Key Points
Young untreated patients with MCL show 13% prevalence of M-CH at baseline.
Large M-CH clones (variant allele frequency ≥ 10%) are associated with MCL progression and worse progression-free and overall survival.
Visual Abstract
Although recent evidence suggests that myeloid clonal hematopoiesis (M-CH) may influence lymphoma clinical outcome, its impact in mantle cell lymphoma (MCL) remains unclear. Here, we report a comprehensive next-generation sequencing–based analysis of the M-CH mutational landscape at baseline and follow-up in patients enrolled in the Fondazione Italiana Linfomi MCL0208 phase 3 trial, evaluating lenalidomide maintenance vs observation after chemoimmunotherapy and autologous stem cell transplantation (ASCT) in untreated young patients with MCL. Overall, 254 of 300 (85%) enrolled patients (median age, 57 years [range, 32-66]) had a baseline sample available for CH analysis. Using stringent criteria, at least 1 mutation involving M-CH candidate genes was described in 34 patients (13%), with DNMT3A being the most frequently mutated gene (54%). After a median follow-up of 7 years, the presence of large CH clones (variant allele frequency of ≥10%) predicted worse progression-free survival (hazard ratio [HR], 2.93; 95% confidence interval [CI] 1.36-6.31; P = .006) and overall survival (HR, 3.02 [1.21-7.55]; P = .018) compared with patients with CH. Importantly, the competing risks analysis demonstrates that the worse clinical outcome associated with M-CH large clones is linked to MCL progression (P < .05). Moreover, large M-CH clones showed longer time to hematological recovery after ASCT than the remaining cohort (P = .026). In conclusion, we showed for the first time that large CH clones might associate with unfavorable clinical impact in patients with MCL. This trial was registered at www.clinicaltrialsregister.eu as EudraCT (2009-012807-25) and www.ClinicalTrials.gov as #NCT02354313.
Introduction
Myeloid clonal hematopoiesis (M-CH) of indeterminate potential has been defined as the presence of a somatic mutation with variant allele fraction (VAF) of ≥2% in a myeloid neoplasm driver gene in a patient lacking a myeloid neoplasm or unexplained cytopenia.1 Because M-CH is an age-related phenomenon, somatic clones are detectable in <1% of individuals aged <40 but rise in frequency with each decade of life.2 Previous studies3-5 suggested an increased risk of developing a hematological malignancy in M-CH carriers. Furthermore, growing evidence has shown an association between M-CH and nonmalignant diseases including stroke, coronary artery disease, peripheral artery disease, and venous thromboembolism.6 Recent evidence suggested that large M-CH clones with VAF of ≥10% better predict the risk of developing hematological cancer and nonmalignant diseases.7,8 Evidence from several types of lymphoma including angioimmunoblastic T-cell lymphoma,9 Hodgkin lymphoma,10 and Waldenström macroglobulinemia11 suggests that M-CH may have a not-negligible impact on clinical outcome. In addition, in several studies involving patients with lymphoma who underwent autologous stem cell transplantation (ASCT), M-CH at the time of ASCT conferred an increased risk of therapy-related myeloid neoplasms (t-MNs).12-14
Mantle cell lymphoma (MCL) is an uncommon subtype of aggressive B-cell non-Hodgkin lymphoma. Although several treatments are at present available, relapses invariably occur, still making MCL an incurable lymphoma. Currently, available M-CH data in MCL are scarce and biased by patients selection,15,16 therefore, we aimed at describing, to our knowledge, for the first time the M-CH mutational landscape both at baseline and after ASCT, as well as at analyzing clinical and outcome correlations in a large series of young patients with MCL enrolled in a prospective clinical trial of the Fondazione Italiana Linfomi (FIL).
Methods
Patient cohort
This study was performed with biological samples collected in the phase 3 FIL MCL0208 trial (ClinicalTrials.gov identifier: NCT027354313).17 Briefly, MCL0208 trial enrolled 300 previously untreated patients with MCL, aged from 18 to 65 years, without clinically significant comorbidities. Patients received 3 cycles of R-CHOP21 (rituximab cyclophosphamide, doxorubicin, vincristine, prednisone), followed by R–high-dose cyclophosphamide (4 g/m2), 2 cycles of R–high-dose cytarabine (2 g/m2 every 3 hours, for 3 days), and ASCT conditioned by using BEAM (carmustine, etoposide, cytarabine, melphalan) or FEAM (fotemustine, etoposide, cytarabine, melphalan) regimen. After ASCT, patients who achieved overall hematological recovery (as defined by neutrophils >1.5 × 109/L and platelets of >60 × 109/L) and showed a complete or partial response, were randomly assigned to observation or lenalidomide (15 mg on days 1-21 every 28 days) for 24 months.17
All patients provided written informed consent for the use of their biological samples for research purposes, in accordance with the institutional review board requirements and the Helsinki Declaration. The clinical trial as well as this study was approved by the ethics committees of all the enrolling centers. All the samples were centralized for scientific analysis in the hematological laboratory of Torino University. Statistical analyses were performed as reported in the supplemental Methods.
Biological samples
Unsorted bone marrow (BM) and peripheral blood (PB) samples were collected at baseline and within 12 months after ASCT based on the preplanned time points of minimal residual disease (MRD) analysis18 and analyzed for M-CH. Further details of DNA extraction, flow cytometry, and mutational and MRD analysis are provided in the supplemental Methods.
CH-targeted sequencing analysis
M-CH mutations were analyzed by next-generation sequencing at IRCCS Policlinico San Matteo Foundation and University of Pavia, Pavia, Italy, using the TruSight myeloid sequencing panel (Illumina, San Diego, CA) including 54 M-CH candidate myeloid genes. Functionally, annotated variants were then filtered based on the information retrieved from public databases, and the expected germ line allele frequency. The remaining variants were finally tagged as oncogenic, based on the information derived from the literature, the catalog of somatic mutations in cancer,19 and in silico prediction effect, as previously described.20
Management of possible overlapping between tumor and M-CH mutations
Because of potential contamination by tumor cells in samples analyzed for M-CH, mutations occurring at baseline involving the most frequently mutated genes in MCL cells were considered as derived from the myeloid lineage only when the VAF was ≥4 times the tumor fraction in the baseline sample as assessed by flow cytometry.9 Among detected mutations, those considered as potentially mimicking M-CH were those involving TP53, TET2, NOTCH1, CDKN2A, BCOR, and KMT2A genes21 At the post-ASCT time point, potentially shared mutations between myeloid and MCL cells were considered as M-CH when not present at the baseline time point (new emerging clones) or when involving MRD-negative samples.
