TO THE EDITOR:
Myelodysplastic syndromes/neoplasms (MDSs) are caused by progressive clonal dominance of hematopoietic stem cells acquiring mutations. Although the assessment of mutations in peripheral blood (PB) is an established standard for monitoring patients with other myeloid malignancies, such as leukemia, the sensitivity and precision of screening and monitoring for somatic mutations in the PB is less certain for patients with MDS and related conditions (such as clonal cytopenia of undetermined signficance [CCUS]).1-9 Published guidelines recommend bone marrow (BM) evaluations for the above,10 an invasive procedure not frequently performed multiple times outside of clinical trials and sometimes not even at diagnosis. From a patient perspective, there would be significant benefit in substitution of an intrusive BM examination with the less invasive PB sample to monitor disease evolution and identify potentially actionable mutations or for when next-generation sequencing (NGS) for mutations is omitted at the time of diagnostic BM sampling. Thus, we compared somatic mutations and variant allele frequencies (VAFs) in paired PB and BM samples to assess the reliability of sequencing PB in lieu of BM as a surrogate for monitoring in the National MDS Natural History Study (National MDS Study; ClinicalTrials.gov identifier: NCT02775383) from the United States. The National MDS Study11 is an observational longitudinal cohort study of cytopenic patients suspected of having a diagnosis of MDS. All patients have PB and BM sampled when entering the study at the same time point. Included individuals were required to have at least 1 variant detected in their baseline BM biopsy sample. Patients diagnosed with CCUS (unique to this report) were prioritized for paired PB sequencing. DNA was extracted from bulk PB using an automated FlexSTAR (AutoGen, Holliston, MA) and from BM cell pellets using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). Targeted exon sequencing of 96 genes supplementary table 1 was performed using our research platform on a NovaSeq 6000 at a mean coverage of >1200× and a mean breadth (bases covered at ≥100×) of >99.9%. Reads were aligned against a patched version of the human reference genome build GRCh38 using BWA-MEM.12 VarScan 2 was used to detect single nucleotide variants and short insertions/deletions with a minimum VAF of 2% and 5%, respectively. Paired BM and PB samples from 40 participants were compared at common germ line single nucleotide polymorphism loci to ensure shared identity; samples from 3 participants with >10% differences at germ line loci were assumed to be mismatched and were excluded13. Somatic variants from 53 genes were manually reviewed to retain likely pathogenic variants. Correlation analysis between BM and PB VAFs was performed using Pearson correlation and linear regression. The study received institutional review board approval at all sites and conducted in accordance with the Declaration of Helsinki.
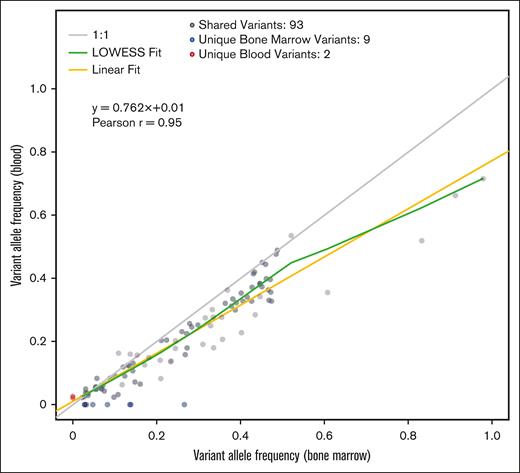
The 37 patients analyzed included 11 (30%) with MDS, 2 (5%) with MDS/MPN, 1 (3%) with acute myeloid leukemia (AML) with <30% blasts, and 23 (62%) with CCUS (Table 1). The median age was 73 years. Only 5 of 37 patients (13%) had circulating PB blasts. Among the 197 total mutations (104 unique variants) identified across all PB and BM samples, the most common were TET2 (28%), SRSF2 (13%), DNMT3A (10%), SF3B1 (8%), and ASXL1 (8%). Most mutations were shared between BM and PB (186/197; 93 mutations found in both PB and BM) resulting in a sensitivity of 91% and positive predictive value of 98% of PB calls when using BM as the gold standard. Two mutations were unique to PB and 9 mutations were unique to BM. The 2 unique PB mutations and diagnoses were (1) ETNK1 (VAF, 2.6%) in a patient with MDS with excess blasts 1 (MDS-EB-1); and (2) CBL (VAF, 2.1%) in a patient with MDS with isolated del(5q). The median VAF was 25% across all mutations, whereas the minimum and median read depth supporting alternate alleles was 40 and 509 reads, respectively (Table 1).
