Key Points
BCR repertoire analysis revealed frequent AID-initiated somatic hypermutation, isotype switching, and immunoglobulin gene fusion transcripts.
The 5′RACE assay identified patients with strongly dominant clonotypes, a greater degree of tumoral infiltration, and poorer outcomes.
Visual Abstract
There is a scarcity of data on the tumor B-cell receptor (BCR) repertoire and lymphoid microenvironment in primary mediastinal B-cell lymphoma (PMBL). We applied 5ʹ rapid amplification of complimentary DNA ends (5′RACE) to tumor RNA samples from 137 patients with PMBL with available gene expression profiling and next-generation sequencing data. We obtained 5′RACE results for 75 of the 137 (54.7%) patients with the following clinical characteristics: median age (range), 33 years (18-64); female, 53.3%; performance status score 0 to 1, 86.7%; stage I to II, 57.3%; first-line treatment with anti-CD20 plus doxorubicin-based chemotherapy, 100%. Among the 60 biopsies that expressed a productive BCR, we highlighted a strong somatic hypermutation profile, defined as <98% identity to the germ line sequence, with 58 (96.7%) patients carrying mutated IgVH. We then identified a subgroup of 12 of the 75 patients (16%) with a worse prognosis (progression-free survival [PFS]: hazard ratio [HR], 17; overall survival [OS]: HR, 21) that was associated with the highest clonal dominance (HCD) status, defined as the dominant clonotype representing >81.1% and >78.6% of all complementarity-determining region 3 sequences for IgVH and IgVL, respectively. When compared with other patients, this subgroup had similar clinical characteristics but a greater median allele frequency for all somatic variants, a decreased BCR diversity, and greater expression of PDL1/PDL2 and MS4A1 genes, suggesting greater tumoral infiltration. We confirmed this poorer prognosis in a multivariate model and in an independent validation cohort in which 6 of 37 (16%) PMBL patients exhibited HCD (PFS: HR, 12; OS: HR, 17).
Introduction
Primary mediastinal B-cell lymphoma (PMBL) is an uncommon and distinct form of large B-cell lymphoma (LBCL) for which the cell of origin (COO) is largely debated. PMBL is a highly curable lymphoma with a survival rate exceeding 80% at 5 years.1,2 The PMBL transcriptional profile is similar to that of classical Hodgkin lymphoma (cHL) but distinct from that of LBCL3 with 3 major signaling pathways, namely JAK-STAT, NF-κB, and the PD1/PDL1/PDL2 axis.4-6 The presumed COO is a thymic medullary B lymphocyte that expresses activation-induced cytidine deaminase (AID).7,8 Unlike most B cells that differentiate in the bone marrow, thymic B cells differentiate from early thymic precursors within the thymus.9-20 In PMBL, heavy and light immunoglobulin genes (IgVH; IgVL) are typically rearranged at the variable (diversity) joining (V(D)J) gene segments with a high burden of somatic hypermutation (SHM) and class switch recombination (CSR), suggesting that the B-cell progenitor is strongly altered by the germinal center (GC) environment during lymphomagenesis.21 Defective expression of surface and cytoplasmic immunoglobulins is also a distinctive feature of PMBL21,22 and cHL.23 To further explore this topic and to better decipher the B- and T-cell receptor (BCR/TCR) repertoires of PMBL, we applied 5ʹ rapid amplification of complimentary DNA ends (5ʹRACE) to samples from a previously described large cohort of patients annotated with clinical and biologic data.24 The aims of this study were to accurately identify and assess the complementarity-determining region 3 (CDR3) diversity and to quantify immunodominant expansions, to describe both the IgVH and IgVL features, to detect fusion transcripts that involve immunoglobulin gene partners, to correlate the CDR3 diversity with previously obtained clinical, imaging, gene expression profiling (GEP), and next-generation sequencing (NGS) data, and to assess patient outcomes based on BCR diversity.
Patients and methods
Patients and data collection
Details of the PMBL Lymphoma Study Association (LYSA) multicenter retrospective study have been published previously.2,24 Briefly, the clinical outcomes of 313 previously untreated patients with PMBL who received 3 standard immunochemotherapy regimens (ACVBP [doxorubicin, cyclophosphamide, vindesine, bleomycin, and prednisone], CHOP14 [cyclophosphamide, doxorubicin, vincristine, and prednisone (delivered every 14 days)], or CHOP21 plus anti-CD20) with or without radiotherapy were assessed between 2007 and 2017 at 25 LYSA centers. From this baseline cohort of 313 patients, 194 patients were centrally reviewed by expert pathologists in the field and were confirmed to have PMBL.24 Extensive clinical and biologic characterization of this cohort based, among others, on data from GEP and NGS analyses of formalin-fixed paraffin-embedded biopsies was conducted and reported previously.24 Patients with remaining available tumor RNA samples were selected for inclusion in this ancillary study. To validate our results, we also applied the 5ʹRACE assay to an independent single-center cohort of 40 patients who were routinely treated with frontline immunochemotherapy for PMBL at the Centre Henri Becquerel (CHB) between 2007 and 2022. The characteristics of the patients included in this cohort were previously described.24 The LYSA scientific committee, the participating LYSA centers and the CHB Internal Review Board (no. 1916B) approved the study. The study was conducted according to the criteria set by the Declaration of Helsinki. Patients’ nonopposition to participation in this study was obtained through a written information note provided before inclusion.
High-throughput BCR and TCR RNA sequencing
The 5′RACE assay was performed as previously described.25,26 Briefly, sequencing libraries for RNA sequencing were prepared using the Template Switching 5′RACE Protocol kit (New England Biolabs, Ipswich, MA) and total RNA (0.5-1 μg) as the template (supplemental Methods). B- and T-cell immunoglobulin rearrangements were investigated using the IgBlast 1.15.0 alignment tool.27 The clonality threshold25 and the Simpson entropy index28,29 were used to describe the 5ʹRACE assay results (supplemental Methods). Samples with fusion transcripts involving IgVH or IgVL identified by 5ʹRACE were validated using conventional reverse transcriptase polymerase chain reaction (RT-PCR) and Sanger sequencing (supplemental Methods).
Statistical analysis
Significant differences in the parameters between different patient subgroups were determined using Fisher exact test for qualitative variables and the Wilcoxon-Mann-Whitney test for nonnormal quantitative variables. To consider multiple comparisons, we performed false discovery rate correction; the corresponding adjusted P values are presented in the tables. The log-rank test was used to assess differences in the overall survival (OS) and progression-free survival (PFS) curves determined using Kaplan-Meier estimates (supplemental Methods). Univariate Cox regression models were developed according to selected relevant variables based on data from the literature and the findings of this study. Selected variables that were significantly associated with the outcome in the univariate analyses were then included in multivariate Cox regression models. Statistical significance was indicated by a P value or adjusted P value of < .05. All statistical analyses were performed using R software version 4.2.1.
