Key Points
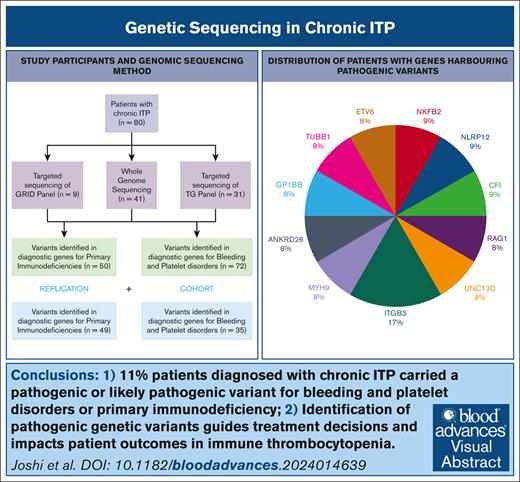
11% patients diagnosed with ITP carried diagnostic grade variants for inherited thrombocytopenia or primary immunodeficiency.
Discovery of pathogenic genetic variants guides treatment decisions and affects patient outcomes in ITP.
Visual Abstract
Immune thrombocytopenia (ITP) is a heterogenous autoimmune disorder diagnosed by excluding other conditions. Misdiagnosis of primary ITP occurs in patients with inherited thrombocytopenia and primary immunodeficiency syndromes. This study investigates whether genetic testing for inherited thrombocytopenia or primary immunodeficiency can enhance diagnostic accuracy in ITP, and guide treatment strategies. We performed whole genome sequencing or targeted panel sequencing on peripheral blood samples in a cohort of 80 participants with chronic ITP, utilizing the ThromboGenomics panel (n = 72) and the Genomics of Rare Immune Disorders panel (n = 50) consisting of genes known to cause bleeding and platelet disorders (BPDG) or primary immuodeficiency genes (PIDG) respectively. A replication cohort of 73 patients underwent clinical genomics testing with either the R90 (BPDG, n = 35) or R15 (PIDG, n = 50) National Health Service Genomics panels. Known pathogenic or likely pathogenic, disease-causing, variants were identified in 9 patients in the first cohort (11%, 95% confidence interval [CI]: 5-20); 7 patients (10%, 95% CI: 4-19) in BPDG and 2 patients (4% CI,1-14) in PIDG. In addition, 26 patients (32.5%) carried variants of uncertain significance. In the replication cohort, 8% (95% CI, 2-20) and 9% (95% CI, 2-23) of patients had a pathogenic variant identified on the R15 (PIDG) or R90 panel (BPDG), respectively. The findings impacted clinical management such as avoidance of immunosuppression (ANKRD26, GP1BB, ETV6, TUBB1, and ITGB3) and eligibility for allogeneic stem cell transplantation (UNC13D). Our findings demonstrate that genomic sequencing identifies diagnostically relevant variants in patients with chronic ITP. Identification of these variants can guide treatment decisions and improve patient outcomes.
Introduction
Immune thrombocytopenia (ITP) is an autoimmune disease of adults and children where reduction in platelet counts results in an increased risk of bleeding. There are no diagnostic tests for ITP, and it is diagnosed through exclusion of other causes of thrombocytopenia. Primary ITP is recognized as platelet count below 100 × 109/L in the absence of any other medical conditions, whereas secondary ITP is associated with autoimmune diseases, infections, malignancy, or primary immunodeficiency syndromes such as common variable immunodeficiency.1
First-line treatment options are steroids and intravenous immunoglobulin and response to these therapies provides further evidence for a diagnosis of ITP. There are many second line treatment options available including thrombopoietin receptor agonists (TPO-RAs), B-cell–depleting therapy, or immune modulators such as SYK inhibitors.2 There is considerable heterogeneity of responses to treatments in patients with ITP and there is an unmet need for a diagnostic test and for prognostic markers.
Some of the heterogeneity of responses could be explained by misdiagnosis and some due to different underlying pathologies. We propose that genetic testing for alternative diagnoses could aid diagnostic accuracy and lead to better treatment stratification.
Inherited thrombocytopenia (IT) is an example of common misdiagnosis; as many as 40% patients with IT are initially misdiagnosed with chronic ITP.3,4 IT results from pathogenic variants in genes implicated in megakaryocyte differentiation and/or platelet formation and clearance. The differentiation of platelets from hematopoietic stem cells involves genes encoding a number of transcription factors and proteins; genetic variants in these genes have been shown to cause IT.5,6 Diagnosis can be made with high throughput sequencing such as the ThromboGenomics (TG) panel test.7 This panel includes 96 diagnostic grade genes curated and approved by the Scientific and Standardisation Committee on Genomics in Thrombosis and Haemostasis of the International Society on Thrombosis and Haemostasis.8 The bleeding and platelet disorder genes (BPDG) on this panel are now available on the R90 panel in the UK National Genomic Test Directory commissioned by the National Health Service (NHS) in England.9
Disorders of immune dysregulation such as common variable immunodeficiency often have accompanying autoimmune manifestations, the most common being ITP.10 Cytopenia typically occurs many years before the immunodeficiency manifests, and constitute the presenting feature in about 10% of patients with primary immunodeficiency (PID).11 The most recent International Union of Immunological Societies expert committee identified 485 genes associated with PID.12,13 Sequencing of these primary immunodeficiency genes (PIDG) to find pathogenic variants can aid in the diagnosis of a PID before other clinical features present.14 These genes have been assessed by external reviewers and the Genomics England clinicians and the majority can be found on the R15 panel in the UK National Genomic Test Directory.15
Here we assess the diagnostic utility of genomic sequencing of BPDG and PIDG in patients with chronic ITP.