Results
Clinical and biological characteristics of the analyzed cohort
Overall, 254 of 300 patients (85%) enrolled in the FIL MCL0208 trial had a sample (227 PB and 27 BM) collected at MCL diagnosis available for M-CH analysis, whereas 191 of 300 (60%) patients had 2 paired samples collected both at diagnosis (168 PB and 23 BM) and within 12 months from ASCT (180 PB and 11 BM). Clinical characteristics of analyzed patients are shown in Table 1 and did not differ significantly from the cohort excluded from this analysis except for lower lactate dehydrogenase levels (greater than the upper limit of normal, 30% vs 48%, respectively; P = .020). No difference was seen in clinical response rate after ASCT between cohorts (complete remission, 91% vs 89%, respectively; P = .5), although analyzed patients showed an advantage in progression-free survival (PFS; 84-month PFS of 46% [95% CI, 40-53] vs 35% [95% CI, 23-53], respectively; P = .034; supplemental Figure 1A) and overall survival (OS; 84-month, OS 72% [95% CI, 67-79] vs 57% [95% CI, 43-75], respectively; P = .031; supplemental Figure 1B), with more patients proceeding to ASCT (87% vs 59%, respectively; P < .001) and being randomized to lenalidomide maintenance (72% vs 50%, respectively; P = .004). After adjusting for age, sex, Mantle Cell Lymphoma International Prognostic Index (MIPI) score, KI67, bulky, BM involvement, and blastoid histology by Cox regression model, patients analyzed for M-CH showed no statistical significant difference in terms of both PFS (hazard ratio [HR], 0.80 [0.51-1.28]; P = .4) and OS (HR, 0.94 [0.51-1.76]; P = .9) compared with the nonanalyzed cohort.
Characteristics of the MCL0208 patient cohort analyzed for M-CH mutations
| Characteristic . | Sample for M-CH analysis available at diagnosis . | Paired samples for M-CH analysis available at diagnosis and after ASCT . | ||||
|---|---|---|---|---|---|---|
| Available, n = 254∗ . | Not available, n = 46∗ . | P value† . | Available, n = 191∗ . | Not available, n = 109∗ . | P value† . | |
| Sex | >.9 | .6 | ||||
| Female | 55 (22%) | 10 (22%) | 43 (23%) | 22 (20%) | ||
| Male | 199 (78%) | 36 (78%) | 148 (77%) | 87 (80%) | ||
| Age, y | .7 | .10 | ||||
| Median (range) | 57 (32, 66) | 58 (28-66) | 56 (32-66) | 58 (28-66) | ||
| Histology | .3 | .13 | ||||
| Blastoid | 20 (7.9%) | 6 (13%) | 13 (6.8%) | 13 (12%) | ||
| Classic | 234 (92%) | 40 (87%) | 178 (93%) | 96 (88%) | ||
| Ki67 index | .12 | .047 | ||||
| <30 | 163 (71%) | 24 (59%) | 128 (73%) | 59 (61%) | ||
| ≥30 | 67 (29%) | 17 (41%) | 47 (27%) | 37 (39%) | ||
| NA | 24 | 5 | 16 | 13 | ||
| LDH above ULN | .020 | .021 | ||||
| ≤ULN | 177 (70%) | 24 (52%) | 137 (72%) | 64 (59%) | ||
| >ULN | 77 (30%) | 22 (48%) | 54 (28%) | 45 (41%) | ||
| MIPI | .3 | .2 | ||||
| High risk | 38 (15%) | 9 (20%) | 28 (15%) | 19 (17%) | ||
| Intermediate risk | 59 (23%) | 14 (30%) | 41 (21%) | 32 (29%) | ||
| Low risk | 157 (62%) | 23 (50%) | 122 (64%) | 58 (53%) | ||
| MIPIc | .2 | .085 | ||||
| High risk | 17 (7.4%) | 7 (17%) | 13 (7.4%) | 11 (11%) | ||
| Intermediate-high risk | 29 (13%) | 6 (15%) | 23 (13%) | 12 (12%) | ||
| Intermediate-low risk | 67 (29%) | 12 (29%) | 44 (25%) | 35 (36%) | ||
| Low risk | 117 (51%) | 16 (39%) | 95 (54%) | 38 (40%) | ||
| NA | 24 | 5 | 16 | 13 | ||
| ECOG PS | .9 | .4 | ||||
| 1-2 | 58 (23%) | 11 (24%) | 41 (21%) | 28 (26%) | ||
| 0 | 196 (77%) | 35 (76%) | 150 (79%) | 81 (74%) | ||
| Stage | .5 | .5 | ||||
| Stage II-III | 14 (5.5%) | 4 (8.7%) | 10 (5.2%) | 8 (7.3%) | ||
| Stage IV | 240 (94%) | 42 (91%) | 181 (95%) | 101 (93%) | ||
| Bulky | .5 | .5 | ||||
| Bulky | 81 (32%) | 17 (37%) | 60 (31%) | 38 (35%) | ||
| Not bulky | 173 (68%) | 29 (63%) | 131 (69%) | 71 (65%) | ||
| TP53 status in tumor cells | >.9 | .3 | ||||
| Mutation or deletion | 28 (17%) | 3 (15%) | 19 (15%) | 12 (21%) | ||
| WT | 138 (83%) | 17 (85%) | 111 (85%) | 44 (79%) | ||
| NA | 88 | 26 | 61 | 53 | ||
| Type of sample at baseline | >.9 | .2 | ||||
| BM | 27 (11%) | — | 23 (12%) | — | ||
| PB | 227 (89%) | — | 168 (88%) | — | ||
| Type of sample within 12 mo from ASCT | >.9 | >.9 | ||||
| BM | 11 (5.8%) | — | 11 (5.8%) | — | ||
