Key Points
We assess how clinical factors (age, sex, platelet count) and training on other cancers influence machine-learning OC detection.
We have developed a highly sensitive machine-learning model dedicated to early OC detection.
Visual Abstract
Ovarian cancer (OC) presents a diagnostic challenge, often resulting in poor patient outcomes. Platelet RNA sequencing, which reflects host response to disease, shows promise for earlier OC detection. This study examines the impact of sex, age, platelet count, and the training on cancer types other than OC on classification accuracy achieved in the previous platelet-alone training data set. A total of 339 samples from healthy donors and 1396 samples from patients with cancer, spanning 18 cancer types (including 135 OC cases) were analyzed. Logistic regression was applied to verify our classifiers’ performance and interpretability. Models were tested at 100% specificity and 100% sensitivity levels. Incorporating patient age as an additional feature along with gene expression increased sensitivity from 68.6% to 72.6%. Models trained on data from both sexes and on female-only data achieved a sensitivity of 68.6% and 74.5%, respectively. Training solely on OC data reduced late-stage sensitivity from 69.1% to 44.1% but increased early-stage sensitivity from 66.7% to 69.7%. This study highlights the potential of platelet RNA profiling for OC detection and the importance of clinical variables in refining classification accuracy. Incorporating age with gene expression data may enhance OC diagnostic accuracy. The inclusion of male samples deteriorates classifier performance. Data from diverse cancer types improves advanced cancer detection but negatively affects early-stage diagnosis.
Introduction
Ovarian cancer (OC) represents a formidable challenge in oncology because of its asymptomatic nature in the early stages, leading to late diagnoses and a mere 5-year overall survival of 50%.1 The absence of effective screenings for average-risk, asymptomatic patients further hinders earlier detection, underlining the critical need for innovative detection methods.2-4 The most specific biomarker for OC, CA-125, has been tested as a screening tool, however, it is not applicable in this setting because of relatively low sensitivity in early OC and a high false-positivity rate, as CA-125 increased levels accompany other conditions, for example, menstruation.5
Recent advancements in liquid biopsies have brought attention to platelets as changes in platelet RNA profiles are detectable at early stages, even during the asymptomatic phases of the disease.6,7 Platelet RNA, in conjunction with artificial intelligence has emerged as a high-accuracy diagnostic tool for OC.8-13 Our recent work showed 76.5% sensitivity at a 99% specificity threshold in OC detection by using logistic regression (LogReg).14 In Gao et al we reported that combining platelet RNA and CA-125 marker resulted in the highest diagnostic accuracy.7 When analyzing platelet RNA profiles alone, a receiver operating characteristic area under the curve (ROC AUC) reached 0.922 in overall OC detection, and 0.879, 0.885, and 0.926 in detecting early-stage, borderline, and nonepithelial ovarian malignancies, respectively.7 The consistency of platelet transcriptome across healthy donors enables precise platelet profiling in disease.15,16
Our study addresses the urgent need for effective OC screening by developing tailored classifiers for distinct demographics. We created 1 classifier optimized for broad population screening with high specificity and another for high-risk groups with high sensitivity. We analyzed factors such as age, sex, and inclusion of other cancer types to assess their impact on classifier performance, aiming to improve sensitivity and specificity. Additionally, we explored the classifiers’ potential to enhance early OC detection through gene ontology (GO) analysis, identifying key transcripts influencing the machine learning models’ decisions.