Results
Patient characteristics
Among the PMBLs confirmed by expert pathologic review (n = 194), 137 (70.6%) patients with available tumor RNA samples were included in this ancillary study (RNA set) (supplemental Table 1). We obtained valid results for 75 of those (54.7%, 5′RACE set). The teverse transcription multiplex ligation-dependent probe amplification quality score was significantly lower in samples for which the 5ʹRACE assay failed (n = 62 [45.3%]; supplemental Figure 1). Patients in the 5ʹRACE set had similar characteristics as those in the RNA set with a median age of 33 years (18-64), 53.3% were female, 86.7% had an Eastern Cooperative Oncology Group performance status score of 0 to 1, 57.3% had stage I to II disease, the median baseline metabolic tumor volume (MTV) was 259.9 cm3 (20.22-1147.52), 72% underwent first-line treatment with anti-CD20 plus ACVBP, 14.7% received CHOP14 treatment, and 13.3% received CHOP21 treatment (Table 1; supplemental Table 1). Previously reported GEP and NGS data from these patients were reused, when available, for correlation with the 5ʹRACE results. Overall, GEP and 5ʹRACE data were obtained for 75 of the 137 (54.7%) patients, and the data from all 3 assays (NGS + GEP + 5ʹRACE) were available for 67 of the 137 (49%) patients (Figure 1).
Characteristics of patients with available 5ʹRACE data (n = 75) in the PMBL LYSA cohort
| Characteristics . | 5′RACE set (n = 75) . |
|---|---|
| Age, median (Q1; Q3, min-max), y | 33 (26; 41.5, 18-64) |
| Female sex | 40 (53.33%) |
| ECOG 0-1 | 65 (86.67%) |
| Ann Arbor stage I-II | 43 (57.33%) |
| Elevated LDH | 61 (81.33%) |
| IPI 0 | 9 (12%) |
| IPI 1-2 | 48 (64%) |
| IPI 3-5 | 18 (24%) |
| Bulky mass ≥10 cm | 45 (60%) |
| Extranodal involvement | 43 (57.33%) |
| First-line treatment | |
| Anti CD20 + ACVBP | 54 (72%) |
| Anti CD20 + CHOP14 | 11 (14.67%) |
| Anti CD20 + CHOP21 | 10 (13.33%) |
| Baseline median MTV (Q1; Q3, min-max), cm3 | 260 (146; 407, 20-1147) |
| MTV ≥360 cm3 | 21 (35%) |
| GEP assay (RT-MLPA) | |
| CD20 expression, median (Q1; Q3, min-max) | 9.56 (4.64; 14.79, 0.75-42.08) |
| High CD20 gene expression (greater than median) | 37 (49.3%) |
| MAL expression, median (Q1; Q3, min-max) | 0.89 (0.35; 3.06, 0.02-21.05) |
| High MAL expression (greater than median) | 37 (49.3%) |
| CD3 expression, median (Q1; Q3, min-max) | 1.92 (1.31; 3.05, 0.22-8.01) |
| Low CD3 expression (less than or equal to median) | 38 (50.7%) |
| CD68 expression, median (Q1; Q3, min-max) | 7.38 (5.63; 8.74, 2.39-24.07) |
| Low CD68 expression (less than or equal to median) | 38 (50.7%) |
| AID expression, median (Q1; Q3, min-max) | 1.61 (0.65; 2.69, 0-18.53) |
| High AID expression (greater than median) | 37 (49.3%) |
| High PDL1/PDL2 expression (PDL1 > 0.402; PDL2 > 9.147) | 22 (29.33%) |
| 5′RACE assay | |
| Presence of a fusion transcript involving IgVH or IgVL | 6 (8%) |
| Characteristics . | 5′RACE set (n = 75) . |
|---|---|
| Age, median (Q1; Q3, min-max), y | 33 (26; 41.5, 18-64) |
| Female sex | 40 (53.33%) |
| ECOG 0-1 | 65 (86.67%) |
| Ann Arbor stage I-II | 43 (57.33%) |
| Elevated LDH | 61 (81.33%) |
| IPI 0 | 9 (12%) |
| IPI 1-2 | 48 (64%) |
| IPI 3-5 | 18 (24%) |
| Bulky mass ≥10 cm | 45 (60%) |
| Extranodal involvement | 43 (57.33%) |
| First-line treatment | |
| Anti CD20 + ACVBP | 54 (72%) |
| Anti CD20 + CHOP14 | 11 (14.67%) |
| Anti CD20 + CHOP21 | 10 (13.33%) |
| Baseline median MTV (Q1; Q3, min-max), cm3 | 260 (146; 407, 20-1147) |
| MTV ≥360 cm3 | 21 (35%) |
| GEP assay (RT-MLPA) | |
| CD20 expression, median (Q1; Q3, min-max) | 9.56 (4.64; 14.79, 0.75-42.08) |
| High CD20 gene expression (greater than median) | 37 (49.3%) |
| MAL expression, median (Q1; Q3, min-max) | 0.89 (0.35; 3.06, 0.02-21.05) |
| High MAL expression (greater than median) | 37 (49.3%) |
| CD3 expression, median (Q1; Q3, min-max) | 1.92 (1.31; 3.05, 0.22-8.01) |
| Low CD3 expression (less than or equal to median) | 38 (50.7%) |
| CD68 expression, median (Q1; Q3, min-max) | 7.38 (5.63; 8.74, 2.39-24.07) |
| Low CD68 expression (less than or equal to median) | 38 (50.7%) |
| AID expression, median (Q1; Q3, min-max) | 1.61 (0.65; 2.69, 0-18.53) |
| High AID expression (greater than median) | 37 (49.3%) |
| High PDL1/PDL2 expression (PDL1 > 0.402; PDL2 > 9.147) | 22 (29.33%) |
| 5′RACE assay | |
| Presence of a fusion transcript involving IgVH or IgVL | 6 (8%) |
ECOG, Eastern Cooperative Oncology Group; IPI, International Prognostic Index; LDH, lactate dehydrogenase; min-max, minimum-maximum; Q, quartile; RT-MLPA, reverse transcription multiplex ligation-dependent probe amplification.
Study flowchart representing the different cohorts of the study based on biologic material availability (RNA set: n = 137; 5′RACE set: n = 75; validation cohort: n = 37).
Study flowchart representing the different cohorts of the study based on biologic material availability (RNA set: n = 137; 5′RACE set: n = 75; validation cohort: n = 37).
