Skip Nav Destination
Close Modal
Update search
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
NARROW
Publications
Format
Subjects
Article Type
Topics
Date
Availability
1-20 of 23861
Follow your search
Access your saved searches in your account
Would you like to receive an alert when new items match your search?
1
Sort by
Journal Articles
A new sUSPect in T-ALL risk stratification
Open Access
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6467–6468.
Published: 2025
Journal Articles
Progressive NK-cell dysfunction and ILC imbalance favor immune evasion in multiple myeloma
Open AccessMaria Teresa Bilotta, Antonio Giovanni Solimando, Vanessa Desantis, Lucia Di Marzo, Andrea Sabatini, Sergio Forcelloni, Alessandro Andriano, Arcangelo Morizio, Antonella Argentiero, Roberto Ria, Linda Quatrini, Lorenzo Moretta, Paola Vacca, Nicola Tumino
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6246–6251.
Published: 2025
Includes: Supplemental data
Images
in Progressive NK-cell dysfunction and ILC imbalance favor immune evasion in multiple myeloma
> Blood Advances
Published: 2025
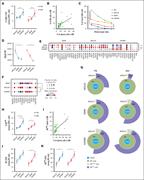
Figure 1. NK cells in MM progression. (A) The plot represents the percentages of NK cells (CD56 + CD3 – ) among CD45 + cells expressed as median values ± standard error of the mean (SEM) in PB and BM of patients with MM among the different stages (MGUS, SMM, and NDMM). A Mann-Whitney t test w... More about this image found in NK cells in MM progression. (A) The plot represents the percentages of NK ...
Images
in Progressive NK-cell dysfunction and ILC imbalance favor immune evasion in multiple myeloma
> Blood Advances
Published: 2025
Figure 2. ILCs in MM progression. (A) Gating strategy for ILC identification and transcription factor expression in different ILC subsets present in BM and PB. (B) The plot represents the percentages of ILCs (Lin – /CD127 + ) expressed as median values ± SEM in PB and BM of patients with MM amon... More about this image found in ILCs in MM progression. (A) Gating strategy for ILC identification and tra...
Journal Articles
Clinical Trials & Observations
Olivier Tournilhac, Kamal Bouabdallah, Solène Lecolant, Maya Hacini, Kamel Laribi, Sébastien Bailly, Thibaut Belmondo, Marie Maerevoet, Loic Ysebaert, Stéphanie Guidez, Steven Le Gouill, Christophe Bonnet, Marc André, Jehan Dupuis, Catherine Thieblemont, Emmanuel Bachy, Nicolas Daguindau, Franck Morschhauser, Sabine Tricot, Charline Moulin, Anne Banos, Roch Houot, Adrien Chauchet, Emmanuel Gyan, Guillaume Cartron, Hassan Farhat, Vincent Camus, Bernard Drenou, Hacene Zerazhi, David Sibon, Emmanuelle Nicolas-Virelizier, Caroline Delette, Sylvia Snauwaert, Sarah Bailly, Richard Delarue, Sylvain Carras, Albane Ledoux-Pilon, Marie-Cécile Parrens, Samuel Griolet, Philippe Gaulard, Marie-Hélène Delfau-Larue, Laurence de Leval, Gandhi Laurent Damaj
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6292–6304.
Published: 2025
Includes: Supplemental data
Journal Articles
Samira Glaeser-Khan, Satoko Ito, Manraj Sra, Rhys Richmond, Robert D. Bona, Harlan M. Krumholz, Stacy E. Croteau, Adam Cuker, George Goshua
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6237–6245.
Published: 2025
Includes: Supplemental data
Journal Articles
Rudra Prasad Dutta, Heng-Huan Lee, Violetta V. Leshchenko, Ravi Prakash Shukla, Fangfang Yan, Yang Liu, H. Ümit Kaniskan, Xing Qiu, Jian Jin, Lapo Alinari, Michael Wang, Samir Parekh
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6267–6278.
Published: 2025
Includes: Supplemental data
Journal Articles
Fifteen years of use of functional imaging in multiple myeloma: where we started and where we are going
Open Access
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6252–6266.
Published: 2025
Journal Articles
Clinical Trials & Observations
Yuting Yan, Bolin Yuan, Tonglu Qiu, Guoqiang Liu, Weihao Chen, Tingyu Wang, Ying Yu, Wenjie Xiong, Gang An, Dehui Zou, Jianyong Li, Liang Wang, Shuhua Yi
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6279–6291.
Published: 2025
Includes: Supplemental data
Journal Articles
Michael Loschi, Marie-Charlotte Villy, Jacques-Emmanuel Galimard, Marie-Mathilde Auboiroux, Nathalie Gachard, Alexandre Perani, Matthieu Duchmann, Laetitia Largeaud, Stéphanie Nguyen, Flore Sicre de Fontbrune, Marie Robin, Ibrahim Yakoub-Agha, Etienne Daguindau, Sylvain Chantepie, Amandine Charbonnier, Delphine Lebon, Sébastien Maury, Hélène Labussiere-Wallet, Anne Huynh, Edouard Forcade, Cristina Castilla-Llorente, Thomas Cluzeau, Lionel Adès, Maud D’aveni, Raynier Devillier, Natacha Maillard, Sami Benachour, Hervé Dombret, Christian Récher, Arnaud Pigneux, Emmanuelle Clappier, Eric Delabesse, Nicolas Duployez, Pascal Turlure, Régis Peffault de Latour, Marie Sébert
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6314–6325.