Methods
Patients were recruited from one tertiary center managing patients with ITP, from 2017 to 2020. Pediatric and adult cases with chronic ITP, defined as persistent platelet count <100 × 109/L for >12 months were included.16 Depending on the testing available to the clinicians at the time, patients either underwent whole genome sequencing (WGS) or targeted sequencing of the diagnostic grade genes associated with either bleeding and platelet disorders via the TG version 3 panel8 or primary immunodeficiencies via the GRID (Genomics of Rare Immune Disorders) panel.17
WGS was performed either via recruitment to the National Institute for Health and Care Research (NIHR) BioResource Rare Diseases consortium or the 100,000 Genomes Project.18 For the patients who underwent WGS, variants were identified in both sets of genes; the PIDG and the BPDG (full list of genes available in supplemental Material Tables 2 and 3).
Written informed consent to obtain samples for genetic research was obtained from all participants. Ethical approval for both the NIHR BioResource and 100,000 Genomes Project was granted by the East of England Cambridge South National Research Ethics Committee (reference numbers 13/EE/0325 and 14/EE/1112, respectively).
For those sequenced through the NIHR BioResource Rare Diseases consortium, WGS was performed by Illumina. Samples were sequenced to 3 read lengths (100 bp, 125 bp, and 150 bp) over the course of the study to a mean read depth of >35×.18 Variant Call Formats (VCFs) were generated by the Isaac variant caller.19 The VCFs were annotated with variant effect predictions from CellBase software.20 The included variants were splice acceptor variants, splice donor variants, stop gained, frameshift variant, stop lost, start lost, inframe insertion, inframe deletion, nonsynonymous missense variants, and 5′ or 3′ untranslated region variants. From these, all rare (Minor Allele Frequency <0.01 from the gnomAD database) and highly conserved (Combined annotation dependent depletion CADD > 10) variants in the target genes were analyzed to determine pathogenicity as per the American College of Medical Genetics criteria. This included annotation with in silico variant prediction tools to determine the effect of variants on protein structure and searching publicly available databases such as ClinVar for reports of pathogenicity. All variants were classified using the American College of Medical Genetics criteria and discussed in multidisciplinary meetings with clinicians and scientists before determining pathogenicity. Variants confirmed in a single allele for a recessive disease were excluded (n = 5). Variants of uncertain significance (VUSs) in PIDG, in which there was no reported association with thrombocytopenia in the literature, were also excluded (n = 10).
Patients who underwent sequencing via 100,000 Genomes,21 GRID,17 or TG panel7 were subject to their own methods of analysis. All variants were annotated using bioinformatics tools such as Ensembl Variant Effect Predictor; this provides comprehensive information on variant effects including impact on protein function, known disease associations and population frequency. All variants were then discussed in multi-disciplinary teams (MDTs) comprising clinicians, geneticists, and bioinformaticians, to reach a consensus on their clinical significance.
Binomial confidence intervals were used for the proportion of pathogenic/likely pathogenic (LP) variants reported. Statistical analysis for association between variant identification and clinical characteristics was performed with the χ2 test.
All genes for which variants were identified through these sequencing efforts were cross-referenced with the Genomics England PanelApp to confirm their consensus diagnostic grade for the phenotype. This ensures that the variants included in the current Genomics England Test Directory, specifically the R90 (BPDG) or R15 (PIDG) panels, making them available for diagnostic testing in clinical settings.15
Since Autumn 2021, clinical genetic testing became available through NHS England. A cohort of 73 patients with chronic ITP underwent testing with either the R90 or R15 panel and we compared these results to our initial cohort to assess reproducibility of our results.
Results
Participant characteristics
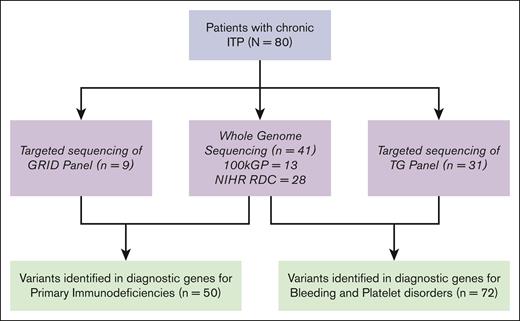
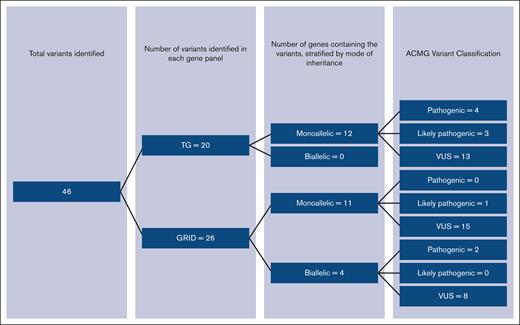
Eighty participants with a diagnosis of chronic ITP were recruited to genetic research studies in the United Kingdom between 2017 and 2020 (26 presenting as children and 54 as adults): 41 underwent WGS through NIHR BioResources or the 100,000 Genomes project; both BPDG and PIDG were analyzed in these participants. Forty participants underwent targeted panel sequencing with either the TG panel (n = 31) testing BPDG or the GRID panel testing PIDG (n = 9). In total, 72 participants underwent variant testing for the BPDG, and 50 participants for the PIDG (Figure 1).