Analysis of the BCR repertoire by 5ʹRACE revealed a high frequency of SHM, CSR, and fusion transcripts involving the immunoglobulin genes
We obtained the following results from the 5ʹRACE set: IgVH clonality, n = 60 (44%); IgVL clonality, n = 59 (43%); IgVH plus IgVL clonality, n = 56 (41%); IgVH polyclonal profile, n = 15 (11%); and IgVL polyclonal profile, n = 16 (11.7%; Table 2; supplemental Table 2). Representative examples of the 5′RACE results are shown in Figure 2.
Clonality assessment of patients with PMBL in the 5′RACE cohort (n = 75)
| IgVH + IgVL clonality assessment (5ʹRACE set, n = 75) . | n (%) . |
|---|---|
| Confirmed B-cell clonality | 63 (84) |
| IgM kappa | 3 (4) |
| IgM lambda | 1 (1.3) |
| IgM polyclonal | 1 (1.3) |
| IgD kappa | 1 (1.3) |
| IgG kappa | 24 (32) |
| IgG lambda | 15 (20) |
| IgG polyclonal | 1 (1.3) |
| IgE kappa | 2 (2.7) |
| IgE polyclonal | 1 (1.3) |
| IgA kappa | 4 (5.3) |
| IgA lambda | 6 (8) |
| IgA polyclonal | 1 (1.3) |
| Polyclonal heavy chain and clonal kappa light chain | 1 (1.3) |
| Polyclonal heavy chain and clonal lambda light chain | 2 (2.7) |
| B-cell clonality not detectable | 12 (16) |
| Polyclonal profile | 12 (16) |
| IgVH + IgVL clonality assessment (5ʹRACE set, n = 75) . | n (%) . |
|---|---|
| Confirmed B-cell clonality | 63 (84) |
| IgM kappa | 3 (4) |
| IgM lambda | 1 (1.3) |
| IgM polyclonal | 1 (1.3) |
| IgD kappa | 1 (1.3) |
| IgG kappa | 24 (32) |
| IgG lambda | 15 (20) |
| IgG polyclonal | 1 (1.3) |
| IgE kappa | 2 (2.7) |
| IgE polyclonal | 1 (1.3) |
| IgA kappa | 4 (5.3) |
| IgA lambda | 6 (8) |
| IgA polyclonal | 1 (1.3) |
| Polyclonal heavy chain and clonal kappa light chain | 1 (1.3) |
| Polyclonal heavy chain and clonal lambda light chain | 2 (2.7) |
| B-cell clonality not detectable | 12 (16) |
| Polyclonal profile | 12 (16) |
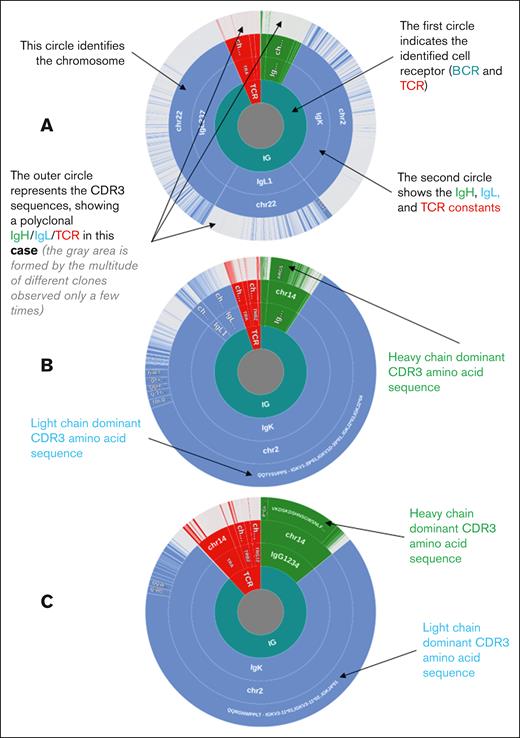
Immunoglobulin and TCR profiles of representative samples (donut charts). The immunoglobulin and TCR sequences are clustered according to the amino acid sequence of CDR3. The BCR or TCR origin (green: IgVH (IgH); blue: IgVL (IgL); red: TCR), identified constant, chromosomal origin, and cluster of identical CDR3 sequences are indicated from the inside to the outside. The 3 samples presented correspond to the 3 different types of 5′RACE results, namely (A) a PMBL case with a polyclonal profile with no dominant clonotype identified. The ratio of B-cell to T-cell (B/T) sequences and the percentage of sequences corresponding to the dominant CDR3s are as follows: B/T ratio, 93.64%; most frequent IgVH CDR3, 0.92%; most frequent IgVL CDR3, 1.48%. (B) A PMBL case with confirmed B-cell clonality, not classified as having the HCD (non-HCD). The clonality threshold was defined as 6.04%; the upper outlier of the distribution of sequencing reads coding the most frequent CDR3 from the heavy and light BCR chains in the absence of tumoral populations. The ratio of B/T sequences and the percentage of sequences corresponding to the dominant CDR3s are as follows: B/T ratio, 94.85%; most frequent IgVH CDR3, 44.74%; most frequent IgVL CDR3, 78.44%. (C) A PMBL case with the HCD in which the dominant clonotype represents >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively. The ratio of B/T sequences and the percentage of sequences corresponding to the dominant CDR3s are as follows: B/T ratio, 88.5%; most frequent IgVH CDR3, 93.35%; most frequent IgVL CDR3, 94.42%. HCD, highest clonal dominance.
Immunoglobulin and TCR profiles of representative samples (donut charts). The immunoglobulin and TCR sequences are clustered according to the amino acid sequence of CDR3. The BCR or TCR origin (green: IgVH (IgH); blue: IgVL (IgL); red: TCR), identified constant, chromosomal origin, and cluster of identical CDR3 sequences are indicated from the inside to the outside. The 3 samples presented correspond to the 3 different types of 5′RACE results, namely (A) a PMBL case with a polyclonal profile with no dominant clonotype identified. The ratio of B-cell to T-cell (B/T) sequences and the percentage of sequences corresponding to the dominant CDR3s are as follows: B/T ratio, 93.64%; most frequent IgVH CDR3, 0.92%; most frequent IgVL CDR3, 1.48%. (B) A PMBL case with confirmed B-cell clonality, not classified as having the HCD (non-HCD). The clonality threshold was defined as 6.04%; the upper outlier of the distribution of sequencing reads coding the most frequent CDR3 from the heavy and light BCR chains in the absence of tumoral populations. The ratio of B/T sequences and the percentage of sequences corresponding to the dominant CDR3s are as follows: B/T ratio, 94.85%; most frequent IgVH CDR3, 44.74%; most frequent IgVL CDR3, 78.44%. (C) A PMBL case with the HCD in which the dominant clonotype represents >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively. The ratio of B/T sequences and the percentage of sequences corresponding to the dominant CDR3s are as follows: B/T ratio, 88.5%; most frequent IgVH CDR3, 93.35%; most frequent IgVL CDR3, 94.42%. HCD, highest clonal dominance.
