFF1, 4 underwent SCT and 3 of 4 patients died at a later time point: 2 from relapse and 1 of toxicity. The remaining 2 patients, who did not undergo SCT, died of relapsed disease. The patient with a KMT2A::MLLT1 gene fusion underwent SCT in the first 6 months after diagnosis and remains alive and relapse-free at last follow-up. MEF2D and ZNF384 rearrangements were noted in 2 and 4 patients, respectively. All 4 patients with ZNF384r (classified as SR) remain alive at last follow-up, with 1 undergoing SCT; 1 of 2 patients with MEF2Dr (classified as AR) died of toxicity at day 54 of the ALL06 protocol, with the other patient alive at last follow-up. Based on cytogenetics alone, 1 patient had low hypodiploidy (classified as AR) and relapsed in the first year, 3 patients had high hyperdiploidy (classified as SR), and 1 was hyperdiploid (48,XY with del9p; classified as AR). Of 3 patients with high hyperdiploidy, 2 remain alive (1 after SCT) and 1 relapsed in the second year but remains alive at last follow-up. The patient with del9p relapsed after SCT and died in the second year after diagnosis. One case was driven by the ETV6::RUNX1 fusion (classified as SR) and remains alive whereas 4 cases were considered “B-other” (NOS genomic classification) with no recurrent driver alteration noted. All of these patients were classified as SR and remain alive at last follow-up.
Of 17 patients with T-ALL, 10 (58.8%) were considered to have AR genomic lesions. This included an STK4::KMT2A fusion (patient died in year 3); PICALM::MLLT10 (n = 1, alive); NUP214::ABL1 (n = 1, alive); and NUP98::RAP1GDS1 (n = 2, both alive after SCT). There were also 2 cases with SET::NUP214 gene fusions, of which 1 had an ETP-ALL immunophenotype previously determined to be AR (both patients died, 1 of relapse, the other of toxicity early in the treatment course). There were an additional 2 cases classified as AR based on the immunophenotype of ETP-ALL alone, without recurrent genomic lesions. Both patients underwent SCT, with 1 patient dying of relapsed disease, the other remains alive. Of 7 SR T-ALL cases, 2 had hyperdiploidy (not high hyperdiploidy), and both are alive; 1 had a TAL1 rearrangement, and 4 were the NOS genomic classification (all remain alive).
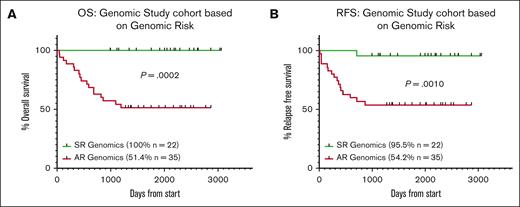
Overall, the OS and RFS of patients with AR genomic lesions were significantly lower than those patients with SR lesions (OS: AR = 51.4% [n = 35] vs SR = 100% [n = 22; P = .0002] and RFS: AR = 54.2% [n = 35] vs SR = 95.5% [n = 22; P = .0010]). Of note, only 1 patient with SR genomic lesions relapsed (remains alive after SCT) on the ALL06 protocol and no others have relapsed or died as of last follow-up (Figure 3A-B).
Patients with SR genomic lesions have significantly better survival outcomes than patients with AR genomic lesions. Kaplan-Meier curves assessed (A) OS and (B) RFS for the total genomic study cohort (57 patients). Statistical significance was determined by the log-rank survival (Mantel-Cox) test.
Patients with SR genomic lesions have significantly better survival outcomes than patients with AR genomic lesions. Kaplan-Meier curves assessed (A) OS and (B) RFS for the total genomic study cohort (57 patients). Statistical significance was determined by the log-rank survival (Mantel-Cox) test.
