Key Points
High CD302 expression is associated with inferior achievement of deep molecular response in patients treated with imatinib and nilotinib.
High CD302 level is associated with enrichment of genes involved in STAT3 pathway, which is associated with tyrosine kinase inhibitor resistance.
Visual Abstract
Achieving a deep molecular response (DMR) is a prerequisite for treatment-free remission in chronic myeloid leukemia (CML) and a key milestone for patients with CML. This study identified patients unlikely to achieve a 5-year DMR through differential expression of cluster of differentiation (CD) genes, and clinical variables at diagnosis. Peripheral blood samples (n = 131) from patients treated with imatinib or nilotinib underwent transcriptomic microarray profiling. The decision-tree analysis delineated 2 distinct poor-risk (PR) cohorts, distinguished by high 3-month BCR::ABL1% (PR-1), or high CD302 expression (PR-2). The 5-years DMR achievement rate was significantly lower in both PR groups than in the good-risk (GR) group in patients treated frontline with imatinib (0% vs 27% vs 83%; P < .0001) or nilotinib (PR-2 vs GR, 17% vs 83%; P = .02). Gene-set enrichment analysis revealed reduced expression of cell cycle–related genes in PR-2, as well as increased metabolism and STAT3 pathway genes, which has previously been linked to leukemic cell persistence and resistance to tyrosine kinase inhibitors. Moreover, PR-2 had a higher frequency of CD34+CD302+ and CD14+CD302+ cells than GR samples. Strategies aimed at targeting STAT3 and/or metabolic pathways associated with high CD302 may provide novel therapeutic approaches that could help improve treatment outcomes and eradicate residual disease.
Introduction
To achieve treatment-free remission (TFR), patients with chronic myeloid leukemia (CML) must first achieve a deep molecular response (DMR). Additionally, patients who successfully achieve DMR rarely experience disease progression and may have a higher probability of survival.1 A number of factors predict for achievement of DMR. For instance, patients who fail to achieve early molecular response (EMR), defined as BCR::ABL1 ≤ 10% international scale (IS) within 3 months of starting treatment, have a lower likelihood of achieving DMR, highlighting the link between EMR, DMR, and TFR eligibility.2,3 EMR is thus a recognized milestone in consensus recommendations for CML management, such as those published by the European LeukemiaNet in 2020.4 Frontline use of higher dose imatinib or more potent tyrosine kinase inhibitors (TKIs) and early achievement of major molecular response (MMR; BCR::ABL1 ≤0.1% IS) are also predictors of DMR.1 However, it should be noted that not all patients who achieve MMR by 12 months will achieve DMR, and identifying additional factors that predict DMR is helpful in patient management. Currently, the association between specific cell surface markers, known as “cluster of differentiation” (CD), and DMR remains unclear. CD markers are an attractive source of potential biomarkers because they are amenable to analysis in most diagnostic laboratories. Studies have previously shown that immunological control may contribute to achievement of molecular responses.5-7 For instance, patients in DMR have restored immune effectors and decreased programmed cell death protein 1 (PD-1) and immune suppressors.5 Therefore, we speculated that the expression of CD markers on circulating cells may serve as a surrogate indicator of the immune milieu and thus be associated with the achievement of DMR. The primary objective of our study is to identify patients with a low probability of achieving DMR at 5 years, using CD markers measured at diagnosis, and identify any associations with known CML biomarkers.
Methods and materials
Patient samples
Blood samples were obtained from patients enrolled in the TIDEL-II trial8 (ACTRN12607000325404). Briefly, patients with chronic phase CML were started on 600 mg imatinib per day. Failure to achieve time-dependent molecular milestones (consistent with optimal targets established in 2013 by the European LeukemiaNet9) led to either an increase in imatinib dose to 800 mg daily or a switch to nilotinib 400 mg twice daily.8 Patients were also switched to nilotinib for imatinib intolerance.8 Mononuclear cells (MNCs) were isolated from peripheral blood collected at diagnosis using density gradient centrifugation. A portion of peripheral blood MNCs were then lysed in TRIzol reagent (Invitrogen, Carlsbad, CA) for RNA stabilization, and the remainder was cryopreserved in RPMI-1640 with 20% fetal bovine serum and 10% dimethyl sulfoxide. Patients with chronic phase CML were started on 300 mg of nilotinib twice daily in the Evaluating Nilotinib Efficacy and Safety in Clinical Trials–Extending Molecular Responses (ENESTxtnd, NCT01254188) study,10 whereas dasatinib 100 mg daily was given in the CML12 DIRECT trial (ACTRN12616000738426), with dose adjustment based on dasatinib trough levels.11,12 Total white cells derived from ENESTxtnd samples were used for microarray profiling. This study was conducted according to the Declaration of Helsinki and approved by all appropriate ethics committees, with written informed consent obtained from all patients.
Variable selection using recursive partitioning and recursive tree (rpart)
A total of 119 diagnostic blood MNC RNA samples from the imatinib-based TIDEL-II trial were subjected to transcriptomic microarray profiling.13 Normalized gene expression was obtained as previously described.13 A total of 394 CD genes classified by the HUGO Gene Nomenclature Committee CD molecular gene group14 were evaluated. However, there were only 357 CD genes available in the microarray. A total of 119 samples were randomly stratified into 2 equal cohorts: the discovery cohort (n = 60) and the validation cohort (n = 59). This stratification ensured that both cohorts had similar rates of DMR achievement. DMR was defined as achieving MR4.5 (BCR::ABL1% [IS] ≤0.0032%) by 5 years. Recursive partitioning trees with 10-fold crossvalidation based on expression of 357 CD genes and 3-months BCR::ABL1% (continuous) were used as variable inputs in the discovery cohort. The tuning parameter for maximum depth was set between 2 and 8. The crossvalidated classification accuracy and κ were used to evaluate the performance of the model in the discovery cohort. The result was further confirmed in the validation cohort. A total of 12 diagnostic total white cell RNA samples from the nilotinib-based ENESTxtnd trial were subjected to transcriptomic microarray profiling.