| PB | 180 (94%) | — | 180 (94%) | — | ||
| NA | 63 | 0 | ||||
| ASCT | <.001 | <.001 | ||||
| ASCT not received | 32 (13%) | 19 (41%) | 0 (0%) | 51 (47%) | ||
| ASCT received | 222 (87%) | 27 (59%) | 191 (100%) | 58 (53%) | ||
| Clinical response after ASCT | .5 | .4 | ||||
| CR | 201 (91%) | 25 (89%) | 170 (89%) | 56 (95%) | ||
| PR | 12 (5.4%) | 3 (11%) | 12 (6.3%) | 3 (5.1%) | ||
| SD or PD | 8 (3.6%) | 0 (0%) | 8 (4.2%) | 0 (0%) | ||
| NA | 33 | 18 | 1 | 50 | ||
| Randomized after ASCT | 182 (72%) | 23 (50%) | .004 | 160 (84%) | 45 (41%) | <.001 |
| Randomized to Lenalidomide | 94 (52%) | 10 (43%) | .5 | 79 (49%) | 25 (56%) | .5 |
| NA | 72 | 23 | 31 | 64 | ||
| Characteristic . | Sample for M-CH analysis available at diagnosis . | Paired samples for M-CH analysis available at diagnosis and after ASCT . | ||||
|---|---|---|---|---|---|---|
| Available, n = 254∗ . | Not available, n = 46∗ . | P value† . | Available, n = 191∗ . | Not available, n = 109∗ . | P value† . | |
| Sex | >.9 | .6 | ||||
| Female | 55 (22%) | 10 (22%) | 43 (23%) | 22 (20%) | ||
| Male | 199 (78%) | 36 (78%) | 148 (77%) | 87 (80%) | ||
| Age, y | .7 | .10 | ||||
| Median (range) | 57 (32, 66) | 58 (28-66) | 56 (32-66) | 58 (28-66) | ||
| Histology | .3 | .13 | ||||
| Blastoid | 20 (7.9%) | 6 (13%) | 13 (6.8%) | 13 (12%) | ||
| Classic | 234 (92%) | 40 (87%) | 178 (93%) | 96 (88%) | ||
| Ki67 index | .12 | .047 | ||||
| <30 | 163 (71%) | 24 (59%) | 128 (73%) | 59 (61%) | ||
| ≥30 | 67 (29%) | 17 (41%) | 47 (27%) | 37 (39%) | ||
| NA | 24 | 5 | 16 | 13 | ||
| LDH above ULN | .020 | .021 | ||||
| ≤ULN | 177 (70%) | 24 (52%) | 137 (72%) | 64 (59%) | ||
| >ULN | 77 (30%) | 22 (48%) | 54 (28%) | 45 (41%) | ||
| MIPI | .3 | .2 | ||||
| High risk | 38 (15%) | 9 (20%) | 28 (15%) | 19 (17%) | ||
| Intermediate risk | 59 (23%) | 14 (30%) | 41 (21%) | 32 (29%) | ||
| Low risk | 157 (62%) | 23 (50%) | 122 (64%) | 58 (53%) | ||
| MIPIc | .2 | .085 | ||||
| High risk | 17 (7.4%) | 7 (17%) | 13 (7.4%) | 11 (11%) | ||
| Intermediate-high risk | 29 (13%) | 6 (15%) | 23 (13%) | 12 (12%) | ||
| Intermediate-low risk | 67 (29%) | 12 (29%) | 44 (25%) | 35 (36%) | ||
| Low risk | 117 (51%) | 16 (39%) | 95 (54%) | 38 (40%) | ||
| NA | 24 | 5 | 16 | 13 | ||
| ECOG PS | .9 | .4 | ||||
| 1-2 | 58 (23%) | 11 (24%) | 41 (21%) | 28 (26%) | ||
| 0 | 196 (77%) | 35 (76%) | 150 (79%) | 81 (74%) | ||
| Stage | .5 | .5 | ||||
| Stage II-III | 14 (5.5%) | 4 (8.7%) | 10 (5.2%) | 8 (7.3%) | ||
| Stage IV | 240 (94%) | 42 (91%) | 181 (95%) | 101 (93%) | ||
| Bulky | .5 | .5 | ||||
| Bulky | 81 (32%) | 17 (37%) | 60 (31%) | 38 (35%) | ||
| Not bulky | 173 (68%) | 29 (63%) | 131 (69%) | 71 (65%) | ||
| TP53 status in tumor cells | >.9 | .3 | ||||
| Mutation or deletion | 28 (17%) | 3 (15%) | 19 (15%) | 12 (21%) | ||
| WT | 138 (83%) | 17 (85%) | 111 (85%) | 44 (79%) | ||
| NA | 88 | 26 | 61 | 53 | ||
| Type of sample at baseline | >.9 | .2 | ||||
| BM | 27 (11%) | — | 23 (12%) | — | ||
| PB | 227 (89%) | — | 168 (88%) | — | ||
| Type of sample within 12 mo from ASCT | >.9 | >.9 | ||||
| BM | 11 (5.8%) | — | 11 (5.8%) | — | ||
| PB | 180 (94%) | — | 180 (94%) | — | ||
| NA | 63 | 0 | ||||
| ASCT | <.001 | <.001 | ||||
| ASCT not received | 32 (13%) | 19 (41%) | 0 (0%) | 51 (47%) | ||
| ASCT received | 222 (87%) | 27 (59%) | 191 (100%) | 58 (53%) | ||
| Clinical response after ASCT | .5 | .4 | ||||
| CR | 201 (91%) | 25 (89%) | 170 (89%) | 56 (95%) | ||
| PR | 12 (5.4%) | 3 (11%) | 12 (6.3%) | 3 (5.1%) | ||
| SD or PD | 8 (3.6%) | 0 (0%) | 8 (4.2%) | 0 (0%) | ||
| NA | 33 | 18 | 1 | 50 | ||
| Randomized after ASCT | 182 (72%) | 23 (50%) | .004 | 160 (84%) | 45 (41%) | <.001 |
| Randomized to Lenalidomide | 94 (52%) | 10 (43%) | .5 | 79 (49%) | 25 (56%) | .5 |
| NA | 72 | 23 | 31 | 64 | ||
Boldface values indicate statistically significant P values (P < .05). CR, complete remission; ECOG PS; Eastern Cooperative Oncology Group performance status; LDH, lactate dehydrogenase; MIPIc, MIPI combined; NA, data not available; PD, progressive disease; PR, partial remission; SD, stable disease; ULN, upper limit of normal; WT, wild-type.
n (%).