Role of genomic risk and MRD at TP1 and TP2 on outcome
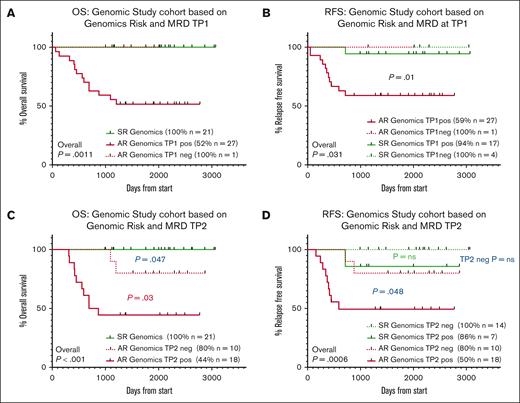
Forty-nine patients had both genomic risk classification and MRD status available for analysis. At TP1 27 of 28 (96.4%) of AR patients were MRDpos and 17 of 21 (81%) of SR patients were MRDpos (P = .15). Patients with AR genomic lesions and TP1 positivity had significantly poorer OS and RFS than those who had SR genomic lesions and any level of MRD positivity (OS: SR/TP1neg, 100% [n = 4] vs SR/TP1pos, 94% [n = 17]); also compared with the 1 AR patient who achieved MRD negativity AR/TP1pos, 59% (n = 27) vs AR/TP1neg, 100% (n = 1), P = .031 (Figure 4A-B).
Patients with AR genomic lesions and MRD positivity have poor outcomes compared with both SR patients and AR patients who achieve MRD negativity. Kaplan-Meier curves assessed (A) OS and (B) RFS for total genomic study cohort patients with TP1 MRD data available (49 patients). (C) OS and (D) RFS for total genomic study cohort patients with TP2 MRD data available (49 patients). Statistical significance was determined by the log-rank survival (Mantel-Cox) test.
Patients with AR genomic lesions and MRD positivity have poor outcomes compared with both SR patients and AR patients who achieve MRD negativity. Kaplan-Meier curves assessed (A) OS and (B) RFS for total genomic study cohort patients with TP1 MRD data available (49 patients). (C) OS and (D) RFS for total genomic study cohort patients with TP2 MRD data available (49 patients). Statistical significance was determined by the log-rank survival (Mantel-Cox) test.
A higher proportion of AR patients, for whom an MRD marker could be determined, demonstrated MRD positivity at TP2 than SR patients (AR: 18/28 (64.3%), SR: 7/21 (33.3%); P = .032). Of 37 patients with B-ALL, 15 of 22 (68.2%) AR patients had residual MRD at TP2 compared with 7 of 15 (46.7%) SR patients (P = .19). In the 12 T-ALL cases, 4 of 6 (66.7%) AR patients had residual MRD at TP2, whereas none of the 6 SR patients had residual MRD (Table 3). Overall, 19 of 26 (73.1%) patients who had TP2 MRD positivity also had AR genomic lesions. In contrast, 7 of 21 (33.4%) patients with SR genomic lesions were TP2 MRDpos. There was a significant difference when OS and RFS were stratified by the combined genomic risk and TP2 MRD status (Figure 4C-D). OS was excellent for patients with SR genomic lesions regardless of their TP2 MRD status (21 patients, 100% survival). For the minority of AR patients who achieved TP2 MRD negativity (10/28 patients, 35.7%), OS was improved compared with those who remained MRDpos (P = .03), although were still inferior to SR patients (80% for AR patients with TP2 MRD negativity, vs 44% who were TP2 MRDpos, P = .047). In contrast, the difference between AR and SR patients with positive TP2 MRD was pronounced with regards to RFS (50% vs 86%, respectively; P = .048). For patients who were TP2 MRDneg, 2 of 10 of those with AR genomic lesions relapsed compared with 0 of 14 patients with SR genomic lesions.