Flow cytometry
Cryopreserved MNCs were prepared for flow cytometric analysis as previously described,15-17 MNCs (0.5 × 106 to 1 × 106 cells) were stained with fixable viability stain (FSV575V, BD Biosciences) according to the manufacturer's instructions. Cells were labeled with antibodies (BD Biosciences) as described in supplemental Table 1, for 45 minutes at 4°C. Data were acquired on a BD FACSymphony flow cytometer and analyzed using FlowJo version 10.6.1.
Statistical analysis
Previously published biomarkers from our group that associated with molecular response outcome such as plasma cytokines,18 genetic variation,19,20 drug transporters expression,21-23BCR::ABL1 rate of decline measurements,24,25 in vivo kinase inhibition,26 and trough imatinib level21 were used to determine the association with the risk groups. The detailed methods are available in supplemental Methods. The association between the risk groups and 2-group comparison was performed using the Mann-Whitney U test or Student t test for continuous variables and χ2 test for categorical variables. The correlation was performed using Pearson product moment. Analysis and graphical plots were performed using GraphPad Prism 9 (GraphPad Software Inc, La Jolla, CA) or R statistical software version 4.1.1.
Results
CD302 expression and 3-months BCR::ABL1% predicts 5-year DMR achievement in patients treated with imatinib
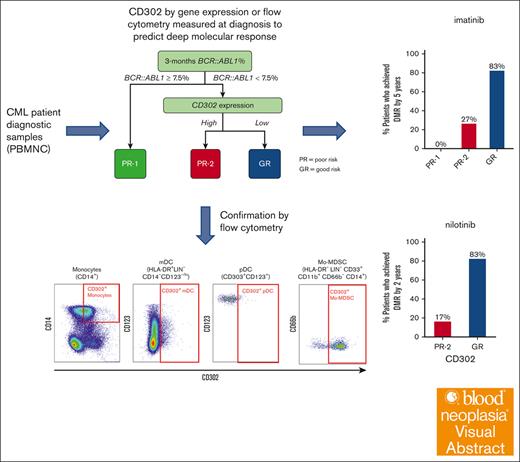
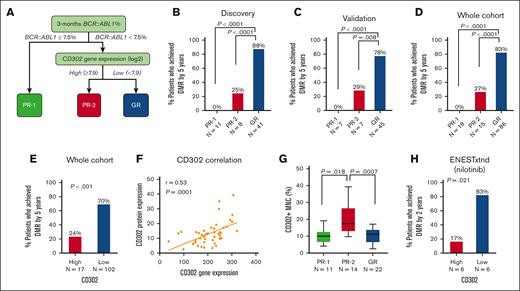
Using the binary rpart method, we performed variable selection to predict DMR achievement using the discovery Table 1 cohort (supplemental Figure 1). CD302 expression and 3-month BCR::ABL1% were identified as the most important variables for predicting DMR achievement. There was no statistically significant correlation between CD302 expression and 3-month BCR::ABL1% (r = 0.01; P = .9). The decision tree analysis identified 3 terminal nodes using the combination of 3-month BCR::ABL1% (Figure 1A). Two distinct poor-risk (PR) groups (PR-1 and PR-2) were identified by the risk model, showing lower probabilities of achieving DMR than the good-risk (GR) group (0% vs 25% vs 88%, P < .0001; Figure 1B).
Patient characteristics in the discovery and validation cohorts
| Variable . | Level . | Discovery . | Validation . |
|---|---|---|---|
| n | 60 | 59 | |
| Median age, y (IQR) | 50.5 (38.3-64.3) | 47.5 (38.0-60.5) | |
| Sex (%) | Female | 27 (45.0) | 20 (33.9) |
| Male | 33 (55.0) | 39 (66.1) | |
| Sokal score (%) | High | 13 (22.8) | 7 (12.7) |
| Intermediate | 19 (33.3) | 19 (34.5) | |
| Low | 25 (43.9) | 29 (52.7) | |
| ELTS score (%) | High | 6 (10.5) | 4 (7.3) |
| Intermediate | 18 (31.6) | 11 (20.0) | |
| Low | 33 (57.9) | 40 (72.7) | |
| BCR::ABL1 transcript type (%) | b2a2 | 26 (43.3) | 21 (35.6) |
| b2a3 | 0 (0.0) | 1 (1.7) | |
| b3a2 | 20 (33.3) | 23 (39.0) | |
| both | 14 (23.3) | 14 (23.7) | |
| EMR status (%) | EMR | 52 (86.7) | 53 (89.8) |
| No EMR | 8 (13.3) | 6 (10.2) | |
| DMR by 5 y (%) | MR4.5 | 38 (63.3) | 37 (62.7) |
| No MR4.5 | 22 (36.7) | 22 (37.3) |
| Variable . | Level . | Discovery . | Validation . |
|---|---|---|---|
| n | 60 | 59 | |
| Median age, y (IQR) | 50.5 (38.3-64.3) | 47.5 (38.0-60.5) | |
| Sex (%) | Female | 27 (45.0) | 20 (33.9) |
| Male | 33 (55.0) | 39 (66.1) | |
| Sokal score (%) | High | 13 (22.8) | 7 (12.7) |
| Intermediate | 19 (33.3) | 19 (34.5) | |
| Low | 25 (43.9) | 29 (52.7) | |
| ELTS score (%) | High | 6 (10.5) | 4 (7.3) |
| Intermediate | 18 (31.6) | 11 (20.0) | |
| Low | 33 (57.9) | 40 (72.7) | |
| BCR::ABL1 transcript type (%) | b2a2 | 26 (43.3) | 21 (35.6) |
| b2a3 | 0 (0.0) | 1 (1.7) | |
| b3a2 | 20 (33.3) | 23 (39.0) | |
| both | 14 (23.3) | 14 (23.7) | |
| EMR status (%) | EMR | 52 (86.7) | 53 (89.8) |
| No EMR | 8 (13.3) | 6 (10.2) | |
| DMR by 5 y (%) | MR4.5 | 38 (63.3) | 37 (62.7) |
| No MR4.5 | 22 (36.7) | 22 (37.3) |
IQR, interquartile range.