The characteristics of patients with identified IgVH clonality (n = 60) differed from those of the patients with a polyclonal IgVH profile (n = 15) as follows: they had a higher median baseline MTV (302.9 vs 180 cm3; P = .04), a higher frequency of high MS4A1/CD20 (60% vs 6.67%; P < .001) and AID (56.7% vs 20%; P = .02) expression, and a higher frequency of low CD3 expression (60% vs 13.3%; P = .001), suggesting greater tumoral infiltration, which was confirmed by a higher percentage of tumor cells when evaluated using histology (80% vs 70%; P = .01; supplemental Table 3). As shown in supplemental Figure 2, B-cell and tumor driver genes represented most of the messenger RNAs (mRNAs) expressed in the cases with confirmed IgVH clonality compared with the high expression of genes associated with the reactive microenvironment in patients with a polyclonal profile. However, the mutational landscape, as assessed by NGS, in patients with a polyclonal profile was not significantly different from that in other patients (supplemental Table 4).
We observed that PMBLs showed a class switch isotype (immunoglobulin G [IgG], IgE, or IgA) in almost all patients. Indeed, among the 60 biopsies that expressed a productive IgVH, 66.7% expressed mainly (>90%) IgG sequences (40 patients), 10% expressed mainly IgM/D sequences (6 patients), 18.3% expressed mainly IgA sequences (11 patients), and 5% expressed mainly IgE sequences (3 patients) (Table 2; supplemental Table 2). Notably, in some cases, particularly for those with the IgG isotype clones, we also observed a few sequencing reads that encoded the dominant CDR3 of other secondary isotypes (IgA, IgE), suggesting an ongoing CSR process in these tumors (supplemental Table 5).
Somatic mutations in the V segments of IgVH/IgVL rearrangements were common. Indeed, among the 60 biopsies that expressed a productive BCR, we observed an SHM profile, and almost all the patients carried mutated IgVH (n = 58 [96.7%]), defined as <98% shared sequence identity with the germ line sequence.30,31 Among them, the median rate of homology with the germ line sequence was 84.9% (supplemental Table 2). Patients with unmutated IgVH (homology rate ≥98%) were rare (n = 2 [3.3%]). Each patient with PMBL had a different IgVH and IgVL CDR3 sequence. The median (interquartile range) sizes of the CDR3 were 14 (11.75-16) and 9 (9-10) amino acids for IgVH and IgVL, respectively (supplemental Table 6). Analyses of the frequencies of variable region of the heavy chain (VH) subfamilies revealed that VH1 (32%) and VH3 (32%) were the most common, followed by VH4 (20%), VH5 (12%), and VH2 (5%) (supplemental Table 7). The VH usage in our cohort was similar to that in patients with other repertoires described in the literature.32-38 Among the light chain subfamilies, VK1 (69%) was dominant in the analyzed sequences. Regarding the usage of VH germ line genes, VH1-30 (12%) and VH1-18 (10%), followed by VH1-46 (7%), VH4-34 (7%), and VH5-51 (7%), were frequently found in the VH repertoires of our cohort. In addition, VK1-39 (29%) and VL1-36 (17%) were the most frequently detected VK and VL repertoires, respectively (supplemental Table 8; supplemental Figure 3).
Finally, we detected fusion transcripts that involved immunoglobulin genes in 11 patients (supplemental Table 9; supplemental Figure 4), including 1 uncommon IgVL/AICDA transcript. Although the exact breakpoint locations remain uncharacterized because of a lack of DNA data, all transcripts were confirmed using RT-PCR and Sanger sequencing (data not shown). The affected genes (CIITA, SOCS1, IgVH, and IgVL) are known targets of somatic mutations in PMBLs.
Identification of a population with the HCD based on the 5ʹRACE analysis and poorer outcomes
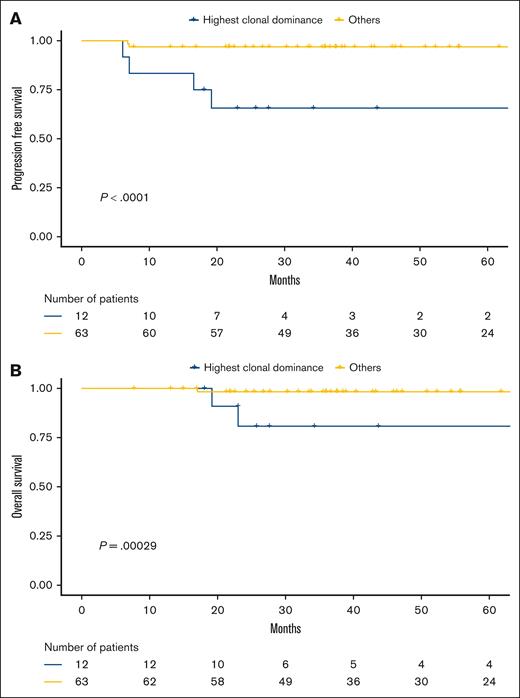
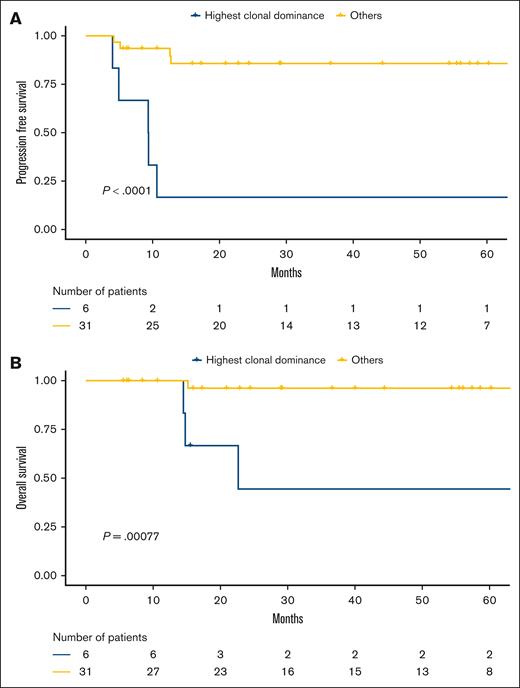
We observed that the distribution of preeminent clonotypes among cases did not follow a normal distribution (supplemental Figure 5). We correlated the B-cell clonal dominance with survival data and identified a population of patients (n = 12/75 [16%]) for whom the dominant clones represented >81.1% and >78.6% of all the IgVH and IgVL CDR3 sequences, respectively. Both criteria had to be met to define highest clonal dominance (HCD), and thresholds were determined by using receiver operating characteristic curve analysis for PFS events (supplemental Figure 6). These patients with HCD had poorer outcomes than the others (PFS: hazard ratio [HR], 17; 3.2-88; Figure 3A; OS: HR, 21; 2.1-210; Figure 3B).