Materials and methods
Study cohorts
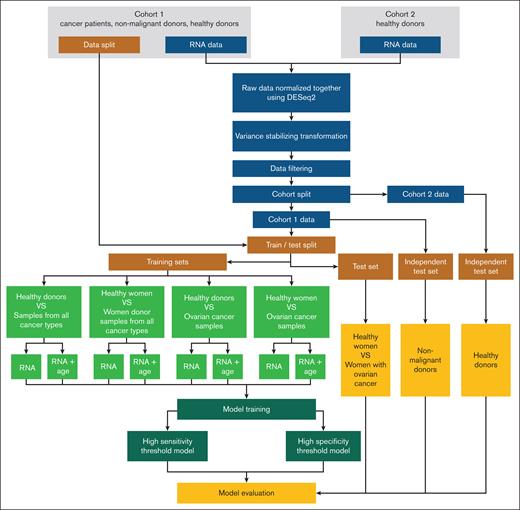
In this study, we used platelet RNA expression data from our previous data set (cohort 1).11 Platelet samples of healthy donors (n = 88 from 38 donors) were derived from the study of Rondina et al and served as an independent data set (cohort 2).16 Cohort 1 comprised a total of 2351 samples from various institutions. It included 390 samples from healthy donors; 144 samples from OC; 1484 samples from patients with other types of cancer (17 cancer types); and 333 samples from donors with nonmalignant conditions, for example angina pectoris, bowel diseases, epilepsy, multiple sclerosis, or pulmonary hypertension (used as an additional independent test set). Samples collected from 1 institution (n = 291) were excluded because of likely laboratory technical errors resulting in hemolysis, platelet activation, and, thus, clotting. Detailed analysis of these samples is presented in Jopek et al.17 Samples with undisclosed donor sex (n = 19) and age (n = 7) were excluded from specific experiments because this information was crucial for training purposes. We also verified how platelet count might affect OC detection, however, the platelet count information was only available for patients recruited at the Medical University of Gdańsk. Hence, platelet count was not included in the main experiments. The assay from cohort 1 originally covered 5346 genes, but not all were present in cohort 2. To ensure consistency and enable robust analyses, we focused on the 5277 genes available in both data sets. Four different modes of training were used: (1) incorporating age as one of the features during training versus training on expression profiles only; (2) training on females only versus training on both sexes; (3) training on OC cases only vs training on all available cancer types; (4) training on females and OC cases vs using data from all available samples. Detailed information on sample numbers and the composition of training and test sets is provided in Table 1.
Sample composition of the training and test sets
| Studied impact . | Performed comparison . | Training set (n) . | Test set (n) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HD . | OC . | Other cancers . | HD . | OC . | |||||||
| M . | F . | M . | F . | M . | F . | M . | F . | M . | F . | ||
| Impact of age | Samples without age as a separate feature | 158 | 110 | – | 33 | 686 | 575 | – | 71 | -– | 102 |
| Samples with age incorporated as a separate feature | 157 | 110 | – | 33 | 682 | 573 | |||||
| Impact of sex | Samples from all cases | 158 | 110 | – | 33 | 686 | 575 | -– | 71 | -– | 102 |
| Samples collected from females only | -– | 110 | -– | 33 | -– | 575 | |||||
| Impact of inclusion of other cancer types | All types of cancer | 158 | 110 | -– | 33 | 686 | 575 | -– | 71 | -– | 102 |
| Only OC cases | 158 | 110 | -– | 33 | -– | – | |||||
| Impact of combination of sex and cancer type | All samples | 158 | 110 | -– | 33 | 686 | 575 | -– | 71 | -– | 102 |
| Only OC and women raining | -– | 110 | -– | 33 | -– | -– | |||||
| Studied impact . | Performed comparison . | Training set (n) . | Test set (n) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HD . | OC . | Other cancers . | HD . | OC . | |||||||
| M . | F . | M . | F . | M . | F . | M . | F . | M . | F . | ||
| Impact of age | Samples without age as a separate feature | 158 | 110 | – | 33 | 686 | 575 | – | 71 | -– | 102 |
| Samples with age incorporated as a separate feature | 157 | 110 | – | 33 | 682 | 573 | |||||
| Impact of sex | Samples from all cases | 158 | 110 | – | 33 | 686 | 575 | -– | 71 | -– | 102 |
| Samples collected from females only | -– | 110 | -– | 33 | -– | 575 | |||||
| Impact of inclusion of other cancer types | All types of cancer | 158 | 110 | -– | 33 | 686 | 575 | -– | 71 | -– | 102 |
| Only OC cases | 158 | 110 | -– | 33 | -– | – | |||||
| Impact of combination of sex and cancer type | All samples | 158 | 110 | -– | 33 | 686 | 575 | -– | 71 | -– | 102 |
| Only OC and women raining | -– | 110 | -– | 33 | -– | -– | |||||
F, female; HD, healthy donors; M, male.
Approval was obtained at ethics committee of units participating in this study.