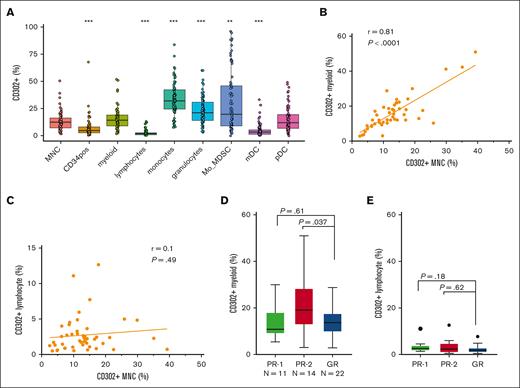
Identification of 2 distinct PR groups defined by 3-months BCR::ABL1% and high CD302 expression. (A) Decision tree selected 2 important variables 3-months BCR::ABL1% and CD302 gene expression to predict DMR (BCR::ABL1<0.0032% by 5 years). (B) Bar plot shows the proportion of patients treated with imatinib in PR-1, PR-2, and GR groups identified by the model using the discovery cohort. (C) Bar plot shows the proportion of patients treated with imatinib PR-1, PR-2, and GR groups identified by the model using the validation cohort. (D) Bar plot shows the proportion of patients treated with imatinib PR-1, PR-2, and GR groups identified by the model using the whole cohort. (E) Bar plot shows the proportion of patients treated with imatinib in high vs low CD302 gene expression using the threshold identified by the model on whole cohort. (F) The correlation between CD302 gene expression and CD302+ protein expression on CD45+ selected MNCs by flow cytometry. (G) Box plot shows the proportion CD302+ MNCs in patients treated with imatinib in PR-1, PR-2, and GR groups. (H) Bar plot shows the proportion of patients treated with nilotinib in high vs low CD302 gene expression based on median cutoff.
Identification of 2 distinct PR groups defined by 3-months BCR::ABL1% and high CD302 expression. (A) Decision tree selected 2 important variables 3-months BCR::ABL1% and CD302 gene expression to predict DMR (BCR::ABL1<0.0032% by 5 years). (B) Bar plot shows the proportion of patients treated with imatinib in PR-1, PR-2, and GR groups identified by the model using the discovery cohort. (C) Bar plot shows the proportion of patients treated with imatinib PR-1, PR-2, and GR groups identified by the model using the validation cohort. (D) Bar plot shows the proportion of patients treated with imatinib PR-1, PR-2, and GR groups identified by the model using the whole cohort. (E) Bar plot shows the proportion of patients treated with imatinib in high vs low CD302 gene expression using the threshold identified by the model on whole cohort. (F) The correlation between CD302 gene expression and CD302+ protein expression on CD45+ selected MNCs by flow cytometry. (G) Box plot shows the proportion CD302+ MNCs in patients treated with imatinib in PR-1, PR-2, and GR groups. (H) Bar plot shows the proportion of patients treated with nilotinib in high vs low CD302 gene expression based on median cutoff.
Using the validation cohort (Table 1), we further confirmed that PR-1 (P < .0001) and PR-2 (P = .008) had statistically significant lower DMR achievement rates than the GR (0% vs 29% vs 78%; Figure 1C-D). The PR-1 (15%) and the PR-2 group (13%) were of similar size. We also demonstrated that CD302 expression alone, without including 3-month BCR::ABL1%, was predictive of DMR (24% vs 70%, P < .001; Figure 1E; supplemental Figure 2).
The PR-1 group was defined as patients with ongoing significant leukemic burden after 3 months of treatment (BCR::ABL1 ≥7.5%), a threshold identified by decision tree as the optimal cutoff for predicting DMR. In contrast, the PR-2 group was identified as having lower leukemic burden at 3 months but higher CD302 gene expression than the PR-1 (P < .01) and the GR (P < .0001) groups in both the discovery and validation cohorts (supplemental Figure 3A-B). There was no statistical difference in 3-month BCR::ABL1% levels between PR-2 and GR in both the discovery (P = .62) and validation cohorts (P = .33; supplemental Figure 3C-D).
Furthermore, to validate the protein expression of the CD302 gene, we performed flow cytometry on CD45+ CML cells. Our analysis revealed significant correlations between CD302 gene expression and CD302+ protein expression on CD45+ CML MNCs (r = 0.53, P = .0001; Figure 1F). Moreover, we observed significantly higher levels of CD302 protein expression on CD45+ CML cells in the PR-2 group compared with both the PR-1 group (P = .018) and the GR group (P = .0007), which corroborated the gene expression findings (Figure 1G). We found that the PR-2 group had significantly higher CD302 protein expression than healthy individuals (normal control; n = 10; P = .0003), but there was no difference in CD302 levels between healthy individuals and PR-1 (P = .31) or GR (P = .48) groups (supplemental Figure 3E). Collectively, these findings provide strong evidence that patients with high CD302 expression exhibit inferior DMR achievement, thus validating the threshold identified by our model.
A comparison between 3-months BCR::ABL1 thresholds 10% and 7.5% for predicting DMR
Overall, 15% of patients (n = 18) had BCR::ABL1 ≥7.5% at 3 months in this cohort. Among these patients, 78% (n = 14) experienced EMR failure (BCR::ABL1 >10% at 3 months), whereas the remaining 22% (n = 4) demonstrated EMR at 3 months but with BCR::ABL1 levels between 7.5% and 10%. Notably, none of these 4 patients achieved DMR by 5 years. It is important to highlight that EMR failure is already recognized as a group with inferior molecular response and clinical outcome.2,3 As such, the primary focus of our study revolves around the patient subgroup denoted as PR-2, characterized by high CD302 expression.