Study participants and genomic sequencing method. 100kGP, 100,000 Genomes Project; NIHR RDC, National Institute of Health Research Rare Diseases Consortium.
Study participants and genomic sequencing method. 100kGP, 100,000 Genomes Project; NIHR RDC, National Institute of Health Research Rare Diseases Consortium.
The clinical phenotype of participants was heterogenous with a mixture of patients with both treatment-refractory phenotypes and relapsing-remitting disease (characteristics presented in Table 1). In total, 11.3% (95% confidence interval [CI], 5.3-20.3) had a pathogenic or LP variant identified within our cohort. A further 32.5% had a VUS (Table 2). Figure 2 shows the variants identified stratified by sequencing method, variants were distributed across the sequencing methods, highlighting the clinical utility of all sequencing techniques.
Participant characteristics: separated by those who underwent sequencing for GRID genes vs TG genes (note overlap as many underwent sequencing of both gene panels)
| . | GRID genes cohort (n = 50) . | TG genes cohort (n = 72) . | All patients (N = 80) . |
|---|---|---|---|
| Adults, n (%) | 35 (70.0) | 53 (73.6) | 54 (67.5) |
| Age at diagnosis, y | 30.0 (1-68) | 31.2 (1-75) | 29.5 (1-75) |
| Male, n (%) | 22 (44.0) | 32 (44.4) | 37 (46.3) |
| White, n (%) | 23 (46.0) | 34 (47.2) | 37 (46.3) |
| Black, n (%) | 5 (10.0) | 7 (9.7) | 7 (8.8) |
| Asian, n (%) | 8 (16.0) | 14 (19.4) | 16 (20.0) |
| Ethnicity not stated, n (%) | 8 (16.0) | 11 (15.3) | 13 (16.3) |
| Other/mixed ethnicity, n (%) | 6 (12.0) | 6 (8.3) | 7 (8.8) |
| Average platelet count nadir, ×109/L | 16.9 | 22.4 | 22.7 |
| Median number of treatment lines | 3.0 | 2.7 | 2.7 |
| Never required treatment, n (%) | 6 (12.0) | 17 (23.6) | 20 (25.0) |
| Complete response to treatment (platelet count >100 × 109 /L), n (%) | 30 (60) | 32 (44.4) | 34 (42.5) |
| Multirefractory phenotype, n (%) | 2 (4.0) | 5 (6.9) | 6 (7.5) |
| Treatment-free response (platelet count >100 × 109 /L more than 1 year since treatment), n (%) | 16 (32.0) | 21 (29.2) | 23 (28.8) |
| Previous splenectomy, n (%) | 7 (14.0) | 11 (15.2) | 13 (16.3) |
| Affected family member, n (%) | 3 (6.0) | 10 (13.9) | 11 (13.8) |
| Autoimmune neutropenia, n (%) | 9 (18.0) | 8 (11.1) | 11 (13.8) |
| Autoimmune hemolytic anemia (Evan syndrome), n (%) | 4 (8.0) | 5 (6.9) | 6 (7.5) |
| Antinuclear antigen positivity, n (%) | 14 (28.0) | 19 (26.4) | 20 (25.0) |
| Presence of other autoimmune condition, n (%) | 18 (36.0) | 25 (34.7) | 28 (35.0) |
| . | GRID genes cohort (n = 50) . | TG genes cohort (n = 72) . | All patients (N = 80) . |
|---|---|---|---|
| Adults, n (%) | 35 (70.0) | 53 (73.6) | 54 (67.5) |
| Age at diagnosis, y | 30.0 (1-68) | 31.2 (1-75) | 29.5 (1-75) |
| Male, n (%) | 22 (44.0) | 32 (44.4) | 37 (46.3) |
| White, n (%) | 23 (46.0) | 34 (47.2) | 37 (46.3) |
| Black, n (%) | 5 (10.0) | 7 (9.7) | 7 (8.8) |
| Asian, n (%) | 8 (16.0) | 14 (19.4) | 16 (20.0) |
| Ethnicity not stated, n (%) | 8 (16.0) | 11 (15.3) | 13 (16.3) |
| Other/mixed ethnicity, n (%) | 6 (12.0) | 6 (8.3) | 7 (8.8) |