HR block therapy and transplantation in the genomic risk groups
As previously described,19 patients with high MRD were scheduled to receive HR therapeutic blocks (supplemental Table 1). Within the genomics cohort, 32 of 57 (56%) patients proceeded to HR therapy, which included 24 of 29 (83%) eligible patients from the AR genomic group and 8 of 22 (36%) of the SR patients. Within the AR patients, 19 of 24 (79%) proceeded to SCT. HR therapy and SCT were most frequent in the KMT2Ar (5/5 eligible patients with B-ALL) and DUX4r (6/7) subgroups. Within the SR group, HR blocks were most prevalent in the patients with hyperdiploidy (3/3, with 2/3 proceeding to SCT) and ZNF384r (4/4, with 1/4 receiving transplantation).
Genomic study cohort outcomes and BMI
We previously published a correlation between high body mass index (BMI; ≥30 kg/m2) and adverse outcomes in the ALL06 cohort, an observation repeated in this analysis.19 In the genomic study cohort, 11 of 57 patients had a high BMI, of whom 9 had AR genomic lesions (supplemental Figure 4). Patients with AR genomic lesions and high BMI appeared to have poor outcomes, with 71.5% experiencing relapse and 86% dying on protocol.
Discussion
The ALL06 study demonstrated that intensive combination chemotherapy designed for pediatric patients with ALL can be delivered safely in patients up to the age of 40 years. A number of recent publications, such as the UKALLXII/ECOG-ACRIN E2993 study in a predominantly adult cohort,39 and several others in children, have highlighted the importance of genomic data in risk stratification in ALL.40-42 The ALL06 substudy presented here adds significantly to this body of data, and, to our knowledge, for the first time, delineates the significance of genomic risk stratification in an AYA cohort (aged 15-39 years) treated with a single, standardized, pediatric-inspired chemotherapy protocol that incorporated MRD-directed treatment intensification.
Significant advances had been made in the last decade in the understanding of genomic drivers of ALL, assisted by availability of NGS and significantly improved bioinformatic interrogation. Using an optimized workflow, we were able to identify the genomic pathology in all but 4 patients with B-ALL and 4 patients with T-ALL (86% successful genomic classification). In previous decades, cases without a clear genomic driver, referred to as “B-other” or “not-otherwise-specified," would have constituted up to 30% of AYA ALL cohorts. We have demonstrated that when cytogenetic, genomic, and immunophenotypic classifications are applied in combination, driver lesions with defined prognostic significance can be elucidated in the majority of B-ALL cases. This may have contributed to a difference between the relative frequency of various genomic lesions in our cohort vs data reported previously. For instance, DUX4r cases were initially reported as rare in AYAs,11,32 although may have been under-reported because of the difficulty in identifying this subtype, using standard diagnostic techniques, and the initial presence of association with ERG deletions. DUX4 is most commonly rearranged with IGH; these lesions are cytogenetically cryptic and difficult to identify even with NGS because of difficulties in bioinformatic analysis secondary to highly homologous sequences.33 With optimized sequencing and bioinformatics, we were able to identify DUX4r in 17.5% of AYA B-ALL cases, supporting a case to establish this as a standalone entity. Furthermore, risk assignment in age groups of patients with DUX4r is less well defined. These patients are reportedly more likely to have high MRD at end of induction, but outcomes in younger patient cohorts treated with intensive risk-adapted approaches appear to be good.43-46 In this AYA cohort, despite an intensive pediatric-inspired approach, a SCT rate of >70% and a death rate approaching 30% was observed, suggesting that in AYA patients DUX4r ALL is not a SR subtype (see supplemental Data).