High CD302 expression associated with inferior DMR in patients treated with nilotinib
We proceeded to investigate whether elevated CD302 expression correlates with DMR within our ENESTxtnd cohort,10 consisting of 12 cases with available gene expression profiling data. None of the patients treated with nilotinib in this study failed EMR at 3 months (no patient in the PR-1 group). We observed that high CD302 expression was associated with a lower likelihood of achieving DMR in patients receiving frontline treatment with nilotinib (17% vs 83%, P = .02; Figure 1H). Using flow cytometry, we further validated our findings on a cohort of patients treated with frontline dasatinib. The results from 25 patients treated with dasatinib are similar to those from patients treated with imatinib or nilotinib. High CD302 expression had lower rate of achieving MR4.5 at 24 months than for patients treated with dasatinib with low CD302 expression (42% vs 77%, P = .036; supplemental Figure 4).
Comparison with existing baseline prognostic scoring methods
To assess the predictive performance of CD302 expression alone measured at diagnosis, we conducted a benchmark comparison with 2 commonly used baseline prognostic scores, Sokal4,27 and ELTS.28 We evaluated the performance of CD302 expression alone in predicting DMR and found that it outperformed both the Sokal and ELTS scores across multiple metrics (supplemental Table 2).
The association of CD302 with published CML biomarkers
To gain a deeper understanding of the drivers underlying the PR nature of the PR-2 patient group compared with GR, we examined the association of CD302 with our published CML biomarkers. We conducted genotyping analysis for ASXL1, BIM, and KIR2DL5B common germ line variants that have been associated with molecular response.20 We discovered that PR-2 exhibited a higher prevalence of KIR2DL5BPOS genotyping when compared with GR (P < .001; Table 2; supplemental Figure 5A). However, there was no statistical association observed between CD302 and BCR::ABL1 transcript type29-31 (P = .66), EMR failure–associated plasma cytokines (tumor growth factor α and interleukin 6 [IL-6]18; P = .5), and prognostic scores Sokal (P = .24) and ELTS (P = .51) when comparing PR-2 with GR groups (supplemental Table 3).
Genotyping of ASXL1, BIM, and KIR2DL5B in the risk groups defined by the model
| Gene name . | Level . | Overall . | GR . | PR-1 . | P value (PR-1 vs GR) . | PR-2 . | P value (PR-2 vs GR) . |
|---|---|---|---|---|---|---|---|
| KIR2DL5B (%) | NEG | 71 (79.8) | 57 (87.7) | 11 (73.3) | .16 | 3 (33.3) | <.001 |
| POS | 18 (20.2) | 8 (12.3) | 4 (26.7) | 6 (66.7) | |||
| ASXL1_rs4911231_risk (%) | Good (T/T) | 38 (32.8) | 30 (35.7) | 1 (5.9) | .015 | 7 (46.7) | .419 |
| Poor (C/C or T/C) | 78 (67.2) | 54 (64.3) | 16 (94.1) | 8 (53.3) | |||
| BIM_rs686952_risk (%) | Good (A/A or A/C) | 66 (56.4) | 50 (58.8) | 8 (47.1) | .371 | 8 (53.3) | .691 |
| Poor (C/C) | 51 (43.6) | 35 (41.2) | 9 (52.9) | 7 (46.7) |
| Gene name . | Level . | Overall . | GR . | PR-1 . | P value (PR-1 vs GR) . | PR-2 . | P value (PR-2 vs GR) . |
|---|---|---|---|---|---|---|---|
| KIR2DL5B (%) | NEG | 71 (79.8) | 57 (87.7) | 11 (73.3) | .16 | 3 (33.3) | <.001 |
| POS | 18 (20.2) | 8 (12.3) | 4 (26.7) | 6 (66.7) | |||
| ASXL1_rs4911231_risk (%) | Good (T/T) | 38 (32.8) | 30 (35.7) | 1 (5.9) | .015 | 7 (46.7) | .419 |
| Poor (C/C or T/C) | 78 (67.2) | 54 (64.3) | 16 (94.1) | 8 (53.3) | |||
| BIM_rs686952_risk (%) | Good (A/A or A/C) | 66 (56.4) | 50 (58.8) | 8 (47.1) | .371 | 8 (53.3) | .691 |
| Poor (C/C) | 51 (43.6) | 35 (41.2) | 9 (52.9) | 7 (46.7) |
A/A, A/A genotype; A/C, A/C genotype; C/C, C/C genotype; T/C, T/C genotype; T/T, T/T genotype.
Demographically, we found that patients with PR-2 were predominantly male as compared with patients with GR (P = .035; supplemental Figure 5B; supplemental Table 3). To determine the independent factors predicting DMR achievement, we performed multivariable analysis. Notably, PR-2 (P = .005) was an independent predictive factor for DMR, even after adjusting for sex, KIR2DL5BPOS, and either Sokal or ELTS scores (supplemental Figure 5C; supplemental Table 4).
The dynamics of the molecular response was different between the PR and GR groups
Studies have shown that patients with a longer BCR::ABL1(IS) halving time, corresponding to a slower initial BCR::ABL1% decline from baseline to 3 months, have an inferior clinical outcome and molecular response achievement.3,32 We analyzed the dynamics of these groups in relation to BCR::ABL1%. As anticipated, PR-1 exhibited a slower decline in BCR::ABL1% or longer BCR::ABL1 halving time because of its high BCR::ABL1% levels at 3 months compared with GR (P < .001; Figure 2A-B). During the first 3 months, no notable difference in the rate of decline of BCR::ABL1% was observed between PR-2 and GR (P = .54; Figure 2B). We observed similar results for the CD302 expression alone group (supplemental Figure 6A). There was no statistically significant correlation between CD302 gene expression and halving time (r = 0.04; P = .71) or BCR::ABL1% expression at diagnosis (r = 0.06; P = .55; supplemental Figure 6B-C).