Demographics for total current cohort including patients with CCUS from the National MDS Study
| Demographic characteristic . | All patients (N = 37) . | Patients with CCUS (n = 23) . |
|---|---|---|
| Female, n (%) | 16 (43.2) | 9 (38.8) |
| Male, n (%) | 21 (56.8) | 14 (61.2) |
| Age, median [IQR] (min-max), y | 73.7 [70.6-79.7] (57.2-88.8) | 74.5 [70.85-80.95] (57.2-88.8) |
| Hemoglobin, median [IQR] (min-max), g/dL | 11 [10.1-12.3] (7.5-15) | 11.8 [10.8-12.6] (7.5-15) |
| WBC ×109/L | 4 000 [2 725-5 575] (1 700-13 570) | 4 000 [2 950-5 200] (1 700-10 100) |
| Plts ×109/L | 115 000 [80 500-188 500] (13 000-428 000) | 114 000 [73 500-181 500] (17 000-327 000) |
| VAF %, median [IQR] (min-max) | 25.3 [10.3-41.4] (2.2-97.9) | 21.7 [7.3-42.1] (2.2-97.9) |
| Median alternate allele read depth [IQR] (min-max) | 509 [186-892] (40-2 608) | 549 [151-916] (49-1 760) |
| MDS, n (%) | 11 (29.7) | n/a |
| MDS with excess blasts-2 (MDS-EB2; 10%-19% blasts), n (%) | 2 (18.2) | n/a |
| MDS with excess blasts-1 (MDS-EB1; 5%-9% blasts), n (%) | 3 (27.3) | n/a |
| MDS with multilineage dysplasia and ring sideroblasts (MDS-RSMLD), n (%) | 1 (9.1) | n/a |
| MDS with single lineage dysplasia and ring sideroblasts (MDS-RSSLD), n (%) | 2 (18.2) | n/a |
| MDS with multilineage dysplasia (MDS-MLD), n (%) | 2 (18.2) | n/a |
| MDS with isolated del(5q), n (%) | 1 (9.1) | n/a |
| MDS, unclassifiable (MDS-U), n (%) | 0 (0) | n/a |
| MDS with single lineage dysplasia (MDS-SLD), n (%) | 0 (0) | n/a |
| Number of Patients with MDS and variants, n (%) | 11 (100) | n/a |
| MDS # variants median [IQR] (min-max) | 4 [3-4] (1-5) | n/a |
| MDS VAF % median [IQR] (min-max) | 24.5 [10.9-36.3] (2.7-48.5) | n/a |
| MDS IPSS-R median [IQR] (min-max) | 2 [2-3.8] (1-4) | n/a |
| AML (<30% blasts), n (%) | 1 (2.7) | n/a |
| CCUS, n (%) | 23 (62.2) | n/a |
| MDS/MPN overlap, n (%) | 2 (5.4) | n/a |
| Demographic characteristic . | All patients (N = 37) . | Patients with CCUS (n = 23) . |
|---|---|---|
| Female, n (%) | 16 (43.2) | 9 (38.8) |
| Male, n (%) | 21 (56.8) | 14 (61.2) |
| Age, median [IQR] (min-max), y | 73.7 [70.6-79.7] (57.2-88.8) | 74.5 [70.85-80.95] (57.2-88.8) |
| Hemoglobin, median [IQR] (min-max), g/dL | 11 [10.1-12.3] (7.5-15) | 11.8 [10.8-12.6] (7.5-15) |
| WBC ×109/L | 4 000 [2 725-5 575] (1 700-13 570) | 4 000 [2 950-5 200] (1 700-10 100) |
| Plts ×109/L | 115 000 [80 500-188 500] (13 000-428 000) | 114 000 [73 500-181 500] (17 000-327 000) |
| VAF %, median [IQR] (min-max) | 25.3 [10.3-41.4] (2.2-97.9) | 21.7 [7.3-42.1] (2.2-97.9) |
| Median alternate allele read depth [IQR] (min-max) | 509 [186-892] (40-2 608) | 549 [151-916] (49-1 760) |
| MDS, n (%) | 11 (29.7) | n/a |
| MDS with excess blasts-2 (MDS-EB2; 10%-19% blasts), n (%) | 2 (18.2) | n/a |
| MDS with excess blasts-1 (MDS-EB1; 5%-9% blasts), n (%) | 3 (27.3) | n/a |
| MDS with multilineage dysplasia and ring sideroblasts (MDS-RSMLD), n (%) | 1 (9.1) | n/a |
| MDS with single lineage dysplasia and ring sideroblasts (MDS-RSSLD), n (%) | 2 (18.2) | n/a |
| MDS with multilineage dysplasia (MDS-MLD), n (%) | 2 (18.2) | n/a |
| MDS with isolated del(5q), n (%) | 1 (9.1) | n/a |
| MDS, unclassifiable (MDS-U), n (%) | 0 (0) | n/a |
| MDS with single lineage dysplasia (MDS-SLD), n (%) | 0 (0) | n/a |
| Number of Patients with MDS and variants, n (%) | 11 (100) | n/a |
| MDS # variants median [IQR] (min-max) | 4 [3-4] (1-5) | n/a |
| MDS VAF % median [IQR] (min-max) | 24.5 [10.9-36.3] (2.7-48.5) | n/a |
| MDS IPSS-R median [IQR] (min-max) | 2 [2-3.8] (1-4) | n/a |
| AML (<30% blasts), n (%) | 1 (2.7) | n/a |
| CCUS, n (%) | 23 (62.2) | n/a |
| MDS/MPN overlap, n (%) | 2 (5.4) | n/a |
IPSS-R, International Prognostic Scoring System; IQR, interquartile range; max, maximum; min, minimum; MPN, myeloproliferative neoplasms; n/a, not applicable.