The HCD correlated with poorer outcomes in the PMBL LYSA cohort. (A) PFS and (B) OS according to clonal dominance status (assessed by 5′RACE) in the 5′RACE set from the PMBL LYSA cohort (n = 75). We identified a subset of 12 of 75 patients (16%) with the HCD (cutoffs: dominant clonotype representing >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively).
The HCD correlated with poorer outcomes in the PMBL LYSA cohort. (A) PFS and (B) OS according to clonal dominance status (assessed by 5′RACE) in the 5′RACE set from the PMBL LYSA cohort (n = 75). We identified a subset of 12 of 75 patients (16%) with the HCD (cutoffs: dominant clonotype representing >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively).
With a median follow-up of 51.3 months (1-152.7), the 2-year PFS rate in the HCD subset was 65.6% (95% confidence interval [CI], 43.2-99.7) vs 96.8% (95% CI, 92.6-100) for the other patients (P < .001), and the 2-year OS rates were 80.8% (95% CI, 60-100) vs 98.3% (95% CI, 95.1-100) for the others (P < .001).
Features of the HCD subset: correlation with clinical, histologic, metabolic imaging, GEP, and NGS data
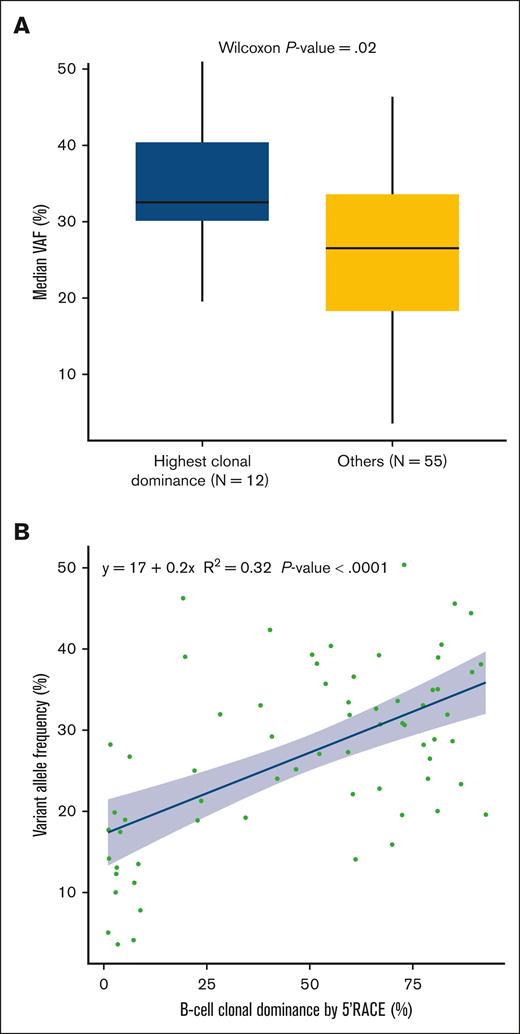
The patients with HCD had comparable baseline characteristics and similar isotypes as the other patients, and the treatments used in this group did not differ from those used in the other patients (Table 3). For these 12 HCD cases, we identified both IgVH and IgVL CDR3 sequences in the dominant clonotype (highlighted in red in supplemental Table 8). There was no correlation between the maximum standardized uptake value and HCD (supplemental Figure 7). However, these patients presented a greater mean AID expression (P = .017; supplemental Figure 8) and a greater median variant allele frequency (VAF) for all somatic variants as identified by NGS (P = .02; Figure 4A). The correlation between the clonal dominance and the median VAF was moderate (R2 = 0.32; P < .001; Figure 4B). Notably, neither the mutational landscape nor the number of somatic variants in patients with HCD was significantly different from that in other patients (supplemental Table 10; supplemental Figure 9). In addition, the 12 patients with HCD exhibited a greater expression of PDL1/PDL2 (66.7% vs 22.2%; P = .043; Table 3). CD20 expression was also moderately correlated with clonal dominance (R2 = 0.25; P < .001; supplemental Figure 10).
Characteristics of different subgroups of patients with PMBL from the 5ʹRACE cohort: patients with PMBL with HCD (n = 12) vs others (n = 63)
| Characteristics . | HCD, n = 12 (16%) . | Others, n = 63 (84%) . | P value . | Adjusted P value . |
|---|---|---|---|---|
| Age, median (Q1; Q3, min-max), y | 40 (25.5; 49.25, 18-57) | 32 (26; 40, 19-64) | .308 | .583 |
| Female sex | 7 (58.33%) | 33 (52.38%) | .762 | .903 |
| ECOG 0-1 | 11 (91.67%) | 54 (85.71%) | 1 | 1 |
| Ann Arbor stage I-II | 6 (50%) | 37 (58.73%) | .752 | .903 |
| Elevated LDH | 10 (83.33%) | 51 (80.95%) | 1 | 1 |
| IPI 0 | 0 (0%) | 9 (14.29%) | .566 | .823 |
| IPI 1-2 | 9 (75%) | 39 (61.9%) | ||
| IPI 3-5 | 3 (25%) | 15 (23.81%) | ||
| Bulky mass ≥10 cm | 8 (66.67%) | 37 (58.73%) | .752 | 0.903 |
| Extranodal involvement | 8 (66.67%) | 35 (55.56%) | .54 | .823 |
| First-line treatment | ||||
| Anti CD20 + ACVBP | 10 (83.33%) | 44 (69.84%) | .888 | .982 |
| Anti CD20 + CHOP14 | 1 (8.33%) | 10 (15.87%) | ||
| Anti CD20 + CHOP21 | 1 (8.33%) | 9 (14.29%) | ||
| Baseline median MTV (Q1; Q3, min-max), cm3 | 369.71 (265.44; 582.42, 166.82-1147.52) | 249.17 (138.09; 405.52, 20.22-1066.3) | .094 | 0.273 |
| MTV ≥360 cm3 | 5 (50%) | 16 (32%) | .298 | .583 |
| GEP assay (RT-MLPA) | ||||
| High CD20 gene expression (greater than median∗) | 8 (66.67%) | 29 (46.03%) | .222 | .547 |
| High MAL expression (> greater than median∗) | 9 (75%) | 28 (44.44%) | .065 | .207 |
| Low CD3 expression (less than or equal to median∗) | 8 (66.67%) | 30 (47.62%) | .346 | .583 |
| Low CD68 expression (less than or equal to median∗) | 8 (66.67%) | 30 (47.62%) | .346 | .583 |
| High AID expression (greater than median∗) | 9 (75%) | 28 (44.44%) | .065 | .207 |
| High PDL1/PDL2 expression (PDL1 > 0.402; PDL2 > 9.147) | 8 (66.67%) | 14 (22.22%) | .004 | .043 |
| 5′RACE assay | ||||
| Presence of a fusion transcript involving IgVH or IgVL | 2 (16.67%) | 4 (6.35%) | .244 | .557 |