Characteristics of imatinib treated PR-2 group compared with GR. (A) The line plot indicates BCR::ABL1% dynamics for PR-1, PR-2, and GR groups. The median BCR::ABL1% at each time point per group was plotted, and the line was fitted by using generalized additive mode smoothing. The shaded gray area for each line indicates 95% confidence intervals. (B) Box plot shows the 3-months BCR::ABL1% halving time (days) in patients treated with imatinib in PR-1, PR-2, and GR groups. (C) Box plot shows the 6-months BCR::ABL1% halving time (days) in PR-1, PR-2 and GR imatinib treated patient groups. (D) Bar plot shows the proportion of inferior achievement of MMR (BCR::ABL1 <0.1% [IS]) at 12 months in patients treated with imatinib classified by PR-1, PR-2, and GR. (E) Volcano plot shows Hallmark pathways that associated with PR-2 when compared with GR based on gene-set enrichment analysis.
Characteristics of imatinib treated PR-2 group compared with GR. (A) The line plot indicates BCR::ABL1% dynamics for PR-1, PR-2, and GR groups. The median BCR::ABL1% at each time point per group was plotted, and the line was fitted by using generalized additive mode smoothing. The shaded gray area for each line indicates 95% confidence intervals. (B) Box plot shows the 3-months BCR::ABL1% halving time (days) in patients treated with imatinib in PR-1, PR-2, and GR groups. (C) Box plot shows the 6-months BCR::ABL1% halving time (days) in PR-1, PR-2 and GR imatinib treated patient groups. (D) Bar plot shows the proportion of inferior achievement of MMR (BCR::ABL1 <0.1% [IS]) at 12 months in patients treated with imatinib classified by PR-1, PR-2, and GR. (E) Volcano plot shows Hallmark pathways that associated with PR-2 when compared with GR based on gene-set enrichment analysis.
One study has shown that the second decline rate of BCR::ABL1 transcript calculated from 3 months to 6 months was associated with clinical outcome but not molecular response.25 Subsequently, we aimed to determine whether differences in the second decline rate existed between the PR-2 and GR groups based on either the percentage of decrease25 or halving time. Our analysis uncovered a statistically significant difference, indicating a longer halving time of BCR::ABL1% in the PR-2 group than in the GR group from 3 to 6 months (P = .007; Figure 2C). The halving time from 3 to 6 months for PR-1 was higher than GR but did not reach statistical significance (P = .09). This result was consistent with using the second decline rate approach25 demonstrating that PR-2 exhibits a slower decrease than the GR group (P = .012; supplemental Figure 6D).
PR-2 group had inferior MMR achievement rate at 12 months
In addition to exhibiting inferior achievement of DMR, both PR groups demonstrated lower rates of MMR achievement at 12 months than the GR group. Specifically, only 46.7% of patients in the PR-2 group achieved MMR at 12 months, which was significantly lower than the GR group (81.4%, P = .004; Figure 2G). Similarly, only 11.1% of patients achieved MMR at 12 months in the PR-1 group, showing a significant difference compared with the GR group (81.4%, P < .0001; Figure 2D).
PR-2 had increased expression in genes involved in STAT3 pathways
To gain further insights into the deregulated pathways in patients with PR-2 compared with GR, we conducted a gene set enrichment analysis using the MSigDB Hallmark database.33,34 We observed an upregulation of genes associated with the IL-6-JAK-STAT3 pathway in the PR-2 group but decreased expression of genes involved in the IL-2-STAT5 and p53 pathways when compared with the GR group (Figure 2E; supplemental Table 5; supplemental Figure 7A). Additionally, our analysis revealed that patients in the PR-2 group exhibited increased gene expression involved in metabolic pathways (oxidative phosphorylation, fatty acid metabolism, and glycolysis) while showing decreased expression in cell cycle/proliferation pathways when compared with patients in the GR group (Figure 2E; supplemental Table 5; supplemental Figure 7B-C).
PR-2 and GR exhibited similar profiles in TKI transporters and in vivo kinase inhibition
To investigate whether the inferior achievement of DMR in the PR-2 group was attributed to differences in TKI uptake or insufficient kinase inhibition, we examined the gene expression levels of well-known TKI associated transporters including ABCB1, ABCG2, and SLC22A1.35,36 However, we found no statistically significant differences in these genes between the PR-2 and GR groups (P = .35, P = .80, and P = .36, respectively; supplemental Table 6). Furthermore, we evaluated OCT-1 activity22 and ABCB1 fold change,23 and again observed no statistically significant functional differences between PR-2 and GR groups (P = .97 and P = .62, respectively; supplemental Table 6). Additionally, we found no statistically significant difference in plasma imatinib trough levels37 at day 21 between PR-2 and GR groups (1600 vs 1680 ng/mL; P = .77; supplemental Table 6).
Regarding kinase inhibition, we compared in vivo kinase inhibition26 at day 7 between PR-2 and GR groups and found no statistically significant difference (22% vs 26%; P = .82; supplemental Table 6). These findings suggest that the inferior achievement of DMR in the PR-2 group is unlikely to be attributed to variations in TKI uptake or efflux, or to inadequate kinase inhibition.
PR-2 had higher frequency of myeloid cells expressing CD302 than in GR
Next, we investigated the association between CD302 and a range of CD markers. The results indicated a positive correlation (r > 0.6; false discovery rate, P < .05; supplemental Figure 8) between CD302 and 9 CD markers including a known myeloid marker CD33. Conversely, a negative association (r > −0.6; false discovery rate, P < .05) was observed for TNFRSF4 (CD134). Additional information regarding the function and expression of these CD markers in immune subsets is elaborated in supplemental Table 7.