Blood processing
For cohort 1, whole blood was collected into EDTA Vacutainer tubes and then processed according to the thromboSeq protocol.18 RNA-sequencing data are available at Gene Expression Omnibus under accession number GSE183635.11 For cohort 2, a streamlined process was implemented to ensure the efficiency of the RNA sequencing. Blood samples were obtained via venipuncture, with citrate tubes (subcohort 1) or acid-citrate-dextrose (subcohort 2) used for collection. Within 30 minutes of phlebotomy, sample processing was initiated. Platelets were isolated using anti-CD45 antigen microbeads (Miltenyi) via the magnetic leukocyte depletion method. RNA extraction was performed using phenol-chloroform extraction (subcohort 1) or a Direct-zol kit (subcohort 2, Zymogen). Subcohorts 1 and 2 were prepared for barcoded sequencing libraries using KAPA Stranded mRNA-Seq Kit (Roche number KK8421) and TruSeq unstranded v2 with poly(A) selection (Illumina number RS-122), respectively. This procedure allowed for a high throughput of samples. Illumina HiSeq 4000 (subcohort 1) or Illumina HiSeq 2000 (subcohort 2) machines were used to sequence libraries with 50 cycles, in a single-end manner, with a depth ranging from 20 to 40 million mapped reads per sample. Fastq files have been deposited in the National Institutes of Health Sequence Read Archive: PRJNA531691.16
Bioinformatic analysis
We combined and then normalized both cohorts using the variance stabilizing transformation by using the DESeq2 package in R.19 The normalization process accounted for the inclusion of various data sources. The human reference genome (hg19) was an annotative reference point in this process. To maintain the integrity and quality of our data set, we omitted samples with fewer than 100 000 total reads and solely incorporated genes backed by confirmed GENCODE status. All samples underwent uniform preprocessing.
Model training
To develop an OC diagnostic tool, we used various machine learning algorithms: random forest, balanced random forest, and XGBOOST. Eventually, we used LogReg, which outperformed all other tested methods in terms of detection accuracy while maintaining the interpretability of the prediction. Detailed model comparison metrics are available in the supplemental Material (Table “Model metrics”). The high-specificity threshold models (meant for OC screening) were tested at a 100% specificity threshold, and the high-sensitivity models were tested at a 100% sensitivity threshold at the test set. While training, we evaluated the impact of various clinical factors on the performance metrics of our classifiers: patient age, patient sex, platelet count, and the incorporation of 16 cancer types other than OC within the training data set. Patient age was included in models as an additional, continuous feature alongside RNA expression levels. The simplified workflow of the performed experiments is presented in Figure 1.
Hyperparameters for all models were fine-tuned using a grid search approach implemented through GridSearchCV, focusing on optimizing ROC AUC. Furthermore, to ensure the reliability of our findings, models were trained using 3 different random seeds. Evaluation of model performance was conducted using a range of metrics, including specificity, sensitivity, accuracy, balanced accuracy, F1 score (harmonic mean of the precision and recall), and ROC AUC. To estimate the precision of these metrics, we used the bootstrap method to calculate 95% confidence intervals. Implementation of the machine learning algorithms and analysis was facilitated using Python (version 3.10) along with essential packages such as scikit-learn (1.2.0), xgboost (1.7.6), imbalanced-learn (0.11.0), numpy (1.22.4), and pandas (1.2.4).20-22 These tools provided the necessary framework for model construction, optimization, and assessment, ensuring the robustness and reproducibility of our findings.
GO analysis
Next, we focused on elucidating the most significant features identified by the LogReg classifier. The analysis aimed to verify whether the model’s predictions are grounded in biologically meaningful features, rather than irrelevant patterns, which could compromise its performance when applied to new data. To achieve this, we conducted pathway analysis on the top 200 variables exhibiting the highest absolute values of model coefficients. We used the enrich GO function from the clusterProfiler package (version 4.10.0) of the R programming language (version 4.3.2).23,24 This function provided insights into the number of genes associated with each pathway among the selected features in relation to the total number of analyzed genes from the GO database. Additionally, it generated P values to assess the statistical significance of gene enrichment within each pathway. Such analyses facilitated the identification of the biological processes in which the most impactful variables for classification are implicated.