In T-ALL, for which risk stratification based on genomic subclassification is not as well defined, we were able to delineate genomic abnormalities in 65% of cases. In the 6 cases of T-ALL without an easily identifiable recurrent genomic driver, iterative analyses against emerging literature may provide an answer in the future.47-49,59
Notwithstanding these limitations, our genomic classification has clear prognostic value. As with other studies, end-of-consolidation MRD was one of the most powerful predictors of outcome in ALL06. Despite universal recognition of the value of MRD stratified therapy,50 relying on MRD response solely for prognostication has one significant disadvantage, this information can only be available after treatment initiation. As such, MRD cannot guide treatment intensification at therapy outset but can only select patients for treatment intensification based on relative treatment insensitivity. Furthermore, in the context of ALL06, early MRD provided only limited information, given that 88% of patients were MRDpos at TP1, and many went on to achieve MRDneg at TP2. Treatment intensification at TP1 for MRD positivity would likely be inappropriate in some of these patients. We have also demonstrated that genomic risk and TP2 MRD response are complementary. In agreement with other studies, AR patients in ALL06 had delayed MRD clearance.51 However, patients with AR genomics who became TP2 MRDneg had significantly better outcomes than patients who remained TP2 MRDpos. This suggests that a more proactive approach to therapeutic escalation may be of value in AR patients. Larger patient numbers are required to be able to explore MRD kinetics and treatment responses of specific genomic subsets. With NGS now becoming more mainstream and bioinformatic pipelines honed to detect recurrent lesions in very short periods of time, it is now highly feasible that genomic information will be available at or before the TP1 (end of induction) MRD is known. Although NGS remains a research tool in many centers, centralized testing is becoming more widely available with several countries now collaborating to form networks to facilitate diagnostic work-up, aiming to have detailed analyses available within 21 days of diagnosis.
Additional work in a larger cohort is required to determine how best to incorporate genomic risk into conventional risk stratification. To date, the majority of treatment protocols rely mainly on cytogenetics and immunophenotype to determine diagnostic subtype in addition to other diagnostic factors such as age, presenting white cell count, and treatment response criteria such as MRD to determine risk stratification. Schemas to integrate genomic information in the context of MRD, such as the pediatric AALL1731 study, featuring a more permissive MRD target for good risk genomic lesions, is relatively uncommon, especially in the AYA setting. None have taken full advantage of the newly discovered genomic subtypes to adapt treatment. This is unsurprising, given the relative lack of genomic information available for AYA disease risk stratification. It may also, in part, reflect significant resource constraints in implementing comprehensive genomic surveys for all ALL diagnoses, and the need to have information in real time to effect treatment decisions. Indeed, the ALL section of the 2022 revision of the WHO classification of hematological neoplasms lists 12 confirmed entities, all based solely on cytogenetics and fluorescence in situ hybridization analyses, except for the newly included BCR::ABL1-like and ETV6::RUNX1-like subtypes.15,52 Additional subtypes, recently described, are classified under the umbrella term “B-ALL with other defined genomic abnormalities” pending additional information regarding the impact on outcome. These include DUX4r, MEF2Dr, ZNF384r, NUTM1r, PAX5alt, and PAX5 p.P80R.9-13,50,51 As with other prognostic markers in ALL, the clinical significance tends to be context dependent. For instance, ETP-ALL was initially discovered as an AR subtype of T-ALL,53,54 and remains so, except in younger patients in whom intensive treatment can be successfully delivered.55 Similarly, the prognostic significance of PAX5 p.P80R may be age dependent, associated with inferior survival in children relative to other subtypes in the same age group, which typically have good prognosis, but is considered a SR marker in adults for whom outcomes are generally less favorable overall.7,9,13
In this and other studies, it is increasingly clear that treatment schemas that apply to children do not equally apply to AYA or adult patients.56,57 Risk stratification will improve with time, but it must be age associated. Furthermore, it may be complicated further by the introduction of Bi-specific T-cell engagers (BiTEs) and chimeric antigen receptor (CAR) T-cells into the front-line armory, given that the responsiveness of different subtypes to immunotherapy may differ from respective chemotherapeutic sensitivities. Future treatment schemas will also have to place increasing importance on patient characteristics, such as BMI. ALL06, similar to other studies, reported an association between body habitus and treatment outcomes, especially in older AYAs.58 Whether this is merely a marker of metabolic syndrome and predictor of treatment toxicity, or indicates a yet unknown association with disease characteristics, is still unclear. Precision approaches, combining all available diagnostic information, and incorporating patient-related factors and MRD status information when available, will optimize ALL survival in a comprehensive approach to improve outcome.