Correlation analyses of the 93 shared mutations between paired PB and BM samples indicated a linear relationship between VAFs detected in PB and BM (Pearson correlation r = 0.95; slope estimate β = 0.762), with an increase in variance observed with increasing VAFs (Figure 1). The deviation from the perfect (gray line) to the observed (yellow line) linear fit indicated that PB VAFs underestimated VAFs in BM by ∼24% overall (Figure 1). For example, a BM VAF of 20% translated to a mean PB VAF of 16% based on the regression fit. A similar trend was found when we repeated the same analysis restricted to only CCUS cases (Pearson r = 0.96; slope estimate x = 0.73). Next, we assessed the relationship between percent of detected BM mutations in PB for varying minimum VAF thresholds. We found that the higher the minimum VAF threshold for BM mutations, the lower the percent of missed mutations in blood. For example, for a minimum of 2%, 10%, and 20% VAFs in BM, the proportions of mutations missed in PB were 8.8%, 3.9%, and 1.7%, respectively.
Scatterplot summarizing the relationship between VAFs for 197 variants (104 unique variants) detected in 37 patients in paired BM and PB samples. The trend lines and Pearson correlation are based on 93 variants that matched between both sample types (gray dots). The blue dots represent variants that were unique to BM (9 variants), and the red dots represent variants unique to PB (2 variants) samples. The yellow line represents the linear regression fit, the gray line represents perfect agreement between VAFs, and the green line represents a locally weighted scatterplot smoothing (LOWESS) trendline.
Scatterplot summarizing the relationship between VAFs for 197 variants (104 unique variants) detected in 37 patients in paired BM and PB samples. The trend lines and Pearson correlation are based on 93 variants that matched between both sample types (gray dots). The blue dots represent variants that were unique to BM (9 variants), and the red dots represent variants unique to PB (2 variants) samples. The yellow line represents the linear regression fit, the gray line represents perfect agreement between VAFs, and the green line represents a locally weighted scatterplot smoothing (LOWESS) trendline.
This real-world prospective population from the National MDS Natural History Study found that PB can be used in most patients to reliably identify somatic mutations in patients with suspected or established MDS and related conditions. The relationship between PB and BM VAFs showed, unsurprisingly, that BM mutations with higher VAFs are more likely to be detected. If the PB contains a mutation, this method of variant detection may be used potentially to complement diagnoses and for monitoring. However, if the mutation is not detected in PB or the VAF is <10%, BM biopsy and testing may be required to identify detectable clonal mutations.
Much of the progress in decreasing cancer morbidity and mortality has come from early detection and prevention efforts. Detection of clonal hematopoiesis (CH), the premalignant state for MDS and AML, offers the potential to identify individuals at greatest risk of developing myeloid malignancies and to shift the paradigm of myeloid malignancy intervention toward prevention.14,15 This is critical, because most patients with myeloid malignancies, the majority of whom are also older,16 succumb to their disease, given the lack of widely available effective treatments.4 Recently, it has become clear that myeloid neoplasms originate with CH, the shared preclinical ancestor.9,10 CH, caused by leukemogenic mutations in hematopoietic cells leading to an expanded clone of blood cells, is a nearly universal feature of aging, and when present these are almost always seen prior to MDS/AML.17 Approximately 10% to 20% of individuals aged ≥70 years have somatic mutations in leukemia drivers detectable at a VAF of ≥2%, with increased testing identifying a larger prevalent population,14,18 particularly given associations with other diseases.19,20 Knowing the accuracy of noninvasive detection and monitoring of mutations is thus of paramount importance, given its widespread use.