| Confirmed IgVH clonality | 12 (100%) | 48 (76.19%) | .109 | .291 |
| Isotype heavy chain | ||||
| IgM | 2 (16.67%) | 3 (6.25%) | .602 | .837 |
| IgD | 0 (0%) | 1 (2.08%) | ||
| IgG | 7 (58.33%) | 33 (68.75%) | ||
| IgE | 0 (0%) | 3 (6.25%) | ||
| IgA | 3 (25%) | 8 (16.67%) | ||
| Confirmed IgVL clonality | 12 (100%) | 47 (74.6%) | .059 | .207 |
| Isotype light chain | ||||
| Kappa | 8 (66.67%) | 27 (57.45%) | .745 | .903 |
| Lambda | 4 (33.33%) | 20 (42.55%) | ||
| Confirmed IgVH and IgVL clonality | 12 (100%) | 44 (69.84%) | .030 | .163 |
| Homology rate with the germ line sequence | ||||
| IgVH, median (Q1; Q3, min-max) | 85.52 (83.43; 86.52, 71.95-97.7) | 85.42 (78.88; 92.35, 64.21-99.37) | .89 | .982 |
| IgVL, median (Q1; Q3, min-max) | 89.42 (86.56; 92.31, 77.5-99.26) | 86.42 (79.82; 92.28, 73.45-99.13) | .413 | .66 |
| Simpson heterogeneity index for IgVH, median (Q1; Q3, min-max) | 0.66 (0.55; 0.71, 0.34-0.79) | 0.14 (0.02; 0.32, 0-0.87) | <.001 | <.001 |
| Simpson heterogeneity index for IgVL, median (Q1; Q3, min-max) | 0.59 (0.54; 0.73, 0.43-0.87) | 0.15 (0.01; 0.49, 0-0.74) | <.001 | <.001 |
| Characteristics . | HCD, n = 12 (16%) . | Others, n = 63 (84%) . | P value . | Adjusted P value . |
|---|---|---|---|---|
| Age, median (Q1; Q3, min-max), y | 40 (25.5; 49.25, 18-57) | 32 (26; 40, 19-64) | .308 | .583 |
| Female sex | 7 (58.33%) | 33 (52.38%) | .762 | .903 |
| ECOG 0-1 | 11 (91.67%) | 54 (85.71%) | 1 | 1 |
| Ann Arbor stage I-II | 6 (50%) | 37 (58.73%) | .752 | .903 |
| Elevated LDH | 10 (83.33%) | 51 (80.95%) | 1 | 1 |
| IPI 0 | 0 (0%) | 9 (14.29%) | .566 | .823 |
| IPI 1-2 | 9 (75%) | 39 (61.9%) | ||
| IPI 3-5 | 3 (25%) | 15 (23.81%) | ||
| Bulky mass ≥10 cm | 8 (66.67%) | 37 (58.73%) | .752 | 0.903 |
| Extranodal involvement | 8 (66.67%) | 35 (55.56%) | .54 | .823 |
| First-line treatment | ||||
| Anti CD20 + ACVBP | 10 (83.33%) | 44 (69.84%) | .888 | .982 |
| Anti CD20 + CHOP14 | 1 (8.33%) | 10 (15.87%) | ||
| Anti CD20 + CHOP21 | 1 (8.33%) | 9 (14.29%) | ||
| Baseline median MTV (Q1; Q3, min-max), cm3 | 369.71 (265.44; 582.42, 166.82-1147.52) | 249.17 (138.09; 405.52, 20.22-1066.3) | .094 | 0.273 |
| MTV ≥360 cm3 | 5 (50%) | 16 (32%) | .298 | .583 |
| GEP assay (RT-MLPA) | ||||
| High CD20 gene expression (greater than median∗) | 8 (66.67%) | 29 (46.03%) | .222 | .547 |
| High MAL expression (> greater than median∗) | 9 (75%) | 28 (44.44%) | .065 | .207 |
| Low CD3 expression (less than or equal to median∗) | 8 (66.67%) | 30 (47.62%) | .346 | .583 |
| Low CD68 expression (less than or equal to median∗) | 8 (66.67%) | 30 (47.62%) | .346 | .583 |
| High AID expression (greater than median∗) | 9 (75%) | 28 (44.44%) | .065 | .207 |
| High PDL1/PDL2 expression (PDL1 > 0.402; PDL2 > 9.147) | 8 (66.67%) | 14 (22.22%) | .004 | .043 |
| 5′RACE assay | ||||
| Presence of a fusion transcript involving IgVH or IgVL | 2 (16.67%) | 4 (6.35%) | .244 | .557 |
| Confirmed IgVH clonality | 12 (100%) | 48 (76.19%) | .109 | .291 |
| Isotype heavy chain | ||||
| IgM | 2 (16.67%) | 3 (6.25%) | .602 | .837 |
| IgD | 0 (0%) | 1 (2.08%) | ||
| IgG | 7 (58.33%) | 33 (68.75%) | ||
| IgE | 0 (0%) | 3 (6.25%) | ||
| IgA | 3 (25%) | 8 (16.67%) | ||
| Confirmed IgVL clonality | 12 (100%) | 47 (74.6%) | .059 | .207 |
| Isotype light chain | ||||
| Kappa | 8 (66.67%) | 27 (57.45%) | .745 | .903 |
| Lambda | 4 (33.33%) | 20 (42.55%) | ||
| Confirmed IgVH and IgVL clonality | 12 (100%) | 44 (69.84%) | .030 | .163 |
| Homology rate with the germ line sequence | ||||
| IgVH, median (Q1; Q3, min-max) | 85.52 (83.43; 86.52, 71.95-97.7) | 85.42 (78.88; 92.35, 64.21-99.37) | .89 | .982 |
| IgVL, median (Q1; Q3, min-max) | 89.42 (86.56; 92.31, 77.5-99.26) | 86.42 (79.82; 92.28, 73.45-99.13) | .413 | .66 |
| Simpson heterogeneity index for IgVH, median (Q1; Q3, min-max) | 0.66 (0.55; 0.71, 0.34-0.79) | 0.14 (0.02; 0.32, 0-0.87) | <.001 | <.001 |
| Simpson heterogeneity index for IgVL, median (Q1; Q3, min-max) | 0.59 (0.54; 0.73, 0.43-0.87) | 0.15 (0.01; 0.49, 0-0.74) | <.001 | <.001 |
HCD cases were defined as those with dominant clonotypes representing >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively. Bold values indicate that statistical significance was reached (P < .05).