| Average platelet count nadir, ×109/L | 16.9 | 22.4 | 22.7 |
| Median number of treatment lines | 3.0 | 2.7 | 2.7 |
| Never required treatment, n (%) | 6 (12.0) | 17 (23.6) | 20 (25.0) |
| Complete response to treatment (platelet count >100 × 109 /L), n (%) | 30 (60) | 32 (44.4) | 34 (42.5) |
| Multirefractory phenotype, n (%) | 2 (4.0) | 5 (6.9) | 6 (7.5) |
| Treatment-free response (platelet count >100 × 109 /L more than 1 year since treatment), n (%) | 16 (32.0) | 21 (29.2) | 23 (28.8) |
| Previous splenectomy, n (%) | 7 (14.0) | 11 (15.2) | 13 (16.3) |
| Affected family member, n (%) | 3 (6.0) | 10 (13.9) | 11 (13.8) |
| Autoimmune neutropenia, n (%) | 9 (18.0) | 8 (11.1) | 11 (13.8) |
| Autoimmune hemolytic anemia (Evan syndrome), n (%) | 4 (8.0) | 5 (6.9) | 6 (7.5) |
| Antinuclear antigen positivity, n (%) | 14 (28.0) | 19 (26.4) | 20 (25.0) |
| Presence of other autoimmune condition, n (%) | 18 (36.0) | 25 (34.7) | 28 (35.0) |
Numbers of patients with confirmed pathogenic, LP, or VUS from the high throughput sequencing
| BPDG, n = 72 . | |||||
|---|---|---|---|---|---|
| . | Pathogenic . | LP . | VUSs . | ||
| Patients (%) | 4 (6) | 3 (4) | 11 (15) | ||
| Monoallelic variant genes | ANKRD26 GP1BB TUBB1 ETV6 | VWF ETV6 ANKRD26 | ITGA2B ANKRD26 TUBB1 | RUNX1 MECOM TBXA2R | GP1BA SLFN14 ITGB2 |
| PIDG, n = 50 | |||||
| Pathogenic | LP | VUSs | |||
| Patients (%) | 1 (2) | 1 (2) | 15 (30) | ||
| Monoallelic variant genes | NOD2 | TLR3 CASP10 PLCG2 TINF2 | NLRP12 NOD2 NFKB2 STAT3 | PARN CTLA4 PIK3CD | |
| Biallelic variant genes | UNC13D | DOCK8 STXBP2 | LRBA | ||
| BPDG, n = 72 . | |||||
|---|---|---|---|---|---|
| . | Pathogenic . | LP . | VUSs . | ||
| Patients (%) | 4 (6) | 3 (4) | 11 (15) | ||
| Monoallelic variant genes | ANKRD26 GP1BB TUBB1 ETV6 | VWF ETV6 ANKRD26 | ITGA2B ANKRD26 TUBB1 | RUNX1 MECOM TBXA2R | GP1BA SLFN14 ITGB2 |
| PIDG, n = 50 | |||||
| Pathogenic | LP | VUSs | |||
| Patients (%) | 1 (2) | 1 (2) | 15 (30) | ||
| Monoallelic variant genes | NOD2 | TLR3 CASP10 PLCG2 TINF2 | NLRP12 NOD2 NFKB2 STAT3 | PARN CTLA4 PIK3CD | |
| Biallelic variant genes | UNC13D | DOCK8 STXBP2 | LRBA | ||
All pathogenic results were confirmed by Sanger sequencing and discussed in MDTs with appropriate clinicians before feeding the results back to the patients. LP findings fulfill the American College of Medical Genetics (ACMG) criteria for this category, but not all have been confirmed by Sanger sequencing and therefore, have not been reported clinically. All VUS categorizations fulfill ACMG criteria but the sequencing is not of clinical standard (except for patients sequenced by 100,000 Genomes in which VUS were reported back to clinicians and patients).
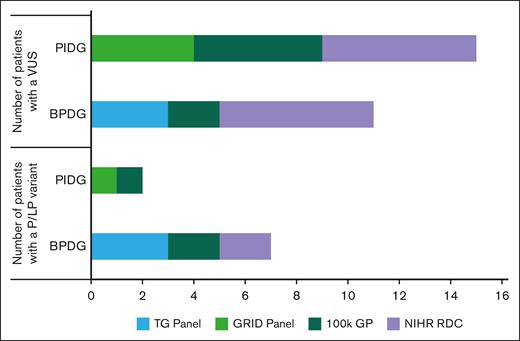
Patients with identified variants, stratified by sequencing method. 100kGP, 100,000 Genomes Project; NIHR RDC, National Institute of Health Research Rare Diseases Consortium; P, pathogenic.
Patients with identified variants, stratified by sequencing method. 100kGP, 100,000 Genomes Project; NIHR RDC, National Institute of Health Research Rare Diseases Consortium; P, pathogenic.