Results
Consideration of platelet count
One of the clinical factors that should be considered in platelet-based biopsy is the platelet count of blood donors. Platelet count is generally higher in women than in men. It decreases with age in both sexes and is known to increase in advanced OC.25-27 Because of the unavailability of platelet count data for cases recruited from institutions other than the Medical University of Gdańsk, we were unable to proceed with model training. To address this limitation, we instead examined the correlation between platelet count and messenger RNA content. Specifically, we conducted comparative analyses between platelet count and various factors, including library size, overall gene expression, translation-related gene expression, and activation-related gene expression (supplemental Material, Table “Training with platelet count”; supplemental Figure 1). Our analysis showed no significant correlation between library size and platelet count (Pearson −0.26; P = .11). This suggests that the total RNA sequenced is independent of platelet count, indicating that RNA content may not scale directly with platelet numbers. Similarly, when we analyzed the general expression level (transcript reads with expression of >0), no statistically significant link was observed (Pearson −0.29; P = .07). Next to general expression, we also explored translation-related genes as our GO analysis indicated their critical role in sample classification. In this particular case, we observed random distribution, with Spearman of −0.02 (P = .9). We were also interested in platelet activation markers (CD40L, B-cell lymphoma 3-encoded protein (BCL3), glycoprotein IIb/IIIa (GPIIb/IIIa), platelet factor 4 (PF4), beta-thromboglobulin (β-TG), P-selectin, integrins αIIbβ3, transforming growth factor β [TGF-β], vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), basic fibroblast growth factor (bFGF), Toll-like receptor 4, and glycoprotein VI [GPVI]). We observed a trend for a higher degree of platelet activation in patients with high platelet count (Pearson of 0.3; P = .07).
Impact of sex
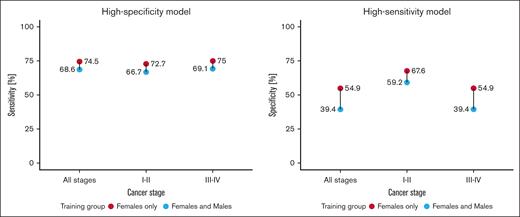
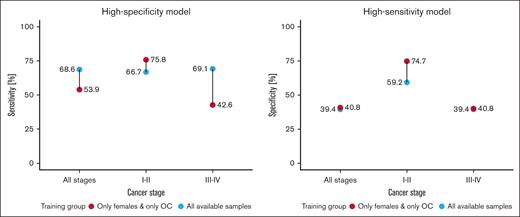
In the next step, we developed training models adjusted to high sensitivity and high specificity across samples collected from women only, as opposed to the data set composed of both sexes. A more specialized data set allowed the model to capture female-specific differences in gene expression, yielding consistently higher results across both high-specificity and high-sensitivity threshold models (Figure 2). Excluding men from the training process increased the model accuracy irrespective of the disease stage. Remarkably, training models on women samples only, also increased the accuracy (from 66.7% to 87.5%) when classifying samples from cohort 2. Similar results were also observed in the high-sensitivity model, in which the exclusion of male samples from the training set boosted the accuracy from 4.2% up to 15.9% in a decision threshold optimized for early cancer detection. The classification accuracy of samples collected from an independent test set of nonmalignant disease donors slightly dropped from 80.6% to 76.5% for the high-specificity model but increased from 7.6% to 20.6% for the high-sensitivity model. A direct comparison of the top 200 features with the highest coefficient importance score between models trained on data from both sexes and only on women revealed a 36.5% intersection, proving that there are differences between RNA expression between men and women, when it comes to cancer profile (supplemental Material, Table “Model features”).
Comparison of cohort 1 classification performance for LogReg model trained with and without males, across early (I-II) and late stages (III-IV). High-specificity model displays sensitivity at 100% specificity, and high-sensitivity model displays specificity as 100% sensitivity.
Comparison of cohort 1 classification performance for LogReg model trained with and without males, across early (I-II) and late stages (III-IV). High-specificity model displays sensitivity at 100% specificity, and high-sensitivity model displays specificity as 100% sensitivity.