Pearson χ2 test; Wilcoxon rank-sum test; Fisher exact test.
DNMT3A is the most frequent M-CH mutation in the MCL0208 cohort
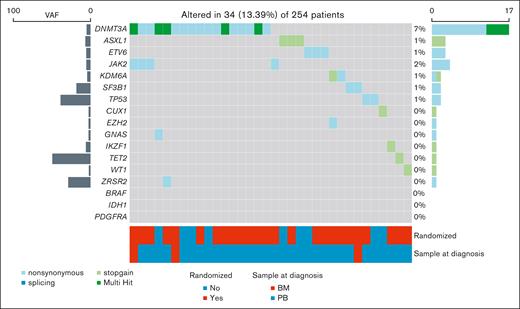
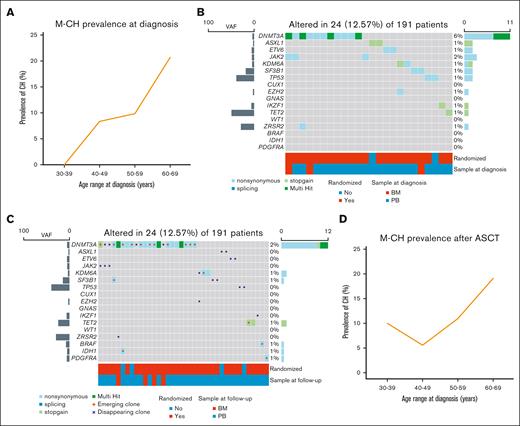
After filtering out variants potentially deriving from MCL infiltration in BM and PB samples (supplemental Table 1), 48 pathogenic mutations involving M-CH candidate genes (including 35 nonsynonymous, 12 stop-gain, and 1 splicing) were detected in 34 of 254 patients (13%, patients carrying at least 1 M-CH mutation with a VAF of ≥2% [M-CH+]; Figure 1). Among patients who are M-CH+, the most frequent mutated genes at baseline were DNMT3A (17/34 [50%]), JAK2 (4/34 [12%]), ASXL1 (3/34 [9%]), and ETV6 (3/34 [9%]). Moreover, focusing the analysis on the 191 patients with paired samples, the M-CH prevalence at diagnosis increased with age (Figure 2A).
Overview of M-CH mutations detected at MCL diagnosis. M-CH mutational landscape of patients with an available sample at MCL diagnosis. M-CH mutations were detected at baseline in 34 of 254 patients (13.4%).
Overview of M-CH mutations detected at MCL diagnosis. M-CH mutational landscape of patients with an available sample at MCL diagnosis. M-CH mutations were detected at baseline in 34 of 254 patients (13.4%).
Prevalence and mutational profile of M-CH clones in MCL patients at diagnosis and after ASCT. Comparison of M-CH prevalence stratified by age group and M-CH mutation types in MCL patients at diagnosis (A-B) and after ASCT (C-D).
Prevalence and mutational profile of M-CH clones in MCL patients at diagnosis and after ASCT. Comparison of M-CH prevalence stratified by age group and M-CH mutation types in MCL patients at diagnosis (A-B) and after ASCT (C-D).
The number of mutations involving genes as DNTM3A and TET2 increased from diagnosis (Figure 2B) during treatment course and follow-up (Figure 2C; supplemental Table 2) whereas mutations involving genes such as ASXL1 and TP53 disappeared after ASCT. Overall, the rising prevalence of M-CH clones with age was also confirmed in post-ASCT samples. (Figure 2D). Of note, 15 of 191 (8%) patients developed new M-CH mutations after ASCT. Interestingly, new mutations affecting M-CH candidate genes as DNTM3A, SF3B1, KDM6A, PDGFRA, IDH1, and BRAF emerged after ASCT. The median M-CH VAF at diagnosis and after ASCT were comparable (4.12% [2.10%-71.96%] and 4.90% [2.30%-43.35%], respectively; P = .4). As supplemental Figure 2 describes, M-CH VAFs mostly showed variable trajectories during follow-up. Interestingly, all TP53 clones disappeared during follow-up.
