TO THE EDITOR:
In myelodysplastic syndromes (MDSs), cytogenetics and risk scores derived from these data are fundamental to prognosticate and treat this disease.1 Owing to the complexity of annotation of cytogenetic strings and the valuable information therein contained, many hours of clinical researchers can be spent parsing through data, selecting abnormalities of interest, and calculating the cytogenetic risk score for each case. Furthermore, given the specialized rules around what is a true abnormality,2 training of personnel requires significant time and resources. Despite such training, annotation errors can still occur.3 To solve this problem, we developed and validated autocyto, a free, web-based tool to calculate cytogenetic risk scores from a cytogenetic string.
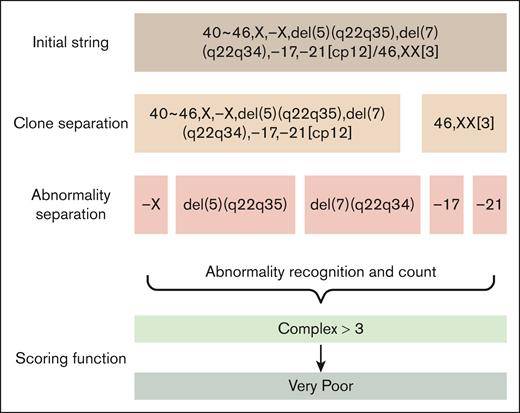
The tool parses cytogenetic strings (eg, 46,XY,del(5)(q23)[20]), extracts abnormalities of interest (eg, del(5)q), and calculates the MDS cytogenetic risk score.1 The tool first transforms the cytogenetic string’s nomenclature from previous nomenclatures to the International System for Human Cytogenomic Nomenclature 2020 if needed. The tool’s operations are outlined in the flowchart in Figure 1. The tool expects processed cytogenetic strings to follow specific formatting: cytogenetic abnormalities should be separated by commas (,), clones by forward slashes (/), and metaphase counts enclosed in square brackets ([20]). As recommended by the International System for Human Cytogenomic Nomenclature,2 singular metaphases (ie, [1]) are excluded and not used to calculate the final score.
Using natural language processing,4 the tool then finds all prognostic abnormalities of interest in MDSs (supplemental Table 1) and creates a binary variable (1 for presence and 0 for absence) of each abnormality (supplemental Table 2). An additional function outputs the metaphases of the dominant clone. Finally, a function calculates the cytogenetic risk score and outputs an integer from 0 (very good) to 4 (very poor). Several safety functions check for missing components, empty rows, or lack of integers. If any of the safety functions is triggered, the tool will ask for a manual check of that case in the final database. Finally, a master function calls all previous functions and provides the final database in .csv format. The tool was fully written in Python 3.12.4 (Python Software Foundation, Wilmington, DE) using libraries pandas,5 numpy,6 re,7 and lifelines.8 We used the streamlit9 library to develop the web application. This study was approved by the institutional review board of The University of Texas MD Anderson Cancer Center.
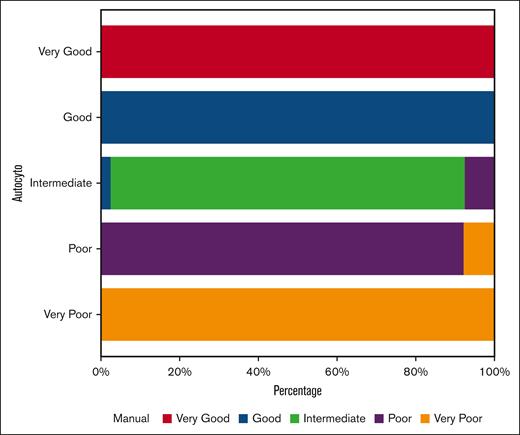
We then tested the tool in a database with manual cytogenetic annotation of 456 patients with MDSs of whom 167 (36.6%) had complex karyotype. The tool asked for a manual check in 21 cases (4.6%). It produced abnormality annotation and cytogenetic risk scores in 435 cases (95.3%). There was a discrepancy between manual and automatic annotation in 46 cases (10%) with 38 (82%) being erroneous manual annotation and 8 (18%) being erroneous tool annotation. The concordance index between manual and autocyto annotations was 0.99 (Figure 2). Kaplan-Meier estimates using manually annotated cytogenetic risk scores vs autocyto annotated cytogenetic risk scores (supplemental Figure 1) showed excellent correlation in survival. For the 456-patient database, the processing time is 0.22 seconds.
The tool expects a comma-delimited file (.csv). Other file types such as excel (.xls or .xlsx) can be easily converted to .csv using the widely available Microsoft Excel (Microsoft Corporation, Redmond, WA). To process a database, the user must type the column header (the name of the column) in the field provided. The user must then upload the .csv file using the Browse files button. This will load the database in the lower part of the website under “Original Data,” and almost immediately a second database will appear as “Processed Data” along with a button Download Processed Data, which will allow the user to download their data to a directory of their choice. All data are deleted once the user closes their browser. The supplemental file (supplemental Figure 1) contains a link to a video for usage demonstration.
Clinical research conclusions depend on the accuracy of data entry and interpretation. This process has been partially improved by clinical data transfer from the medical record for discrete data points such as laboratory values, dates, or diagnosis codes. However, data entry errors can happen in up to 20% of clinical research databases and the main reasons continue to be errors in annotation and interpretation.10 Tools exist for data integrity assessment such as impossible dates (death dates before diagnosis) and for impossible values (eg, “stage I MDS”), but these are rarely used within clinical trials11 and almost never used in retrospective data analyses, which represent more than 70% of clinical research found in PubMed.12
Cytogenetic strings have multiple potential annotation pitfalls: at the time of the abnormality annotation by the cytotechnologist, at the time of transcription into the clinical record, and at the time of transcription into the clinical database. Unfortunately, given that most cytogenetic reports are uploaded as a document, instead of a specific field value, this last step is almost always manual. Furthermore, the current nomenclature of cytogenetics includes multiple clicks and multiple special character keystrokes that make transcription more error prone. Artificial intelligence technologies are being developed to automate this process.13
To calculate the International Prognostic Scoring System or its revised and molecular versions,14,15 clinical researchers with different levels of training extract each cytogenetic abnormality from the medical record manually. This process includes direct observation of the cytogenetic string, manual extraction, and calculation of each score for each patient. Errors notwithstanding, this process is tedious, takes long hours, and takes longer to verify for each patient. In MDS clinical research, cytogenomic data have the highest prognostic value and data integrity is critical for reporting. Accurate clinical research database annotation is mandatory to be able to extract insights from retrospective data and inform hypotheses or treatment practices. We found that discordance between manual and autocyto annotations was caused by manual errors in more than 80% of cases, highlighting the need for the automation of cytogenetic string analysis. Our tool can reduce research workhours in a user-friendly interface. The tool can be accessed at https://autocyto.streamlit.app.
Contribution: S.U. designed the tool and wrote the manuscript; G.T. reviewed the tool’s nomenclature assessment for accuracy; and K.S.C., Z.L., A.B., and G.G.-M. reviewed the tool’s performance and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Samuel Urrutia, Division of Oncology, Washington University School of Medicine, 660 S Euclid Ave, St Louis, MO 63104; email: surrutia@wustl.edu.
References
Author notes
The data used to validate the tool are not available. The source code and the application are freely available online at https://github.com/eotf/autocyto, and the application may be used directly at https://autocyto.streamlit.app.
The full-text version of this article contains a data supplement.