Impact of inclusion of other cancer types
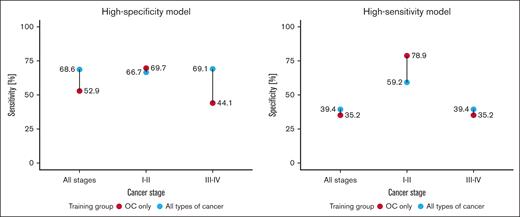
Training of high-specificity models only on samples from OC vs from all cancer types showed that the former generally scored much lower in overall OC detection sensitivity but performed higher in early-stage OC detection (Figure 3). Restricting patients with cancer in the training data set to OC also increased the classification accuracy of samples from nonmalignant controls for a high-specificity threshold model from 80.6% to 91.8% and samples from cohort 2 from 66.7% to 89.9%. In the high-sensitivity threshold models, in which the early-stage specificity was the highest of all experiments, restricting the training data set to samples only from OC decreased the accuracy of nonmalignant controls from 7.6% to 15.8%, and cohort 2 samples from 4.2% to 6.7%. A direct comparison of the top 200 features of the highest coefficient importance score between both models revealed only a 13% intersection, suggesting that OC has a highly distinct platelet profile.
Comparison of cohort 1 classification performance for LogReg model trained on all available cancer samples and on data only from patients with OC, across early (I-II) and late stages (III-IV). High-specificity model displays sensitivity at 100% specificity; and high-sensitivity model displays specificity as 100% sensitivity.
Comparison of cohort 1 classification performance for LogReg model trained on all available cancer samples and on data only from patients with OC, across early (I-II) and late stages (III-IV). High-specificity model displays sensitivity at 100% specificity; and high-sensitivity model displays specificity as 100% sensitivity.
Optimization for early-stage detection
Refining the insights and data from previous experiments by reducing the training data set to samples only from asymptomatic healthy women and patients with OC resulted in the most accurate and highly specific early-stage OC detection model (Figure 4). This model also exhibited the highest classification accuracy in nonmalignant controls and cohort 2: 92.4% and 88.3%, respectively. The highly sensitive model scored 36.% classification accuracy for cohort 2% and 17.6% for nonmalignant samples.
Comparison of cohort 1 classification performance for LogReg model trained on all available data, and on data limited to healthy female donors and patients with OC, across early (I-II) and late stages (III-IV). High-specificity model displays sensitivity at 100% specificity; and high-sensitivity model displays specificity as 100% sensitivity.
Comparison of cohort 1 classification performance for LogReg model trained on all available data, and on data limited to healthy female donors and patients with OC, across early (I-II) and late stages (III-IV). High-specificity model displays sensitivity at 100% specificity; and high-sensitivity model displays specificity as 100% sensitivity.
Impact of age
Incorporating age as an additional feature into the training set enhanced the performance of the highly specific and highly sensitive models in OC detection across late-stage cancer in the case of the highly specific model, and across all available samples in the case of the highly sensitive model for the cohort 1 test set (Figure 5A). When training models on samples only from women (Figure 5B), the inclusion of age as a feature resulted in an improvement in the highly specific model, with cohort 2 accuracy rising from 87.5% to 91.7%, and symptomatic control accuracy increasing from 76.5% to 79.4%. For samples solely involving OC cases (Figure 5C), the highly specific model saw a notable decrease in performance, with cohort 2 accuracy falling to 68.8% from 87.5%, and symptomatic control accuracy plummeting from 91.8% to 46.6%. For samples belonging to healthy women and OC cases (Figure 5D), the highly specific model maintained robust performance, with cohort 2 accuracy slightly improving from 88.25% to 89.6%.
Comparison of cohort 1 classification performance for LogReg model trained with and without additional information on patient age, across early (I-II) and late stages (III-IV). (A) Data set based on all available samples; (B) data set based on samples only from women; (C) data set based on samples only from OC; (D) data set based on samples only from healthy women and OC cases.
Comparison of cohort 1 classification performance for LogReg model trained with and without additional information on patient age, across early (I-II) and late stages (III-IV). (A) Data set based on all available samples; (B) data set based on samples only from women; (C) data set based on samples only from OC; (D) data set based on samples only from healthy women and OC cases.
The per-stage metrics in Figure 5, show that the inclusion of age did not enhance the model’s performance for early-stage OC detection but increased the model’s ability to classify at late-stage of cancer. Although the incorporation of age generally increases the performance of the classifier, the uneven age distribution among healthy donors and patients with cancer warrants caution when interpreting these particular results (supplemental Material, “Age”).