Large M-CH clones predict MCL progression and OS
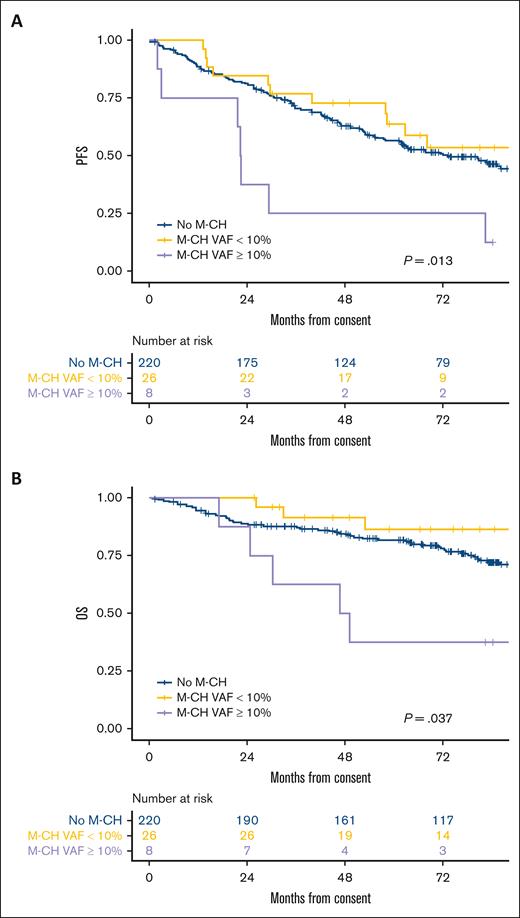
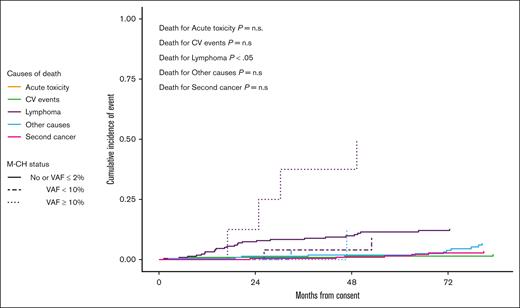
Baseline characteristics were superimposable between patients who are M-CH+ and those patients without M-CH mutations (M-CH−), except for higher median age in those who are M-CH+ (60 years [46-66] vs 56 years [32-66], respectively; P = .011; supplemental Table 1). After a median follow-up of 7 years, the presence of M-CH did not significantly affect PFS (P = .457, supplemental Figure 3A) or OS (P = .514, supplemental Figure 3B). However, an M-CH VAF threshold of 10% predicted shorter PFS (P = .006; Figure 3A) and OS (P = .006; Figure 3B). Specifically, Cox regression analysis showed that large clonal hematopoiesis of indeterminate potential clones (VAF of ≥10%, n = 8) conferred higher risk of progression (HR, 2.93 [1.36-6.31]; P = .006) and death (HR, 3.02 [1.21-7.55]; P = .018) compared to M-CH- patients. After applying propensity score adjustment for baseline factors (age, sex, MIPI, Ki67, bulky, BM involvement, and blastoid subtype), patients affected by large M-CH clones still showed shorter PFS (HR, 2.27 [1.03-5.01]; P = .042) and a trend toward worse OS (HR, 1.87 [0.71-4.87]; P = .203). Importantly, the competing risks analysis demonstrates that the worse clinical outcome exhibited by patients carrying large clones is linked to MCL progression (P < .05; Figure 4) rather than acute or late toxicity events or secondary cancers occurrence.
A 10% M-CH VAF cutoff identifies patient with different PFS and OS. (A) PFS. (B) OS.
A 10% M-CH VAF cutoff identifies patient with different PFS and OS. (A) PFS. (B) OS.
Patients who wereM-CH+ with a VAF of ≥10% at baseline have increased incidence of relapse mortality when compared with other patients. At 84 months of median follow-up, patients who were M-CH+ with a VAF of >10% compared with who were M-CH+ with a VAF between 2% and 10% or who were M-CH– presented higher incidence of relapse mortality (P < .05). n.s., not significant.
Patients who wereM-CH+ with a VAF of ≥10% at baseline have increased incidence of relapse mortality when compared with other patients. At 84 months of median follow-up, patients who were M-CH+ with a VAF of >10% compared with who were M-CH+ with a VAF between 2% and 10% or who were M-CH– presented higher incidence of relapse mortality (P < .05). n.s., not significant.
Interestingly, large M-CH clones were associated with higher baseline lymphocytes values (median, 13 vs 2 cells per μL; P = .01) and lower platelets count (median: 129 × 109/L vs 191 × 109/L; P = .035) than patients without M-CH clones or with small clones (supplemental Table 4). In addition, patients with large M-CH clones were also characterized by less favorable MIPI (high risk, 62% vs 13%; P = .003) and combined MIPI (high risk, 57% vs 6%; P = .002) scores. Conversely, CH clones identified after ASCT did not affect patient outcome (72 months after ASCT PFS, 49% [41%-58%] vs 56% [39%-81%], respectively; P = .53, and 72 months after ASCT OS, 75% [68%-83%] vs 82% [68%-100%], respectively; P = .81; supplemental Figure 4), independently from the size of the clones (supplemental Figure 5).
M-CH affects hematological recovery after ASCT
Despite that no significant differences emerged between patients who are M-CH+ and those who are M-CH− at baseline in terms of hematological toxicity after induction chemotherapy or overall hematological recovery after ASCT (Table 2), interestingly, patients who are M-CH+ exhibited a near-significant trend to a delayed thrombocytes recovery after ASCT compared with the remaining cohort (median days from ASCT to platelet count of ≥20 × 109/L: 15 vs 13, respectively; P = .053). This trend was even more relevant among patients with large M-CH clones vs small or no clones (median days: 16 vs 13, respectively; P = .026; supplemental Table 3) and in patients with M-CH detected after ASCT (median days: 16 vs 13, respectively; P = .016), as shown in Table 2. Nonetheless, no difference in median number of lenalidomide cycles or dose reduction and no higher incidence of secondary cancers was observed between M-CH+ and M-CH– subgroups. Of 254 patients analyzed for M-CH, 7 patients (2.7%) developed a secondary hematological cancer (5 cases of myelodysplastic neoplasms, 1 case of acute myeloid leukemia, and 1 case of acute lymphoblastic leukemia). The 6 cases of t-MNs were observed at a median time from ASCT of 36.5 months (21-63). Interestingly, only 1 patient who developed myelodysplastic neoplasms after ASCT (without lenalidomide maintenance, time from ASCT to t-MNs was 39 months) had 2 newly arising M-CH mutations after autologous transplantation, namely DNTM3A and IDH1 (VAF of 6.53% and 5.03%, respectively). Retrospective analysis of the baseline sample from the patient showing DNTM3A and IDH1 emerging M-CH mutations also identified a small DNTM3A clone (VAF of 0.6%) at the time of MCL diagnosis, previously excluded from analysis because it was below the 2% VAF threshold. Moreover, no small IDH1 clones were detected at the time of MCL diagnosis.