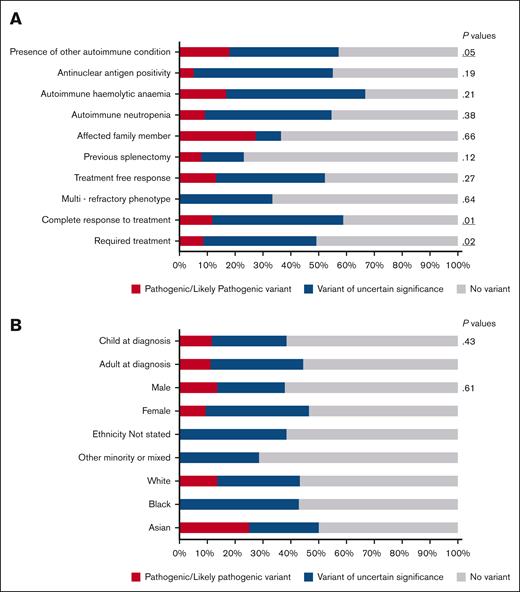
The demographics of the patients identified to have pathogenic variant or VUS are shown in Figure 3B. There were no age or sex differences; patients with variants were distributed across demographics of age and sex (Figure 3). Patients of mixed ethnicities had less variants identified than those of White, Black, or Asian ethnicity (Figure 3B).
Variant association with clinical and demographic variables. (A) Clinical characteristics separated by cohort of those with a variant identified and those without. Clinical characteristics include P values for χ2 test of association. (B) Variant detection stratified by demographic variables.
Variant association with clinical and demographic variables. (A) Clinical characteristics separated by cohort of those with a variant identified and those without. Clinical characteristics include P values for χ2 test of association. (B) Variant detection stratified by demographic variables.
When comparing clinical characteristics, more variants were identified in patients who respond to treatment, rather than multirefractory patients (Figure 3A). Patients more likely to have a variant were those who had a complete response to treatment with a platelet count of >100 × 109/L (P = .01), patients who require treatment at some point during the disease course (P = .02) and patients with another autoimmune condition (P = .05). The presence of family history, antinuclear antigen positivity or another autoimmune cytopenia were not significantly associated with a variant (Figure 3A).
BPDG
A pathogenic or LP variant was identified in 10% (CI, 4-19) of patients tested for BPDG (Figure 4). Four patients had a heterozygous pathogenic variant (ANKRD26, ETV6, GP1BB, and TUBB1) and 3 patients had heterozygous LP variant (ANKRD26, ETV6, and VWF; Table 2). All variants identified were monoallelic (Figure 5).
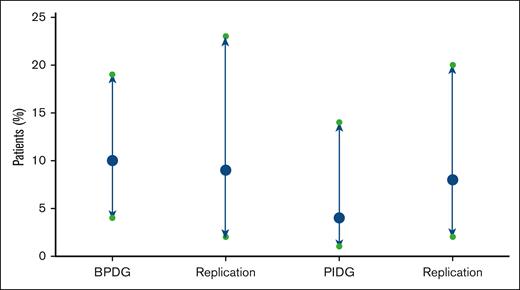
Proportion of patients (%) with pathogenic or LP variants identified with binomial CIs in the test and replication cohorts.
Proportion of patients (%) with pathogenic or LP variants identified with binomial CIs in the test and replication cohorts.
Distribution of variants identified by gene and ACMG classification. ACMG, American College of Medical Genetics.
Distribution of variants identified by gene and ACMG classification. ACMG, American College of Medical Genetics.
Two of the 4 participants with pathogenic variants (GP1BB and TUBB1) had a family history of thrombocytopenia at the time of diagnosis, 1 child and 1 adult (Table 3). Two of the participants with IT had complete responses (platelet count >100 × 109/L) to treatment with TPO-RAs and had no response to immunosuppression, and 3 participants did not have severe enough disease to warrant treatment. The participant with the GP1BB variant resulting in autosomal dominant thrombocytopenia had a partial response (platelet count >30 × 109/L but <100 × 109/L) to TPO-RAs and short-lived response to steroids (Table 3). Diagnosis of Bernard-Soulier disease was established with flow cytometry confirming the lack of CD42b/GP1b. This changed management as the patient was given platelet transfusions for procedures following the molecular diagnosis (Table 3).