GO pathway analysis
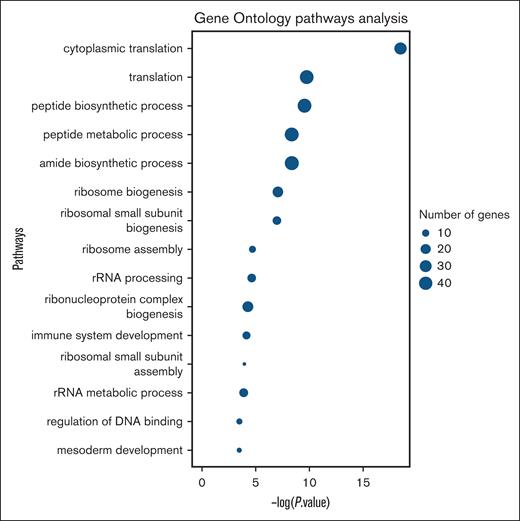
Based on the features (transcripts) that scored the highest importance in early-stage OC detection, we used GO pathway analysis on the dysregulated genes in the analyzed platelet RNA. This allowed us to examine the biological insights of the most accurate model for early-stage OC detection. As outlined in Figure 6, the GO analysis highlights the biological processes with the most significant representation of genes from the top 200 features (transcripts) compared with the entire gene set. The majority of these genes were found to be associated with ribosomal functions, reinforcing the model’s ability to identify biologically relevant markers. Additionally, we separately analyzed the expression of genes in healthy women and patients with OC, sorting them by importance level in our most accurate model (supplemental Material, Table “Gene ontology analysis”). The top-10 significant features included 8 upregulated genes in patients with OC: kinase suppressor of Ras 1 (KSR1), histone deacetylase 8 (HDAC8), NF-κB inhibitor α (NFKBIA), myosin light chain 2 (MYL2), tumor necrosis factor, α-induced protein 3 (TNFAIP3), cathepsin W (CTSW), chromobox protein homolog 7 (CBX7), and 5′-aminolevulinate synthase 2 (ALAS2); and 2 downregulated genes in patients with OC: ribosomal protein L23a (RPL23A), and ribosomal protein S15a (RPS15A). Reticulation marker selectin P (SELP) and platelet activation markers: CD40L, BCL3, GPIIb/IIIa, PF4, β-TG, integrins αIIbβ3, TGF-β, VEGF, PDGF, bFGF, Toll-like receptor 4, and GPVI) with the possible exception of BCL3 (ranked 73rd), all scored below the 200 analyzed genes.
Platelet RNA GO analysis of 200 most impactful transcripts (based on the highest importance score of these 200 features) for the most sensitive early-stage cancer detection model. rRNA, ribosomal RNA.
Platelet RNA GO analysis of 200 most impactful transcripts (based on the highest importance score of these 200 features) for the most sensitive early-stage cancer detection model. rRNA, ribosomal RNA.
Discussion
By assessing the impact of age, sex, and training on various cancer types on the performance of our models, we have provided a comprehensive analysis of the critical role these variables play in enhancing the accuracy and efficacy of platelet-based liquid biopsy diagnostics. By integrating age and RNA expression data, we achieved improved classification results for samples of the late-stage OC but decreased classification accuracy for early-stage OC. In terms of sex, training exclusively on female data allows the model to capture representations of the disease that are more aligned with female physiology, enhancing sensitivity and accuracy in identifying relevant patterns. Although the incorporation of platelet count as a feature remains inconclusive because of limited sample size, we hypothesize that the algorithm trained for early-stage cancer primarily focuses on translation-related transcripts, which appear to be more directly indicative of cancer presence than clinical factors. Furthermore, based on our GO analysis and feature importance, the developed final machine-learning model only slightly accounts for 1 platelet activation–related feature (BCL3) and does not account for platelet reticulation (SELP). Although advanced age is a risk factor in cancer incidence and it has been reported that platelet count decreases with age, its influence on the molecular profile of early-stage OC may be less pronounced.28,29 Our results align with the recent work of Eledkawy et al, in which 39 biomarkers were combined with age, sex, and ethnicity.30 The training data set composition experiments revealed that the inclusion of male samples had an adverse effect on the model’s performance. This validated our hypothesis that sex-specific analysis enhances diagnostic accuracy in female cancers, highlighting differences in platelet RNA expression between sexes. Although these differences are minor, they significantly influence machine learning model development.15 Our experiments also demonstrated that models trained exclusively on OC samples are notably more sensitive in early-stage OC detection than those trained on broader data sets. These results underscore the importance of appropriate training data sets for developing precise diagnostic tools.