Overview of chemotherapy- and lenalidomide-related toxicity according to M-CH status at MCL diagnosis and after ASCT
| Characteristic . | M-CH analyzed at diagnosis . | M-CH analyzed after ASCT . | ||||
|---|---|---|---|---|---|---|
| Patients who are M-CH− at diagnosis, n = 220∗ . | Patients who are M-CH+ at diagnosis, n = 34∗ . | P value† . | Patients who are M-CH− after ASCT, n = 167∗ . | Patients who are M-CH+ after ASCT, n = 24∗ . | P value‡ . | |
| Hematological toxicity after first R-CHOP | .2 | .2 | ||||
| G1-G2 | 35 (16%) | 3 (8.8%) | 21 (13%) | 6 (25%) | ||
| G3-G4 | 30 (14%) | 8 (24%) | 22 (13%) | 4 (17%) | ||
| No toxicity | 154 (70%) | 23 (68%) | 124 (74%) | 14 (58%) | ||
| NA | 1 | 0 | ||||
| Hematological toxicity after second R-CHOP | .4 | .7 | ||||
| G1-G2 | 35 (16%) | 3 (8.8%) | 21 (13%) | 4 (17%) | ||
| G3-G4 | 24 (11%) | 6 (18%) | 19 (11%) | 3 (12%) | ||
| No toxicity | 160 (73%) | 25 (74%) | 127 (76%) | 17 (71%) | ||
| NA | 1 | 0 | ||||
| Hematological toxicity after third R-CHOP | .6 | .3 | ||||
| G1-G2 | 40 (18%) | 4 (12%) | 26 (16%) | 5 (21%) | ||
| G3-G4 | 28 (13%) | 6 (18%) | 20 (12%) | 5 (21%) | ||
| No toxicity | 151 (69%) | 24 (71%) | 121 (72%) | 14 (58%) | ||
| NA | 1 | 0 | ||||
| Hematological toxicity after R-CTX | .2 | .013 | ||||
| G1-G2 | 14 (6.7%) | 0 (0%) | 10 (6.0%) | 2 (8.3%) | ||
| G3-G4 | 115 (55%) | 23 (70%) | 88 (53%) | 19 (79%) | ||
| No toxicity | 81 (39%) | 10 (30%) | 69 (41%) | 3 (12%) | ||
| NA | 10 | 1 | ||||
| Hematological recovery after ASCT | 136 (87%) | 19 (100%) | .13 | 117 (89%) | 15 (94%) | >.9 |
| NA | 64 | 15 | 35 | 8 | ||
| Days from ASCT to neutrophils ≥ 0.5 × 109/L | .11 | .5 | ||||
| Median (range) | 10 (6-70) | 11 (3-19) | 10 (7-70) | 11 (3-13) | ||
| NA | 46 | 9 | 17 | 5 | ||
| Days from ASCT to neutrophils ≥ 1.0 × 109/L | .11 | .7 | ||||
| Median (range) | 11 (4-132) | 12 (10-27) | 11 (4-132) | 11 (10-14) | ||
| NA | 56 | 11 | 25 | 8 | ||
| Days from ASCT to PLTS of ≥20 (×109/L) | .053 | .016 | ||||
| Median (range) | 13 (3-145) | 15 (6-54) | 13 (3-145) | 16 (7-29) | ||
| NA | 44 | 10 | 18 | 4 | ||
| Days from ASCT to PLTS of ≥50 (×109/L) | .6 | .3 | ||||
| Median (range) | 18 (6-726) | 20 (14-45) | 19 (6-726) | 21 (12-50) | ||
| NA | 67 | 15 | 35 | 9 | ||
| Randomized patients | 157 (71%) | 25 (74%) | .8 | 138 (83%) | 22 (92%) | .4 |
| Randomized to lenalidomide | 74 (34%) | 8 (24%) | .2 | 61 (37%) | 6 (25%) | .3 |
| Lenalidomide completed cycles, n | .5 | .4 | ||||
| Median (range) | 20 (0-24) | 17 (0-24) | 20 (0-24) | 16 (0-24) | ||
| NA | 136 | 24 | 96 | 16 | ||
| Lenalidomide dose reduction (%) | .3 | .3 | ||||
| Median (range) | 34 (0-100) | 57 (0-100) | 37 (0-100) | 26 (0-100) | ||
| NA | 136 | 24 | 96 | 16 | ||
| Skin secondary cancer | 2 (0.9%) | 0 (0%) | >.9 | 1 (0.6%) | 0 (0%) | >.9 |
| Secondary cancer (no skin) | 19 (8.6%) | 1 (2.9%) | .5 | 14 (8.4%) | 1 (4.2%) | .7 |
| Therapy-related hematological cancer | 7 (3.2%) | 0 (0%) | .6 | 6 (3.6%) | 1 (4.2%) | >.9 |
| Characteristic . | M-CH analyzed at diagnosis . | M-CH analyzed after ASCT . | ||||
|---|---|---|---|---|---|---|
| Patients who are M-CH− at diagnosis, n = 220∗ . | Patients who are M-CH+ at diagnosis, n = 34∗ . | P value† . | Patients who are M-CH− after ASCT, n = 167∗ . | Patients who are M-CH+ after ASCT, n = 24∗ . | P value‡ . | |
| Hematological toxicity after first R-CHOP | .2 | .2 | ||||
| G1-G2 | 35 (16%) | 3 (8.8%) | 21 (13%) | 6 (25%) | ||
| G3-G4 | 30 (14%) | 8 (24%) | 22 (13%) | 4 (17%) | ||
| No toxicity | 154 (70%) | 23 (68%) | 124 (74%) | 14 (58%) | ||
| NA | 1 | 0 | ||||
| Hematological toxicity after second R-CHOP | .4 | .7 | ||||
| G1-G2 | 35 (16%) | 3 (8.8%) | 21 (13%) | 4 (17%) | ||
| G3-G4 | 24 (11%) | 6 (18%) | 19 (11%) | 3 (12%) | ||
| No toxicity | 160 (73%) | 25 (74%) | 127 (76%) | 17 (71%) | ||
| NA | 1 | 0 | ||||
| Hematological toxicity after third R-CHOP | .6 | .3 | ||||
| G1-G2 | 40 (18%) | 4 (12%) | 26 (16%) | 5 (21%) | ||
| G3-G4 | 28 (13%) | 6 (18%) | 20 (12%) | 5 (21%) | ||
| No toxicity | 151 (69%) | 24 (71%) | 121 (72%) | 14 (58%) | ||
| NA | 1 | 0 | ||||
| Hematological toxicity after R-CTX | .2 | .013 | ||||
| G1-G2 | 14 (6.7%) | 0 (0%) | 10 (6.0%) | 2 (8.3%) | ||
| G3-G4 | 115 (55%) | 23 (70%) | 88 (53%) | 19 (79%) | ||
| No toxicity | 81 (39%) | 10 (30%) | 69 (41%) | 3 (12%) | ||
| NA | 10 | 1 | ||||
| Hematological recovery after ASCT | 136 (87%) | 19 (100%) | .13 | 117 (89%) | 15 (94%) | >.9 |
| NA | 64 | 15 | 35 | 8 | ||
| Days from ASCT to neutrophils ≥ 0.5 × 109/L | .11 | .5 | ||||
| Median (range) | 10 (6-70) | 11 (3-19) | 10 (7-70) | 11 (3-13) | ||
| NA | 46 | 9 | 17 | 5 | ||
| Days from ASCT to neutrophils ≥ 1.0 × 109/L | .11 | .7 | ||||
| Median (range) | 11 (4-132) | 12 (10-27) | 11 (4-132) | 11 (10-14) | ||
| NA | 56 | 11 | 25 | 8 | ||
| Days from ASCT to PLTS of ≥20 (×109/L) | .053 | .016 | ||||
| Median (range) | 13 (3-145) | 15 (6-54) | 13 (3-145) | 16 (7-29) | ||
| NA | 44 | 10 | 18 | 4 | ||
| Days from ASCT to PLTS of ≥50 (×109/L) | .6 | .3 | ||||
| Median (range) | 18 (6-726) | 20 (14-45) | 19 (6-726) | 21 (12-50) | ||
| NA | 67 | 15 | 35 | 9 | ||