ECOG, Eastern Cooperative Oncology Group; IPI, International Prognostic Index; LDH, lactate dehydrogenase; min-max, minimum-maximum; Q, quartile; RT-MLPA, reverse transcription multiplex ligation-dependent probe amplification.
Median values of each marker, defined in the overall population included in the 5′RACE set, are described in Table 1.
Description of the median VAF determined by NGS and clonal dominance assessed by 5′RACE in patients with available data for both variables (n = 67 in 5′RACE set). (A) Box plot displaying the median VAF comparison between patients with the HCD (n = 12) and others (n = 55). (B) Correlation between the degree of B-cell clonal dominance (defined by the percentage of CDR3 sequences occupied by the dominant clone) according to 5′RACE and the median VAF of somatic variants detected by NGS (5′RACE set).
Description of the median VAF determined by NGS and clonal dominance assessed by 5′RACE in patients with available data for both variables (n = 67 in 5′RACE set). (A) Box plot displaying the median VAF comparison between patients with the HCD (n = 12) and others (n = 55). (B) Correlation between the degree of B-cell clonal dominance (defined by the percentage of CDR3 sequences occupied by the dominant clone) according to 5′RACE and the median VAF of somatic variants detected by NGS (5′RACE set).
As shown in Figure 5, we observed a lower percentage of sequences that encoded in-frame TCR rearrangements in the HCD biopsies than in the other cases, suggesting a numerically lower degree of T-cell infiltration in these tumors (median proportion of TCR reads, 6.5% [range, 1.46%-16.5%] vs 8.79% [range, 0.11%-74.5%] for others; P = .4). As shown in supplemental Figures 11 and 12, genes associated with B-cell malignancies (including MS4A1/CD20, CD19, CD23, KI67, and STAT6) represented the majority of mRNAs expressed in the HCD cohort compared with genes associated with the reactive microenvironment (including CD3, CD5, CD8, GATA3, FOXP1) that were highly expressed in the others (non-HCD cases). Unsupervised clustering analysis revealed that the HCD tumors were mainly PDL1high/PDL2high and strongly expressed tumor driver genes. In addition, these patients exhibited decreased expression of T-cell–associated markers (supplemental Figures 13 and 14). Furthermore, we compared the GEP of markers associated with activated and exhausted T-cell signatures between the HCD and non-HCD groups and found significantly lower CD8 (P = .04) and GZMB (P = .02) expression in HCD cases. Other activated T-cell markers (PRF1, CXCL13, TBET, IFN-γ, CXCR5, and ICOS) and exhausted T-cell markers (CTLA4, LAG3, and PD1) showed no significant differences (supplemental Figures 15 and 16).
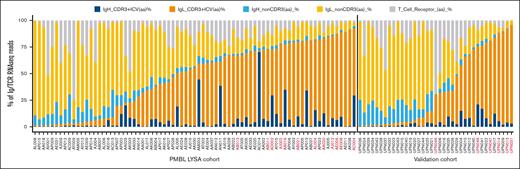
BCR and TCR repertoire diversity in the 2 patient sets. The frequencies of immunoglobulin heavy (H) and light (L) clonotypes are depicted for each sample, along with the contribution of TCR sequences. The samples were sorted based on the percentage of immunodominant clones (IgH + IgL dominant CDR3) and by cohort as follows: the PMBL LYSA cohort (5′RACE set) and the validation cohort. Samples with the HCD as defined by 5′RACE are indicated in red.
BCR and TCR repertoire diversity in the 2 patient sets. The frequencies of immunoglobulin heavy (H) and light (L) clonotypes are depicted for each sample, along with the contribution of TCR sequences. The samples were sorted based on the percentage of immunodominant clones (IgH + IgL dominant CDR3) and by cohort as follows: the PMBL LYSA cohort (5′RACE set) and the validation cohort. Samples with the HCD as defined by 5′RACE are indicated in red.
We also evaluated the lymphoma cellularity on hematoxylin- and eosin-stained slides and observed that the median tumoral infiltration was higher in HCD cases than in others (median, 80% [range, 50%-90%] vs 70% [range, 30%-95%]; P = .38; supplemental Figure 17A). We also observed a minimal correlation between the proportion of pathologist-estimated malignant cells and clonal dominance (R2 = 0.1; P = .003; supplemental Figure 17B). We found no correlation between CD3+ and/or CD5+ T-cell quantification (histology) and the frequency of sequences that coded in-frame TCR rearrangements (supplemental Figure 18). In addition, there was a weak inverse correlation between CD3+ and/or CD5+ T-cell quantification and clonal dominance (supplemental Figure 19). These results suggest that the HCD subgroup had lower infiltration by reactive B and T cells.
Univariate and multivariate survival analyses
According to univariate analysis, the following variables were associated with a shorter PFS: HCD (HR, 17; 95% CI, 3.2-88; P < .001) and AID expression (HR, 1.2; 95% CI, 1-1.3; P = .032; Table 4). There were more patients in the PDL1high/PDL2high subgroup who lacked 5′RACE results than in other subgroups (supplemental Table 11). Therefore, we could not analyze the prognostic impact of combining the 2 variables because of the risk of bias associated with attrition. A multivariate Cox regression model that included HCD and AID expression indicated that the former was an independent prognostic factor for inferior PFS (HR, 14.6; 95% CI, 2.46-86.8; P = .003; Table 4).