When comparing our findings with the existing OC detection methods, we noted significant advancements. The deep learning–based imPlatelet tool by Pastuszak et al exemplifies the potential of platelet-based diagnostics, achieving up to 88% specificity but at a lower (95%) sensitivity (ROC AUC = 0.95).8 In our previous work based on LogReg, we reached 72% sensitivity at a 99% specificity threshold in early-stage OC detection. Gao et al, by combining the platelet RNA and CA-125, reached 85.3% specificity at 86.4% sensitivity across samples from various stages, and 73.2% sensitivity at an 86.0% specificity in the case of early-stage detection in OC classification involving the combination of platelet RNA and CA-125.7 This was not feasible in our case because of the lack of CA-125 marker data for asymptomatic controls.
Another liquid biopsy type used for OC detection is cell-free DNA (cfDNA). Cohen et al developed a blood-based CancerSEEK diagnostic test, which comprised protein biomarker concentrations and cfDNA (circulating tumor DNA), reaching a remarkable 98% OC detection level and 70% mean detection for other cancer types.31,32 However, this test for samples collected from patients with stage I disease showed only a 40% sensitivity. Wong et al33 and Rahaman et al34 achieved 96.6% and 99.2% sensitivity, respectively, at a 99% specificity for cancer detection and ∼99% sensitivity for OC detection using the semi-naïve Bayesian machine-learning model (CancerA1DE) and average 1-dependent estimators model (CancerEMC) on the CancerSEEK data set. The former study used 39 biomarkers and considered patient sex, whereas the latter used only 15 biomarkers combined with age, ethnicity, and sex. Another multicancer screening test, Galleri (GRAIL), scored a 67% OC detection rate at 99% specificity in the early stage of its development.35 The recent Pathfinder study shed more light on how this test could be performed on a broader scale.36,37 Additionally, Katchman et al reported 45% sensitivity at 98% specificity in serous OC using a panel of tumor antigen-associated autoantibodies.38
An in-depth analysis of the models’ performance revealed several key dysregulated genes influencing their decisions. Identifying these features is vital for validating the model’s biological relevance and enhancing its diagnostic accuracy. The dysregulated genes, such as KSR1, HDAC8, and NFKBIA, are closely linked to critical cancer-related processes, proliferation, immune evasion, and tumor progression.15,38-42 Recognizing these genes validates the model’s effectiveness and highlights the importance of feature selection in uncovering biological background of RNA changes.
The identified prominent biological processes are closely linked to ribosomal functionality. Similar ribosomal-related processes in Bongiovanni and Santamaria et al were reported to be characteristic of reticulated platelets.43 Moreover, in Best et al the number of reticulated platelets in patients with non–small cell lung cancer correlated with platelet counts.44 Although we cannot confirm that platelet count and SELP expression (reticulation marker) play a role in our highly sensitive model, we do observe upregulation of genes directly related to cancer and strong downregulation ribosomal genes RPL23A and RPS15A. However, as the RNA content in platelets is shaped by a variety of complex factors, including changed expression in megakaryocytes, direct and indirect interactions between cancer cells and platelets, interactions with the tumor microenvironment, and the dynamic process of RNA uptake and release as platelets circulate through the bloodstream, it is challenging to attribute specific RNAs to individual processes. The final RNA content in platelets likely represents a cumulative outcome of these interactions. Beyond ribosome-related processes, immune system development also plays a key role, particularly in cancer through its roles in immune surveillance and evasion.45