Next, CD302 protein expression was assessed across CML cell populations: MNC (CD45+), myeloid cells (CD33+CD11b+), CD34+ stem/progenitor cells, lymphocytes (CD33−CD11b−CD3+/CD19+/CD56+), granulocytes (CD33+CD11b+CD15+), myeloid dendritic cells (mDCs: HLA-DR+LIN−CD14−CD123−/low), plasmacytoid DCs (pDCs: CD123+CD303+), monocytic myeloid-derived suppressor cells (Mo-MDSC: HLA-DR−LIN−CD33+CD11b+CD66b−CD14+), and monocytes (CD14+). Notably, among these populations assessed, monocytic cells (monocytes, Mo-MDSC) showed the highest expression of CD302 (Figure 3A).
CD302 expressed predominantly in myeloid than lymphoid compartment. (A) The landscape of CD302+ protein expression across stem and progenitor cells and myeloid and lymphoid cells by flow cytometry. (B) The correlation between CD302+CD45+ selected MNCs and CD302+ myeloid cells (CD33+CD11b+). (C) The correlation between CD302+CD45+ selected MNCs and CD302+ lymphoid cells (CD33−CD11b−CD3+/CD19/+CD56+). (D) The box plot shows the frequency of CD302+ on myeloid cells by flow cytometry in PR (PR-1 and PR-2) and GR groups. (E) The box plot shows the frequency of CD302+ on lymphoid cells by flow cytometry in PR-1, PR-2, and GR groups.
CD302 expressed predominantly in myeloid than lymphoid compartment. (A) The landscape of CD302+ protein expression across stem and progenitor cells and myeloid and lymphoid cells by flow cytometry. (B) The correlation between CD302+CD45+ selected MNCs and CD302+ myeloid cells (CD33+CD11b+). (C) The correlation between CD302+CD45+ selected MNCs and CD302+ lymphoid cells (CD33−CD11b−CD3+/CD19/+CD56+). (D) The box plot shows the frequency of CD302+ on myeloid cells by flow cytometry in PR (PR-1 and PR-2) and GR groups. (E) The box plot shows the frequency of CD302+ on lymphoid cells by flow cytometry in PR-1, PR-2, and GR groups.
There was a statistically significant correlation between CD302 protein expression in CML CD45+ cells and CD302+ CD33+CD11b+ myeloid cells (r = 0.81, P < .0001; Figure 3B) but not on CD3+ T lymphocytes (r = 0.1, P = .49; Figure 3C). A similar correlation was observed when comparing CD302 gene expression in myeloid (r = 0.39, P = .007) and lymphoid cells (r = 0.03; P = .82; supplemental Figure 9A-B). Furthermore, PR-2 exhibited a higher frequency of myeloid cells expressing CD302 when compared with the GR group (P = .037; Figure 3D). Importantly, this increase in CD302 expression was not attributed to higher myeloid absolute counts in the PR-2 group (P = .39; supplemental Figure 10A). Conversely, there were no significant differences in either the frequency of lymphocytes expressing CD302 (P = .62; Figure 3E) or their absolute counts (P = .57; supplemental Figure 10B) between the PR-2 and GR groups.
PR-2 had a higher frequency of CD34+CD302+ and CD14+CD302+ cell populations compared with GR
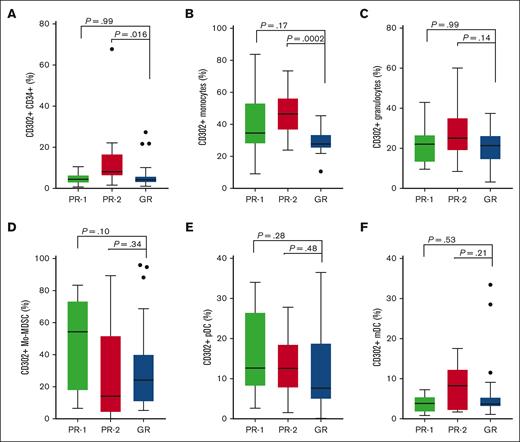
Our next objective was to compare the protein expression of CD302 in the PR-2 and GR groups across different cell populations, including Mo-MDSCs, monocytes, CD34+, pDCs, mDCs, and granulocytes, using flow cytometry. An example of gating strategy is shown in supplemental Figure 11. Our analysis revealed statistically significant higher frequencies of CD302 expressing CD14+ monocytes (P = .0002) and CD34+ cells (P = .016) in the PR-2 group compared with the GR group (Figure 4A-B). The observed increase in CD302 expression in CD34+ cells and monocytes was not because of a rise in absolute CD34+ cell counts (P = .13) or monocyte counts (P = .33) when comparing PR-2 with GR (supplemental Figure 12). There were no statistically significant differences in other populations including granulocytes (P = .14), Mo-MDSCs (P = .34), pDCs (P =.48), and mDCs (P = .21) when comparing PR-2 and GR groups (Figure 4C-F). Next, we assessed PD-1 expression as a surrogate marker for immune exhaustion on immune effector cells by flow cytometry and observed a correlation between the frequency of CD302+CD45+ MNCs and PD-1+CD3+ T cells (r = 0.51, P = .0001), PD-1+CD8+ T cells (r = 0.33, P = .015), and PD-1+CD4+ T cells (r = 0.35, P = .01; supplemental Figure 13)
PR-2 had higher expression of CD302+ on CD14+ and CD34+ cell populations. CD302+ protein expression was assessed by flow cytometry. The box plots show the proportion of CD302+ in PR (PR-1 and PR-2) and GR groups for (A) stem and progenitor cells (CD34+), (B) monocytes (CD14+), (C) granulocytes (CD33+CD11b+CD15+), (D) Mo-MDSCs (HLA-DR−LIN−CD33+CD11b+CD66b−CD14+), (E) pDCs (CD123+CD303+), and (F) mDCs (HLA-DR+LIN−CD14−CD123−/low).