Clinical features of patients with identified pathogenic variants
| Gene and mode of inheritance . | Age at diagnosis of ITP, y . | Family history of thrombocytopenia . | Ethnicity . | Platelet count nadir, ×109/L . | Treatment for ITP . | Effect on management . | Reference . |
|---|---|---|---|---|---|---|---|
| ETV6 p.Arg369Trp Dominant missense variant | 47 | No | White | 34 | Limited/no response to splenectomy, azathioprine, steroids Complete response to romiplostim | Patient had gone through many ineffective immunosuppressive agents before romiplostim was licensed. | 22 |
| ANKRD26 c.-116C>G Dominant 5′UTR variant | 30 | No | Asian | 45 | Not required | Give platelets for procedures, monitor for malignancy | 23 |
| GP1BB p.Leu16Pro Dominant missense variant | 1 | Yes (maternal thrombocytopenia in pregnancy) | White | 13 | Short-lived response to steroids, partial response to eltrombopag and romiplostim | Use of HLA-matched platelet transfusions | 24 |
| TUBB1 p.Asn347Lys Dominant missense variant | 62 | Yes (daughter has thrombocytopenia) | White | 62 | Not required | Give platelets for procedures | 7 |
| UNC13D p.Val210Tr[fs]Ter39 p.Leu409Pro Recessive variants: 1 frameshift, 1 missense | 19 | No | White | 3 | Complete response to steroids and Mycophenolate Mofetil | Pathogenic variants only confirmed after patient developed symptoms of HLH aged 26 y (sequencing was re-examined). Diagnosis led to allogeneic stem cell transplant curative treatment. | 25 |
| Gene and mode of inheritance . | Age at diagnosis of ITP, y . | Family history of thrombocytopenia . | Ethnicity . | Platelet count nadir, ×109/L . | Treatment for ITP . | Effect on management . | Reference . |
|---|---|---|---|---|---|---|---|
| ETV6 p.Arg369Trp Dominant missense variant | 47 | No | White | 34 | Limited/no response to splenectomy, azathioprine, steroids Complete response to romiplostim | Patient had gone through many ineffective immunosuppressive agents before romiplostim was licensed. | 22 |
| ANKRD26 c.-116C>G Dominant 5′UTR variant | 30 | No | Asian | 45 | Not required | Give platelets for procedures, monitor for malignancy | 23 |
| GP1BB p.Leu16Pro Dominant missense variant | 1 | Yes (maternal thrombocytopenia in pregnancy) | White | 13 | Short-lived response to steroids, partial response to eltrombopag and romiplostim | Use of HLA-matched platelet transfusions | 24 |
| TUBB1 p.Asn347Lys Dominant missense variant | 62 | Yes (daughter has thrombocytopenia) | White | 62 | Not required | Give platelets for procedures | 7 |
| UNC13D p.Val210Tr[fs]Ter39 p.Leu409Pro Recessive variants: 1 frameshift, 1 missense | 19 | No | White | 3 | Complete response to steroids and Mycophenolate Mofetil | Pathogenic variants only confirmed after patient developed symptoms of HLH aged 26 y (sequencing was re-examined). Diagnosis led to allogeneic stem cell transplant curative treatment. | 25 |
One patient with an ETV6 LP variant was a child whose variant was also identified in a sibling with mild thrombocytopenia and the unaffected mother through cascade testing. This is clinically relevant because if they require treatment for thrombocytopenia they can avoid immunosuppression. Variants in ETV6 are also associated with an increased risk of malignancy, so all carriers can now be monitored. The patient with the LP variant in VWF had more bleeding clinically than would be expected for platelet counts between 30 × 109/L to 50 × 109/L. Unfortunately, he was lost to follow-up and therefore, the pathogenicity of the variant could not be confirmed.
One of the adults with an ANKRD26 associated IT presented with a phenotype typical for this variant (mild chronic thrombocytopenia), the second presented later in life with features typical of ITP and responded to a TPO-RA. This heterogeneity in disease presentation even by different variants in the same gene highlights the difficulty in correlating clinical features with pathogenicity in patients with thrombocytopenia.
A VUS in a BPDG was identified in a further 11 (15%) patients (Table 2). Thirteen different VUSs were identified within these 11 patients (Figure 4); 1 patient had 3 different VUS in MECOM, TBXA1R, and GP1BA. It should be highlighted that these variants require investigation with familial segregation analysis or functional studies to confirm pathogenicity and could just represent benign variation.
PIDG
A pathogenic or LP variant was found in 2 (4% CI, 0.5-14) patients; biallelic variants in UNC13D and a monoallelic variant in NOD2 (Table 2).
One patient originally had only 1 recessive pathogenic variant identified in the PID panel (UNC13D). She presented with ITP at age 16 and responded well to first-line steroids but progressed to relapsing-remitting ITP responsive to mycophenolate. Seven years after her initial diagnosis of ITP, she presented with florid hemophagocytic lymphohistiocytosis (HLH) 6 weeks after her second dose of the AstraZeneca COVID-19 vaccine. The variants were reinvestigated, and Sanger sequencing of the family confirmed compound heterozygous variants in UNC13D, 1 known pathogenic variant, and 1 rare VUS. On further exploration, the VUS segregated with the disease in the family and was therefore reclassified as pathogenic. After 12 months of life-threatening inflammatory disease, she underwent a successful allogeneic hematopoietic stem cell transplant.
One patient with NOD2 variant presented with inflammatory bowel disease at birth, 3 years before the diagnosis of ITP. NOD2 variants are associated with an increased risk of inflammatory bowel disease and autoinflammation.26 This patient also had a VUS in NLRP12, in which dominant pathogenic variants for an autoinflammatory syndrome have been reported.
Twenty-three VUS were identified across 15 patients tested for the PIDG (Figure 4). There were 8 VUS identified with a biallelic mode of inheritance (Figure 4) in 3 genes: STXBP2, DOCK8, and LRBA (Table 2). Phasing of these variants to determine if they are in trans and therefore result in compound heterozygous disease was not performed. The gene with the highest number of identified VUS was PLCG2 (Figure 6). Four (8%) patients had ≥1 variant in different genes identified. These all require further investigation to determine pathogenicity. All variants identified in each patient can be found in the supplemental Material alongside the 100,000 Genomes Project allele frequency, CADD scores, and number of pathogenic ClinVar submissions.