The study has some limitations. First, the lack of publicly available platelet RNA data restricts testing and may affect model robustness and generalizability. Although differences in blood processing exist between cohorts, they likely enhanced model reliability, as consistent performance across data sets suggests robustness to sample variability. Second, including age as a feature may introduce bias because of uneven age distribution between patients with OC and healthy donors, affecting model performance across age groups. Splitting cohorts by age (eg, >55 years) could address this but was not feasible here because of differing age distributions. However, we do believe that incorporating age in a balanced data set would be of value for classifiers’ development. As many studies reported the platelet count as the prognostic factor for various cancer types,46-49 we trained models combining platelet counts with RNA data but observed no statistically significant performance improvement, likely because of the small sample size. The high number of variables included in the model may dilute key features, limiting the model’s ability to detect subtle patterns. Although additional features like platelet count could improve accuracy, our sample size prevents us from confirming this. Third, our method relies on many genes (5277 transcripts), limiting compatibility with quantitative reverse transcription polymerase chain reaction, although decreasing sequencing costs make it feasible. Finally, comparing methods across studies is difficult because of differing benchmarks. Many studies focus on distinguishing healthy donors from patients with cancer, overlooking the harder task of separating nonmalignant donors. Sensitivity, specificity trade-offs, and varying metrics add to the complexity of creating universally effective diagnostic tools for OC detection. Future studies could focus on enhancing our model by the inclusion of platelet count along with the other features or integrating additional biomarkers such as CA-125, which has shown promise as a separate feature in our previous study.7 Additionally, combining our approach with cfDNA analysis might further enhance early-stage detection rates, as demonstrated by the success of multimodal tests such as CancerSEEK.32 Studying sex-related differences between other types of cancer might be also a promising lead. Exploring these integrative strategies could provide more comprehensive diagnostic tools and ultimately lead to better outcomes for patients with OC.
Conclusion
Refining the platelet RNA-based model training data set by including patient sex and sample localization of origin plays a significant role in OC detection. Limiting the training data set to asymptomatic women and patients with OC yielded the most sensitive model for early-stage detection, namely a cancer-screening model. Including patient age information alongside gene expression data improves overall classification. Including the platelet count information was inconclusive because of the limited sample size. Although the downregulation of ribosome-related processes in platelets from patients with OC and upregulation of genes KSR1, HDAC8, and NFKBIA plays a significant role in machine-learning prediction of OC, the role of platelet activation (CD40L, BCL3, GPIIb/IIIa, PF4, β-TG, P-selectin, integrins αIIbβ3, TGF-β, VEGF, PDGF, bFGF, Toll-like receptor 4, and GPVI) and platelet reticulation markers (SELP) is less pronounced.
Acknowledgments
The authors acknowledge the support of the specialists employed at the Centre of Biostatistics and Bioinformatics (Małgorzata Grešner, Kamil Myszczyński, and Anna Koelmer) as well as the administrative staff (Małgorzata Madej).
This research was supported by the SONATA grant of the National Science Centre (2018/31/D/NZ5/01263), LIDER XI grant 0059/L-11/2019 of the National Center for Research and Development, and the statutory grant ST-23 (Department of Oncology and Radiotherapy, Medical University of Gdańsk, Poland).
Authorship
Contribution: M.A.J., A.S., and M.T.R. conceptualized and designed the study; M.A.J. and K.P. performed data processing; M.A.J. and M.S. performed the experiments and analysis; M.A.J., A.S., M.S., K.P., M.T.R., and A.J.Ż. interpreted the results; M.A.J. and M.S. visualized the data; M.A.J., A.S., M.S., K.P., M.T.R., S.Ł.S., and J.J. wrote the draft of the manuscript; A.S., M.T.R., and J.J. supervised the study; and A.S., J.J., and A.J.Ż. provided funding.
Conflict-of-interest disclosure: M.T.R., A.J.Ż, M.S., K.P., J.J., A.S., and M.A.J. have patents related to using platelet RNA for diagnostics.
Correspondence: Anna Supernat, Laboratory of Translational Oncology, Intercollegiate Faculty of Biotechnology of the University of Gdańsk and the Medical University of Gdańsk, ul. Dębinki 1, 80-211 Gdańsk, Poland; email: anna.supernat@gumed.edu.pl.
References
Author notes
The study is based on publicly available platelet RNA data stored in the Gene Expression Omnibus database (accession number: GSE183635) and in the National Institutes of Health Sequence Read Archive (PRJNA531691).
The full-text version of this article contains a data supplement.