Published: 2025
Includes: Supplemental data
Journal Articles
Bhavna Bhasin-Chhabra, Tao Wang, John E. Levine, Shalini Shenoy, Miguel-Angel Perales, Asad Bashey, Hershel Raff, Wael Saber
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6394–6401.
Published: 2025
Includes: Supplemental data
Journal Articles
Dual anti-CD19/22 and anti-BCMA CAR T-cell therapy in a patient with multiple myeloma and secondary B-ALL
Open AccessMalte Roerden, Luca Hensen, Britta Besemer, Friederike Schwartz, Kristina Reuss, Alisha Weiss-Haug, Lennart Harland, Franziska Ginsberg, Christoph Faul, Wolfgang Bethge, Claudia Lengerke
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6340–6344.
Published: 2025
Journal Articles
A novel isoform of tensin-1 promotes actin filament assembly for efficient erythroblast enucleation
Open AccessArit Ghosh, Megan Coffin, Dimitri M. Diaz, Sarah Barndt, Vincent P. Schulz, Patrick G. Gallagher, Su Hao Lo, Velia M. Fowler
Journal:
Blood Advances
Blood Adv (2025) 9 (24): 6356–6369.
Published: 2025
Includes: Supplemental data
Journal Articles
Images
in Fifteen years of use of functional imaging in multiple myeloma: where we started and where we are going
> Blood Advances
Published: 2025
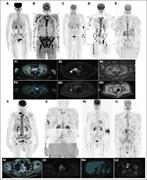
Figure 1. Disease patterns by 18 F-FDG–PET/CT and WB-DW-MRI. (A) DD by 18 F-FDG–PET/CT; (B) DD by WB-DW-MRI; (C) focal on DD by 18 F-FDG–PET/CT; (D) focal on DD by WB-DW-MRI; (E) micronodular disease by WB-DW-MRI; (F) focal disease by 18 F-FDG–PET/CT before (i) and after (ii) treatment; (G-H... More about this image found in Disease patterns by 18 F-FDG–PET/CT and WB-DW-MRI. (A) DD by 18 F-FDG–PE...
Images
in Fifteen years of use of functional imaging in multiple myeloma: where we started and where we are going
> Blood Advances
Published: 2025
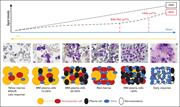
Figure 2. Relation between ADC values, signal intensity in DWI sequences, and BM cellular density during disease course and response to therapy. MGUS, monoclonal gammopathy of undetermined significance. Adapted from Dutoit et al 92 and Ormond et al. 93 More about this image found in Relation between ADC values, signal intensity in DWI sequences, and BM cell...
Images
in Monoclonal gammopathy defines distinct clinical subsets in chronic lymphocytic leukemia across therapeutic eras
> Blood Advances
Published: 2025
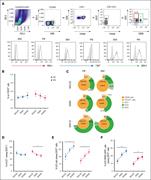
Figure 1. The distribution of MG in CLL and its relationship to IGHV gene. (A) Distribution of different subtypes of MG. (B) The distribution of lambda and kappa light-chain use in different types of paraproteinemia. (C) Proportion of patients with mutated IGHV in different MG group. (D) Rel... More about this image found in The distribution of MG in CLL and its relationship to IGHV gene. (A) Dis...
Images
in Monoclonal gammopathy defines distinct clinical subsets in chronic lymphocytic leukemia across therapeutic eras
> Blood Advances
Published: 2025
Figure 2. Gene mutation spectrum detected by next-generation target sequencing. (A) Gene mutation rate in MG-positive and MG-negative patients with CLL. (B) Mutation rates in patients with different types of paraproteinemia. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001. More about this image found in Gene mutation spectrum detected by next-generation target sequencing. (A) ...
Images
in Monoclonal gammopathy defines distinct clinical subsets in chronic lymphocytic leukemia across therapeutic eras
> Blood Advances
Published: 2025
Figure 3. Survival analysis of MG-positive and MG-negative patients with CLL. (A) TTFT. (B) PFS. (C) OS. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001. More about this image found in Survival analysis of MG-positive and MG-negative patients with CLL. (A) TT...
Images
in Monoclonal gammopathy defines distinct clinical subsets in chronic lymphocytic leukemia across therapeutic eras
> Blood Advances
Published: 2025
Figure 4. Subgroup survival analysis of patients with MG stratified by IGHV mutation status. (A) The proportion of patients with MG in each risk group of CLL-IPI. (B-C) PFS and OS in patients with CLL with mutated IGHV . (D-E) PFS and OS in patients with CLL with unmutated IGHV . More about this image found in Subgroup survival analysis of patients with MG stratified by IGHV mutatio...
1
Advertisement intended for health care professionals