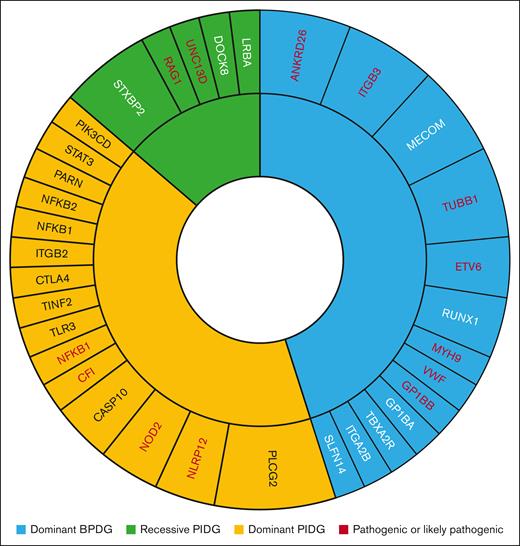
Number of patients (correlates to size of segment) with variants identified per gene and mode of inheritance in test and replication cohorts. Any genes with pathogenic or LP variants are in red font.
Number of patients (correlates to size of segment) with variants identified per gene and mode of inheritance in test and replication cohorts. Any genes with pathogenic or LP variants are in red font.
Replication cohort
A replication cohort (n = 73) was tested to confirm reproducibility of our results (Figure 5). The patients in this cohort underwent testing through the UK NHS Genomic Medicine service with either the R15 PID panel (n = 49) or the R90 bleeding and platelet disorders panel (n = 35) between October 2021 and December 2022. In total 10% (CI, 3.9-19.8) of patients carried a pathogenic or LP variant, compared to 11% (CI, 5.3-20.3) in our original study population. Comparison of the BPDG and PIDG cohort separately can be found in Figure 5. Four patients tested with the R15 panel were found to have pathogenic variants in genes RAG1, NLRP12, NFKB1, and CFI (Figure 6). Three patients had a pathogenic variant identified on the R90 panel in genes MYH9 and ITGB3 (Figure 6).
Discussion
ITP is a heterogenous disease characterized by the absence of specific diagnostic or prognostic tests. This results in misdiagnoses and inappropriate treatments, leaving many patients cycling through multiple treatment types before responding and a subset of patients who remain refractory to all treatments. Because ITP is historically considered an acquired disease, genetic testing is not often performed, apart from screening for inherited disease. For example, genetic sequencing for IT is recommended by the International Consensus guidelines2 for investigation of ITP in children where the thrombocytopenia has been present since early in life or there is a strong family history.
In this pilot study, we demonstrate that genomic sequencing in adults and children with chronic ITP has influenced clinical management in >10% patients, by identifying pathogenic or LP variants. These findings corrected previous misdiagnoses of IT and revealed cases of ITP secondary toPID. Treatment responsive disease and the presence of other autoimmune conditions were associated with variant identification, indicating that these patients should be prioritized for genetic sequencing. This confirms that the presence of an underlying PID does not correlate with more refractory ITP.
Multiple variants were identified in 6% of the patients. This highlights how much there is still to understand about the multifactorial nature of ITP and the complex interaction between genetic variants and the environment.27 A limitation of whole blood sequencing is its inability to detect somatic variation potentially contributing to the disease, such as clonally expanded CD8 T cells recently implicated in ITP pathogenesis.28 Autoimmune diseases have a complex etiology, highlighted by the large number of genomic loci identified by Genome Wide Association Studies (GWAS).29 The interplay between genetic variation, cellular phenotype, immunophenotype, and presentation of the disease is reflected in the variety of phenotypes in patients with ITP.27 GWAS in ITP are very limited, and only 1 reported 3 significant loci associated with ITP, but none of these were within the coding genome.30 Gene association studies have reported variants in CTLA4 and TUBB1 to be associated with ITP.31,32
The discovery of pathogenic variants in genes like RUNX1, ETV6, and ANKRD26 indicates a specific syndrome of thrombocytopenia with a predisposition to hematological malignancies.33 For these patients, their disease was misdiagnosed as ITP and therefore, future management was modified to include monitoring of blood counts, along with prompt investigation of results outside normal parameters. Many of these patients had been treated with TPO-RAs, despite limited evidence supporting their use in IT. A recent literature review identified sparse clinical trials or case reports that demonstrated the use of TPO-RAs in ITs. While TPO-RAs appear effective for bridging compared to surgery or stem cell transplantation in some cases, their use in conditions predisposing to myelodysplastic syndromes or malignancy requires caution due to potential risks of accelerated disease progression and thrombosis.34 Given these findings, association between response to TPO-RAs and IT should be investigated in future studies.