In conclusion, patients with cytopenia present diagnostic challenges,21,22 and monitoring recommendations to detect clonal evolution in these patients are not standardized.2,23 Our data confirm similar findings seen in other larger data sets.8,9,24 It is noted that some reports may note higher correlations between PB and BM VAFs most likely due to a higher sensitivity of their NGS analyses. However, we note that sensitivity of NGS for reliably detecting variants with lower VAFs increases with sequencing coverage and different sequencing platforms have different error profiles. Thus, our results need to be interpreted in the context of our sequencing coverage level (1200×) and sequencing technology used (NovaSeq 6000). This coverage does correspond to the average read depth used by most laboratories, allowing for extrapolation of our results to the clinic.
Often, our clinical approaches may be insufficient to capture an event in a way that allows for meaningful intervention before progression or can also be overly prescriptive with frequent marrow biopsies, which are burdensome. NGS technology is now widely available, can reliably be performed on the PB as shown here, and is becoming more affordable. Consequently, testing is often performed as part of the routine evaluation of cytopenia, necessitating guidance to inform about its accuracy, and when a BM biopsy may be needed.25,26 Our results show that although PB underestimates true VAFs and is missing up to 9% of BM variants in the lowest VAF range, it can be used as a surrogate for BM with VAFs ≥10% with a reduced loss of sensitivity (<4%). The linear regression equation we provided in this article may be used to correct PB VAF estimates obtained via sequencing alone. This report has 23 patients with CCUS evaluated, which is distinct from other similar publications. PB NGS analyses can frequently and nearly painlessly be performed in these patients, and thus, rising VAFs could result in earlier indication for performing a second BM biopsy, which then might confirm the diagnosis of MDS at an earlier time point than if one waited for deepening cytopenia. Although this might not result in earlier treatment for a patient, it would allow expectation management for potentially increased disease kinetics. Longer term follow-up of this and similar cohorts will allow for additional clinical guidance.
Acknowledgments: The National Myelodysplastic Syndrome (MDS) Natural History Study has been supported by US federal government contracts (HHSN268201400003I and HHSN268201400002I) from the National Heart, Lung, and Blood Institute (NHLBI) and additional funding by the National Cancer Institute to clinical centers.
This manuscript has been reviewed by the NHLBI MDS study for scientific content and consistency of data interpretation with previous NHLBI MDS study publications.
Contribution: A.E.D., J.B.G., and M.A.S. designed the research, analyzed data, and wrote the manuscript; T.L.J. processed and analyzed data, S.N.S., N.K.G., G.A.A., E.P., H.J.D., T.A.B., J.J.L., R.S.K., S.D.G., W.S., R.B., R.C.L., M.J.W., S.S., and N.D. contributed to study design and interpretation of the data; and all authors contributed to preparation of the manuscript and approved its content.
Conflict-of-interest disclosure: A.E.D. served as chair of a data safety monitoring committee for Geron; holds consultancy roles for Geron, Novartis, and Bristol Myers Squibb (BMS); and holds memberships on boards or advisory committees in Geron, Novartis, BMS, and Takeda. R.C.L. reports consultancy fees from Takeda, bluebird bio, and Thermo Fisher Scientific. R.B. reports employment and stock in a private company, Aptose Biosciences; serves as chair of data safety monitoring committee in Gilead and Epizyme; holds membership on board or advisory committee in Silence Therapeutics; and research funding from Takeda. T.A.B. reports stock in a private companies BMS, AstraZeneca, Epizyme, and Heron Therapeutics; and holds memberships on boards or advisory committees in BMS, MorphoSys, Karyopharm, and Cardinal Health. E.P. reports research funding from BMS and Kura; honoraria from Taiho and Blueprint; and research funding from Incyte. R.S.K. reports honoraria from Novartis, Geron, and Acceleron; and honoraria and speakers bureau fees from Agios, AbbVie, Jazz Pharmaceuticals, and BMS. M.A.S. reports memberships on boards or advisory committees of Novartis, Takeda/Millenium, and BMS. The remaining authors declare no competing financial interests.
Correspondence: Amy E. DeZern, Hematologic Malignancies, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, 1650 Orleans St, CRB1 Room 3M87, Baltimore, MD 21287; email: adezern1@jhmi.edu.
References
Author notes
Data have been submitted to The NHLBI Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC) in compliance with the National Heart, Lung, and Blood Institute data sharing policy. The Resource Request Portal is available to the public at: https://thenationalmdsstudy.net/mds-study-information.
The full-text version of this article contains a data supplement.