PR-2 had higher expression of CD302+ on CD14+ and CD34+ cell populations. CD302+ protein expression was assessed by flow cytometry. The box plots show the proportion of CD302+ in PR (PR-1 and PR-2) and GR groups for (A) stem and progenitor cells (CD34+), (B) monocytes (CD14+), (C) granulocytes (CD33+CD11b+CD15+), (D) Mo-MDSCs (HLA-DR−LIN−CD33+CD11b+CD66b−CD14+), (E) pDCs (CD123+CD303+), and (F) mDCs (HLA-DR+LIN−CD14−CD123−/low).
Discussion
Achieving a DMR is a clinically significant goal for patients with CML.38 DMR reflects a deeper and more complete suppression of leukemic cells, indicating a favorable treatment response.1 Studies have suggested that patients who achieve a sustained DMR have improved long-term outcomes in terms of overall survival and disease progression.1 Although DMR itself may not be a direct predictor of survival, it serves as a marker for a deeper treatment response associated with better disease control and reduced risk of progression to advanced stages of CML.
Moreover, achieving a sustained DMR allows some patients to discontinue treatment, which can alleviate the burden of long-term TKI use and its associated side effects. In a recent meta-analysis involving 12 trials focused on TKI discontinuation, trials with a molecular criterion of MR4.5 or better for selecting patients for TKI discontinuation had higher rates of TFR at 24 months than those with a criterion of MR4 (57.2% vs 50.5%), respectively.39
We identified 2 distinct groups with a lower probability of attaining DMR: PR-1 and PR-2, defined respectively by 3-months BCR::ABL1% value and CD302 gene expression. Failure to achieve EMR (BCR::ABL1 ≤10% at 3 months) has long been known to correlate with failure to achieve DMR,40 and this relationship was confirmed in the PR-1 group. Interestingly, we found that another group of patients, PR-2, may very well have achieved their early targets but may still fail to achieve DMR. These patients are identified by high CD302 expression. Previous CML studies41-43 investigating TKI resistance did not identify CD302 as a differentially expressed gene when comparing nonresponders with responders, defined by the achievement of complete cytogenetic response at 12-months. This observation is in line with our results because the PR-2 subgroup with high CD302 expression falls within the subset of patients who achieved complete cytogenetic response but not deeper responses. Additionally, we demonstrated that patients with BCR::ABL1 ≥7.5% at 3 months exhibited low CD302 expression similar to that exhibited by the GR group.
CD302 is a C-type lectin receptor involved in cell adhesion and migration. It is expressed in myeloid populations as well as in blasts and leukemic stem cells (LSC) in acute myeloid leukemia.44 Further analysis indicated that this increased expression in our cohort was largely confined to CD33+ myeloid cells, specifically to monocytes and CD34+ cells. The role of CD302 on monocytes, immune cells, and CD34+ populations remain to be defined. Investigating the interplay between CD302 expression and CD34+ as well as CD14+ monocytic cells in the context of CML persistence would provide valuable insights into the underlying mechanisms and potential therapeutic targets.
We observed an enrichment of KIR2DL5BPOS in patients with high CD302 expression. KIR2DL5B encodes an inhibitory “orphan” KIR (killer-cell immunoglobulin-like receptor) whose ligand remains unknown.45 However, its absence may enhance the efficiency of natural killer cell (NK)-mediated killing of LSC. The expression of KIR2DL5 has been described in CD8+ T cells and a CD56dim NK subset.46 In CML, KIRD2DL5A and KIRD2DL5B genotype has been associated with inferior molecular response with upfront TKI treatment, and delayed second deep molecular remission on TKI rechallenge after molecular relapse on a trial of stopping TKI.19,47-49 This suggests that a proportion of patients with PR-2 with KIR2DL5BPOS may have inefficient NK-mediated killing of leukemic cells.
Sopper et al demonstrated that a high level of CD62L-expressing CD4+ T cells correlates with a significantly higher rate of achieving MR4 and MR4.5 than lower levels of CD62L in patients treated with nilotinib.50 Regarding soluble CD62L (sCD62L) measured in plasma, there was a significantly higher proportion (56%) of patients with low sCD62L who achieved MR4 at month 18 than only 11% of patients with high sCD62L levels treated with nilotinib.50 This observation is further supported by data from a cohort of patients treated with imatinib, in which 62% of those with low sCD62L achieved MR4 compared with only 32% of those with high sCD62L.50 In our study, high CD302 expression was identified in patients who achieved EMR but failed to achieve MR4.5. Nevertheless, further investigation is needed to determine whether assessment of both CD302 and CD62L could significantly enhance DMR prediction in CML.
Based on gene-set enrichment analysis,33 our study revealed an increased expression of genes involved in the STAT3 signaling pathway in PR-2 compared with GR. Several studies have shown that persistent activation of STAT3 signaling can contribute to the survival and persistence of leukemic cells as well as treatment resistance.51-54 For instance, in a subset of LSCs, cell-extrinsic STAT3 activation upon Bcr-Abl inhibition was demonstrated, with JAK1 identified as the key kinase responsible for this activation.51 Combined inhibition of Bcr-Abl and JAK1 significantly induced apoptosis even in quiescent LSCs. This finding suggests stem cell persistence in CML is mediated by extrinsically activated JAK1-STAT3 signaling.51 Furthermore, in TKI-persistent CML cells, STAT3 activation promotes metabolic reprogramming, leading to increased reliance on glycolysis over oxidative phosphorylation.52 Targeting STAT3 or its downstream metabolic pathways, such as glycolysis, has emerged as a promising strategy to sensitize TKI-persistent CML cells to treatment and potentially eradicate them.52 This provides critical insight into alternative therapeutic targets for eliminating TKI-resistant LSCs. Eiring et al showed that activation of STAT3 was crucial in BCR::ABL1 kinase-independent TKI resistance.54 In primary cells from patients with CML with BCR::ABL1 kinase-independent TKI resistance, BP-5-087, a potent STAT3 SH2 domain inhibitor, restored TKI sensitivity suggesting its potential clinical utility in CML.