| Randomized patients | 157 (71%) | 25 (74%) | .8 | 138 (83%) | 22 (92%) | .4 |
| Randomized to lenalidomide | 74 (34%) | 8 (24%) | .2 | 61 (37%) | 6 (25%) | .3 |
| Lenalidomide completed cycles, n | .5 | .4 | ||||
| Median (range) | 20 (0-24) | 17 (0-24) | 20 (0-24) | 16 (0-24) | ||
| NA | 136 | 24 | 96 | 16 | ||
| Lenalidomide dose reduction (%) | .3 | .3 | ||||
| Median (range) | 34 (0-100) | 57 (0-100) | 37 (0-100) | 26 (0-100) | ||
| NA | 136 | 24 | 96 | 16 | ||
| Skin secondary cancer | 2 (0.9%) | 0 (0%) | >.9 | 1 (0.6%) | 0 (0%) | >.9 |
| Secondary cancer (no skin) | 19 (8.6%) | 1 (2.9%) | .5 | 14 (8.4%) | 1 (4.2%) | .7 |
| Therapy-related hematological cancer | 7 (3.2%) | 0 (0%) | .6 | 6 (3.6%) | 1 (4.2%) | >.9 |
Boldface values indicate statistically significant P values (P < .05). G1-G2, grade 1 or grade 2 toxicity; G3-G4, grade 3 or grade 4 toxicity; NA, data not available; PLTS, platelets; R-CTX, rituximab in combination with cyclophosphamide.
n (%).
Pearson χ2 test; Fisher exact test; Wilcoxon rank-sum test.
Fishers exact test; Pearson χ2 test; Wilcoxon rank-sum test.
Discussion
In this study, we characterized the mutational landscape of M-CH throughout induction chemoimmunotherapy and ASCT in a large subset of young patients with MCL enrolled in the FIL MCL0208 phase 3 trial. We dynamically explored the potential correlations between M-CH clones, patient clinical features, survival, and toxicities. To our knowledge, this is, to our knowledge, the first study investigating this issue in paired samples collected in a prospective trial because previous evidence referring to M-CH in patients with MCL only relied on MRD-negative cohorts after ASCT15 or small series of relapsed/refractory cases.16
First, although a formal comparison was not the focus of our work, we found higher age-adjusted baseline M-CH prevalence in patients with MCL than previously reported in people unselected for cancer or hematological phenotypes.3,4 Interestingly, we found shorter PFS and OS in patients with large M-CH clones (VAF ≥ 10%) at time of diagnosis. To filter out potential bias, we conducted a competing risk analysis, confirming MCL progression as the primary cause of increased mortality in these patients. Additionally, we provided a propensity score analysis to validate the adverse effect of large M-CH clones on PFS, mitigating any potential confounding factors introduced by baseline MCL prognostic characteristics. Despite the differences in VAF threshold chosen by different authors to define M-CH clones, the demonstrated correlation between M-CH and PFS in patients with MCL is in line with those previously exhibited in cohort of relapsed/refractory patients from the PHILEMON trial.16 Speculatively, as clonal hematopoiesis may modulate the function of several hematopoietic cells in the osteo-hematopoietic niche,22 our findings might suggest a role for M-CH in shaping the tumor microenvironment in a MCL-permissive manner. Indeed, in contrast to T-cell lymphomas,9 M-CH mutations in genes such as DNMT3A do not appear to also be present in MCL cells,21 suggesting a role of tumor-extrinsic mechanisms for M-CH clones. However, the possibility that both processes are expressions of a common underlying permissive aberration cannot be ruled out by our findings. Although we were not able to assess an increased hematological toxicity in patients harboring large M-CH mutations, the lower baseline median platelet counts and the longer time intervals to platelets engraftment after ASCT observed in patients with large M-CH clones might also point toward the idea of a perturbed microenvironment with impaired hematopoietic function. Of note, the delayed platelet recovery after ASCT in patients harboring M-CH clones appears to be consistent with previously published patient series.23 Interestingly, the availability of paired samples collected both at the time of MCL diagnosis and after ASCT allowed us to assess that 7% of patients developed de novo M-CH clones after ASCT. Moreover, as expected,24 we observed a variable evolution of M-CH clone sizes throughout treatment course, with new clones emerging and previously existing clones reducing under the pressure of cytotoxic chemotherapy. Speculatively, it is possible that chemotherapy and ASCT also led to a reduction of M-CH clones, including TP53 below the threshold of detectability, resulting in their disappearance. In addition, neither of the 2 patients with M-CH TP53 clones received lenalidomide after ASCT, which is known to confer a selective advantage to TP53-mutant hematopoietic progenitor cells.25 Notably, we did not observe higher incidence of hematological secondary cancers in patients with M-CH: in detail, only 1 M-CH+ case developed a myelodysplastic syndrome after ASCT. This was driven by the expansion of a DNMT3A clone and the acquisition of an emerging IDH1 mutation, which was not detectable at the time of MCL diagnosis. In agreement to our findings, Eskelund et al15 reported that only 1 of 4 patients who developed t-MNs in their cohort of 149 patients with MCL was a carrier of a M-CH clone.