All patients receive counseling regarding cascade testing of family members. This highlights the importance of informed consent before carrying out these tests. Patients must understand the implications of finding pathogenic variants both for themselves and their families, and this has led to guidance for clinicians on informed consent from the International Society of Thrombosis and Haemostasis.35 As genetic testing becomes more common in the diagnostic workup of ITP, clinicians must be appropriately trained to obtain consent, informing patients of these risks. Another ethical consideration is the appropriate age for testing children, given that the results may have lifelong implications.
The TG panel demonstrated a diagnostic yield of 47.8% in patients with thrombocytopenia,7 higher than our diagnostic yield of 25% (total number of patients with a pathogenic or LP variant or detected VUS). This discrepancy likely reflects differences in patient phenotypes, with the TG study focusing on patients with macrothrombocytopenia or IT features, whereas our cohort consisted of chronic ITP patients. Nonetheless, our finding that 10% of patients had an underlying IT indicates the utility for this test in the ITP diagnostic pathway. Variants in the BPDG such as TUBB1 have previously been shown to affect response to ITP treatment,32 and given the number of VUS identified in these genes, the association with treatment response should be prioritized for future research.
The identification of PIDG, such as those implicated in post-vaccine HLH and variants in NOD2, further illustrates the role of genetics in ITP pathogenesis. ITP is an unusual presentation of HLH but identifying biallelic variants responsible for familial HLH transformed patient care in 1 case, enabling timely diagnosis and successful allogeneic stem cell transplantation. This case highlights that ITP can also present in other primary immunodeficiencies, not just common variable immunodeficiency. The patient with the NOD2 mutation, who also had inflammatory bowel disease but lacked other NOD2-associated symptoms such as synovitis, exemplifies the incomplete penetrance often observed in single-gene disorders.36
Genomics England have developed gene panels for both bleeding and platelet disorders and PID which are curated based on multiple external reviewers’ consensus on the evidence for pathogenicity and include genes from historical gene panels such as the TG panel and GRID panel.15 These panels are now accessible to all NHS patients during diagnostic workup and this raises the questions of when to perform genetic testing as part of investigation of a patient with ITP. In the investigation of rare inherited anemias, the diagnostic yield is >50% and these are recommended during investigation of rare anemias in the latest guidelines.37 Conversely, new guidelines in the investigation of Evans syndrome38 have discouraged the use of molecular studies for primary immunodeficiencies unless there is specific clinical suspicion.
This study provides evidence supporting the utility of genetic diagnosis in patients with ITP, although the criteria for selecting patients who would benefit most, is still unclear. Given the high number of VUSs identified, further research is required to understand the role of these variants in the pathogenesis of ITP. As more patients with VUSs are identified, multidisciplinary team meetings involving clinicians, geneticists, and bioinformaticians will be crucial for interpreting results and determining the implications for clinical management.
Conclusion
This study shows that genetic diagnosis can significantly improve patient outcomes and guide treatment decisions for individuals with chronic ITP. Our findings identified a rare pathogenic or LP variant in 11% of patients and at least 1 VUS in a further 32.5% of patients. These results emphasize the pivotal role of genomic sequencing in uncovering misdiagnosed IT or PID among patients with chronic ITP and therefore, in improving patient outcomes. However, the high prevalence of VUSs highlights the need for further research to elucidate their possible role in disease pathogenesis. Future efforts should focus on integrating genomic data with functional studies to better understand the molecular mechanisms underlying ITP. By refining targeted sequencing approaches, we can enhance diagnostic accuracy and provide more personalized and effective treatment strategies for patients with chronic ITP.
Acknowledgments
The authors thank National Institute for Health and Care Research (NIHR) BioResource volunteers for their participation, and acknowledge NIHR BioResource centers and National Health Service (NHS) Trusts and staff for their contribution. The authors thank the NIHR, NHS Blood and Transplant, and Health Data Research UK as part of the Digital Innovation Hub Programme. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care).
The 100,000 Genomes Project is funded by the NIHR and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the NHS as part of their care and support. The research was supported by the Imperial NIHR Biomedical Research Centre.
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
The open access fee was paid by the Imperial College London Open Access Fund.
Authorship
Contribution: N.J. designed the study, collected, and analyzed the data, interpreted results, and wrote the manuscript; N.C. designed the study, interpreted results, and reviewed the manuscript; H.L.-A., K.D., and I.S. performed bioinformatic analysis and manuscript review; C.V. and D.P. collected samples and clinical data; and A.H. and C.A. collected samples and clinical data for the replication cohort.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nehal Joshi, Centre for Haematology, Department of Immunology and Inflammation, Imperial College London, Commonwealth Building, Hammersmith Hospital Campus, London W12 0NN, United Kingdom; email: nehal.joshi16@imperial.ac.uk.
References
Author notes
Original sequencing data may be accessed upon reasonable request to the National Institute for Health and Care Research (NIHR) BioResource (dac@bioresource.nihr.ac.uk). The genotype data of all the NIHR BioResource participants are available from the European Genome-phenome Archive at the European Molecular Biology Laboratory European Bioinformatics Institute (accession number EGAD00001004519).
The full-text version of this article contains a data supplement.