Strategies aimed at targeting STAT3 pathways associated with high CD302 levels may provide novel therapeutic approaches in CML. Studies have shown that inhibition of STAT3 prevented CML cells from developing TKI persistence, and sensitized the persistent CML cells to TKI treatment.51,52 These approaches have the potential to restore immune surveillance, aid in eliminating minimal residual disease, and enhance treatment outcomes.
Limitations of our study were the small sample size in the group treated with TKIs and a lack of confirmatory functional analysis establishing the link between CD302 expression and achieving DMR through STAT3 signaling. Additionally, using single-cell RNA sequencing would be advantageous for comprehensively characterizing the temporal dynamics of CD302 expression after TKI treatment, particularly across various immune cell populations, to identify specific cell types wherein CD302 expression persists despite TKI therapy. Despite limitations, CD302 shows promise as a predictive biomarker for achieving DMR after TKI treatment, thereby offering a promising avenue for personalized therapeutic strategies, particularly in patients with good initial molecular responses who do not achieve DMR with current approaches.
Acknowledgments
T.P.H. received support from the National Health and Medical Research Council of Australia (APP1135949) and has the financial support of Cancer Council SA’s Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health. The original TIDEL-II study was sponsored by the Australasian Leukaemia and Lymphoma Group. This project is supported by Christoford Bequest (HSCGB05-20). N.S. received scholarship funding from the Royal Adelaide Hospital Research Foundation Dawes Scholarship. S.B. received support from the National Health and Medical Research Council of Australia (APP1117718, APP1138935, and APP1027531), from the Ray and Shirl Norman Research Trust and the Cancer Council SA’s Beat Cancer Project on behalf of its donors, and the State Government of South Australia through the Department of Health. D.Y. is a clinical investigator of the Cancer Council SA’s Beat Cancer Project.
Authorship
Contribution: C.H.K. contributed key concepts and methodology, designed and planned data analysis, analyzed data, interpreted data, and wrote the manuscript; Y.I. and J.C. performed experiments and critically reviewed the manuscript; V.S. and P.D. performed sample processing and critically reviewed the manuscript; N.S. and S.B. provided clinical information and critically reviewed the manuscript; D.Y. designed and conducted the TIDEL-II trial and coordinated the correlative studies, interpreted data, and critically reviewed the manuscript; A.S.M.Y. contributed key concepts and methodology, interpreted data, and assisted in writing the manuscript; and T.P.H. designed and conducted the TIDEL-II and ENESTxtnd trials and coordinated the correlative studies, contributed key concepts and methodology, interpreted data, and assisted in writing the manuscript.
Conflict-of-interest disclosure: N.S. received research funding from Novartis, and honoraria from Takeda. S.B. is a member of the advisory boards of Qiagen and Cepheid; received honoraria from Qiagen, Novartis, and Cepheid; and received research funding from Novartis and Cepheid. D.Y. received research funding from Novartis and Bristol Myers Squibb; and received honoraria from Novartis, Takeda, Pfizer, and Amgen. A.S.M.Y. is a member of the advisory board of Novartis; received research funding from Novartis, Bristol Myers Squibb, and Celgene; and received honoraria from Novartis and Bristol Myers Squibb. T.P.H. received research funding from Novartis and Bristol Myers Squibb; and received honoraria from Novartis, Bristol Myers Squibb, and Takeda. The remaining authors declare no competing financial interests.
Correspondence: Timothy P. Hughes, Precision Cancer Medicine Theme, South Australian Health & Medical Research Institute, North Terrace, Adelaide, SA 5000, Australia; email: tim.hughes@sahmri.com.
References
Author notes
Microarray data are available at Gene Expression Omnibus under accession number GSE130404.
The data that support the findings of this study are available upon reasonable request, from the corresponding author, Timothy P. Hughes (tim.hughes@sahmri.com).
The full-text version of this article contains a data supplement.

![Characteristics of imatinib treated PR-2 group compared with GR. (A) The line plot indicates BCR::ABL1% dynamics for PR-1, PR-2, and GR groups. The median BCR::ABL1% at each time point per group was plotted, and the line was fitted by using generalized additive mode smoothing. The shaded gray area for each line indicates 95% confidence intervals. (B) Box plot shows the 3-months BCR::ABL1% halving time (days) in patients treated with imatinib in PR-1, PR-2, and GR groups. (C) Box plot shows the 6-months BCR::ABL1% halving time (days) in PR-1, PR-2 and GR imatinib treated patient groups. (D) Bar plot shows the proportion of inferior achievement of MMR (BCR::ABL1 <0.1% [IS]) at 12 months in patients treated with imatinib classified by PR-1, PR-2, and GR. (E) Volcano plot shows Hallmark pathways that associated with PR-2 when compared with GR based on gene-set enrichment analysis.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodneoplasia/1/2/10.1016_j.bneo.2024.100014/2/m_bneo_neo-2023-000125-gr2.jpeg?Expires=1767746065&Signature=Iw1oCApuZWCy5Mq9vOPVuFMrtoqmsqnVfLzDRLivDEVdJhwWgG-20aTmSRUa0gGoQWKMMqlu~SHBDgxJyTzqUTx8mgQfMr9ZJ2DM2z3UUn9coNs92UI-WT2VNhRGHk0sz9HN199zNd5ZTYgFCx-nk0kGBopCHqaB3ueODLZrJeGk-bWVEIHg9sCfhF3KRrWdTR5TkaqokxCK4amNJBYuTtcTQPbYqYnEk8T-8kZCupNcOxXNH-z17QnASSZx4ZmmIYdJAwyVUWZMK6zgTpGQXEbudYJzkQp07z0pSh-93XI10gnIsjrhYFKzg3i9eZStKmCqlcxnN5hz3XKLAolrlw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)