Collectively, these data suggest that most of t-MNs appear not to be related to the expansion of M-CH detectable at the time of MCL diagnosis but rather to newly occurrent myeloid clones consequent to genotoxic therapies. However, it is possible that some of the M-CH clones were present at a very low level before the start of chemotherapy and could only be detected by ultra-sensitive sequencing techniques.
The main strengths of this study derive from the large and evenly treated cohort provided by MCL0208 clinical trial and the long follow-up of the study. However, potential limitations are to be considered. One limitation might be related to the flow-based algorithm applied to rule out possible, even minimal, lymphoma contamination in the samples analyzed for M-CH: this issue might have underpowered our observations, especially in the PB in which M-CH analysis was performed on neutrophils separated by Ficoll and, thus, in samples already highly representative of the myeloid lineage. In addition, samples from early-progressing patients are lacking in our series and this might have introduced a bias in the retrospective selection of patients for M-CH analysis.
Taken together, our findings offer novel insights in MCL biology by dynamically characterizing the M-CH landscape in these patients. We propose that large M-CH clones may exert a not-negligible role in lymphoma progression, potentially affecting patient outcome. Further efforts are required to elucidate how M-CH might influence the tumor microenvironment and eventually affect the survival of MCL cells and their response to treatment. In conclusion, this work sheds light onto the emerging role of large M-CH clones in shaping MCL clinical progression, paving the way to new future studies focused on the potential causal mechanisms by which M-CH may influence MCL outcome.
Acknowledgments
The authors thank all patients and investigators involved in MCL0208 clinical trial.
This work was supported by grants from the Leukemia & Lymphoma Society (grant no. MCL 7005-24); Progetto di Ricerca Sanitaria Finalizzata 2021 (RF-2021-12371972, CUP G13C21001540001), Torino, Italy; Fondi di Ricerca Locale, Università degli Studi di Torino, Torino, Italy; Associazione DaRosa, Torino, Italy; the Gilead Fellowship Program 2019, Milano, Italy; Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy (investigator grant no. 20125; AIRC 5 × 1000 project number 21267); Cancer Research UK (C355/A26819); and Fundación Científica de la Asociación Española contra el Cáncer and AIRC under the accelerator award program.
Authorship
Contribution: S.R. and A.G. were equally responsible for conducting the search, assembling and analyzing data, interpreting results, and writing the manuscript; E.G., M.G., and B.A. contributed to molecular analysis and manuscript writing; A.M.C. contributed to results interpretation; A.E. performed propensity score analysis; E.A., V.S., F.C., F.B., B.P., D.V., M.M., A. Pascarella, A. Palmas, C.P., E.L., M.G.C., M. Merli, M.P., C.B., M.B., V.R.Z., and M.G.d.S. provided biological samples for the study, contributed to result interpretation, and reviewed the manuscript; B.B. supervised data analysis and provided feedback on the report; E.R. contributed to next generation analysis and results interpretation; M.L. contributed to the design of the study, supervised the conduction of the research, and contributed to results interpretation and manuscript writing; L.M. and S.F. designed the study and were equally responsible for supervising the conduction of the research, results interpretation, and manuscript revision; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: V.R.Z. serves on the advisory board of Kite/Gilead and Takeda; is a consultant for Roche; received research funding from Kite/Gilead; is a member of the speakers bureau for Janssen, Lilly, and Takeda; and received travel support from BeiGene, Janssen, Roche, and Takeda. M.G.d.S. received research grants from Gilead Sciences and AstraZeneca; serves on the advisory board of Janssen, Roche, Gilead Sciences, Lilly, and Takeda; received institutional payments from Janssen and AbbVie; and received travel support from Roche, AbbVie, Janssen, Gilead, and Takeda. M.L. reports consulting fees from AstraZeneca, BeiGene, Janssen, and Lilly; reports honoraria for lectures, presentations, speakers bureaus, manuscript writing, or educational events from AstraZeneca, BeiGene, Janssen, and Lilly; and reports participation on a data safety monitoring board with Acerta. S.F. is a consultant for Janssen, EUSA Pharma, AbbVie, and Sandoz; serves on the advisory board of Janssen, EUSA Pharma, Recordati, Incyte, Roche, Astra Zeneca, and Italfarmaco; received speaker’s honoraria from Janssen, EUSA Pharma, Recordati, Lilly, BeiGene, Gilead, and Gentili; and received research funding from Gilead, BeiGene, and Morphosys. E.R. owns shares in enGenome s.r.l. The remaining authors declare no competing financial interests.
Correspondence: Simone Ferrero, Department of Molecular Biotechnologies and Health Sciences, University of Torino, Via Genova 3, 10126 Torino, Italy; email: simone.ferrero@unito.it.
References
Author notes
S.R. and A.G. contributed equally to this study.
L.M. and S.F. are joint last authors.
Data generated and/or analyzed during this study are available upon reasonable request from the corresponding author, Simone Ferrero (simone.ferrero@unito.it).
The full-text version of this article contains a data supplement.