Acknowledgments
The authors thank the patients and their families who participated in ALL06. The authors also thank the following investigators and their institutions for participation in this study: Sushrut Patil and Andrew Wei, The Alfred Hospital, Melbourne; Emma Palfreymann and Philip Crispin, Canberra Hospital, Canberra; Matthew Wright and Julian Cooney, Fiona Stanley Hospital, Perth; Campbell Tiley, Mark Dean and Brenton Wylie, Gosford Hospital, Sydney; Michael Harvey, Liverpool Hospital, Sydney; Devinder Gill, Princess Alexandra Hospital, Brisbane; Susan MacCallum and Mark Hertzberg, Prince of Wales Hospital, Sydney; Agnes Yong, Peter Bardy and Ian Lewis, Royal Adelaide Hospital, Adelaide; Luke Coyle, Keith Fay, Chris Arthur and Ian Kerridge, Royal North Shore Hospital, Sydney; Stephen Larsen, Christina Brown, Liane Khoo, and Harry Iland, Royal Prince Alfred Hospital, Sydney; Bradley Auguston, Dustin Hall, Rebecca Howman, Carolyn Grove and Dejan Radeski, Sir Charles Gairdner Hospital, Perth; Amanda Johnston, Kenneth Micklethwaite, Ian Bilmon, David Gottlieb and Warwick Benson, Westmead Hospital, Sydney; Kim Cartwright, Peter Presgrave and Gurdeep Parmar, Wollongong Hospital, Sydney; Antoinette Anazodo and Anthea Ng, Cancer Centre for Children, The Children’s Hospital at Westmead, Sydney. The authors also acknowledge the help of ALLG coordinators Jenny Nguyen, Lisa Rowley, Amanda Jager and Dan Engeler, Nicola Venn from Childrens’ Cancer Institute of Australia, Chris Frampton from the University of Otago, and Ian Irving from Haematology and Oncology Clinic of Australia.
This work was supported by The Barr Family Foundation, Servier and the Clinical Oncological Society of Australia Youth Cancer Networks Program funded by the Australian Federal Department of Health and Ageing.
The authors acknowledge the contribution of our friend and colleague, the late James D’Rozario, to the successful completion of the ALL06 study.
Authorship
Contribution: D.T.Y., L.N.E., K.B., M.G., and D.L.W. designed and conducted the study, analyzed the data, and wrote the manuscript; J.R. performed bioinformatic analyses; S.L.H., B.J.M., E.C.P., C.E.S., R.S., and M.J.H. performed scientific studies and reviewed the manuscript; M.P.O., T.T., J.K., S.M., and L.D.-P. provided samples and scientific insight, and critically reviewed the manuscript.
Conflict-of-interest disclosure: M.G. has served on advisory boards for Amgen, Pfizer, Servier, and Jazz Pharmaceuticals; and has received trial and research support from Amgen and Servier. D.T.Y. reports honoraria from Pfizer, Novartis, Bristol Myers Squibb, and Amgen. D.L.W. reports honoraria from Amgen. The remaining authors declare no competing financial interests.
Correspondence: Deborah L. White, Precision Cancer Medicine Theme, South Australian Health and Medical Research Institute, North Terrace, Adelaide, SA 5000, Australia; email: deborah.white@sahmri.com.
References
Author notes
D.T.Y. and L.N.E. are joint first authors.
M.G. and D.L.W. are joint senior authors.
RNA-sequencing data have been deposited in the European Genome-Phenome Archive under accession number EGAS50000000752. Additional data used in this study have been deposited into European Genome-Phenome Archive under a previous project (accession number EGAS00001006460).
Data are available on request from the corresponding author, Deborah L. White (deborah.white@sahmri.com).
The full-text version of this article contains a data supplement.