Univariate and multivariate analyses of the prognostic factors associated with PFS and OS in the 5′RACE cohort from the PMBL LYSA cohort
| PFS . | n . | Univariate analysis . | Multivariate analysis . | |||
|---|---|---|---|---|---|---|
| HR (95% CI) . | P value . | n . | HR (95% CI) . | P value . | ||
| Treatment (ref = R-ACVBP) | ||||||
| R-CHOP14 | 65 | Infinite | ||||
| R-CHOP21 | 64 | 1 (0.12-8.4) | 1 | |||
| LDH greater than ULN | 75 | 0.52 (0.1-2.7) | .43 | |||
| IPI 3-5 | 75 | 2.1 (0.47-9.7) | .32 | |||
| MTV ≥360 cm3 | 60 | 2.8 (0.47-17) | .26 | |||
| PDL1high/PDL2high | 75 | 1.7 (0.38-7.8) | .48 | |||
| HCD (>81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, 5′RACE) | 75 | 17 (3.2-88) | .00087 | 75 | 14.6 (2.46-86.8) | .00317 |
| IgVH clonal dominance (continuous variable, 5′RACE) | 75 | 1 (1-1.1) | .034 | |||
| IgVL clonal dominance (continuous variable, 5′RACE) | 75 | 1 (0.99-1.1) | .15 | |||
| Highest IgVH clonal dominance (>81.1% of all CDR3 sequences for IgVH, 5′RACE) | 75 | 9.9 (1.9-51) | .0065 | |||
| Highest IgVL clonal dominance (>78.6% of all CDR3 sequences for IgVL, 5′RACE) | 75 | 5.2 (1-27) | .05 | |||
| AID gene expression | 75 | 1.2 (1-1.3) | .032 | 75 | 1.04 (0.88-1.2) | .6563 |
| AID gene expression greater than median | 75 | 6.1 (0.73-51) | .095 | |||
| Proportion of tumoral infiltration (histology) | 74 | 1 (0.95-1.06) | .86 | |||
| OS | n | Univariate analysis | Multivariate analysis | |||
| HR (95% CI) | Pvalue | n | HR (95% CI) | Pvalue | ||
| Treatment (ref = R-ACVBP) | ||||||
| R-CHOP14 | 65 | Infinite | ||||
| R-CHOP21 | 64 | 2 (0.2-19) | .56 | |||
| LDH greater than ULN | 75 | 0.63 (0.065-6.1) | .69 | |||
| IPI 3-5 | 75 | 2.5 (0.35-18) | .36 | |||
| MTV ≥360 cm3 | 60 | Infinite | ||||
| PDL1high/PDL2high | 75 | 2.1 (0.3-15) | .45 | |||
| HCD (>81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, 5′RACE) | 75 | 21 (2.1-210) | .0094 | 75 | 11.4 (1-128.8) | .0496 |
| IgVH clonal dominance (continuous variable, 5′RACE) | 75 | 1 (0.99-1.1) | .09 | |||
| IgVL clonal dominance (continuous variable, 5′RACE) | 75 | 1 (0.98-1.1) | .32 | |||
| Highest IgVH clonal dominance (>81.1% of all CDR3 sequences for IgVH, 5′RACE) | 75 | 13 (1.3-120) | .03 | |||
| Highest IgVL clonal dominance (>78.6% of all CDR3 sequences for IgVL, 5′RACE) | 75 | 6.5 (0.67-62) | .11 | |||
| AID gene expression | 75 | 1.3 (1.1-1.5) | .0067 | 75 | 1.2 (1-1.5) | .1467 |
| AID gene expression greater than median | 75 | Infinite | ||||
| Proportion of tumoral infiltration (histology) | 74 | 1 (0.93-1.07) | .9 | |||
| PFS . | n . | Univariate analysis . | Multivariate analysis . | |||
|---|---|---|---|---|---|---|
| HR (95% CI) . | P value . | n . | HR (95% CI) . | P value . | ||
| Treatment (ref = R-ACVBP) | ||||||
| R-CHOP14 | 65 | Infinite | ||||
| R-CHOP21 | 64 | 1 (0.12-8.4) | 1 | |||
| LDH greater than ULN | 75 | 0.52 (0.1-2.7) | .43 | |||
| IPI 3-5 | 75 | 2.1 (0.47-9.7) | .32 | |||
| MTV ≥360 cm3 | 60 | 2.8 (0.47-17) | .26 | |||
| PDL1high/PDL2high | 75 | 1.7 (0.38-7.8) | .48 | |||
| HCD (>81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, 5′RACE) | 75 | 17 (3.2-88) | .00087 | 75 | 14.6 (2.46-86.8) | .00317 |
| IgVH clonal dominance (continuous variable, 5′RACE) | 75 | 1 (1-1.1) | .034 | |||
| IgVL clonal dominance (continuous variable, 5′RACE) | 75 | 1 (0.99-1.1) | .15 | |||
| Highest IgVH clonal dominance (>81.1% of all CDR3 sequences for IgVH, 5′RACE) | 75 | 9.9 (1.9-51) | .0065 | |||
| Highest IgVL clonal dominance (>78.6% of all CDR3 sequences for IgVL, 5′RACE) | 75 | 5.2 (1-27) | .05 | |||
| AID gene expression | 75 | 1.2 (1-1.3) | .032 | 75 | 1.04 (0.88-1.2) | .6563 |
| AID gene expression greater than median | 75 | 6.1 (0.73-51) | .095 | |||
| Proportion of tumoral infiltration (histology) | 74 | 1 (0.95-1.06) | .86 | |||
| OS | n | Univariate analysis | Multivariate analysis | |||
| HR (95% CI) | Pvalue | n | HR (95% CI) | Pvalue | ||
| Treatment (ref = R-ACVBP) | ||||||
| R-CHOP14 | 65 | Infinite | ||||
| R-CHOP21 | 64 | 2 (0.2-19) | .56 | |||
| LDH greater than ULN | 75 | 0.63 (0.065-6.1) | .69 | |||
| IPI 3-5 | 75 | 2.5 (0.35-18) | .36 | |||
| MTV ≥360 cm3 | 60 | Infinite | ||||
| PDL1high/PDL2high | 75 | 2.1 (0.3-15) | .45 | |||
| HCD (>81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, 5′RACE) | 75 | 21 (2.1-210) | .0094 | 75 | 11.4 (1-128.8) | .0496 |
| IgVH clonal dominance (continuous variable, 5′RACE) | 75 | 1 (0.99-1.1) | .09 | |||
| IgVL clonal dominance (continuous variable, 5′RACE) | 75 | 1 (0.98-1.1) | .32 | |||
| Highest IgVH clonal dominance (>81.1% of all CDR3 sequences for IgVH, 5′RACE) | 75 | 13 (1.3-120) | .03 | |||
| Highest IgVL clonal dominance (>78.6% of all CDR3 sequences for IgVL, 5′RACE) | 75 | 6.5 (0.67-62) | .11 | |||
| AID gene expression | 75 | 1.3 (1.1-1.5) | .0067 | 75 | 1.2 (1-1.5) | .1467 |
| AID gene expression greater than median | 75 | Infinite | ||||
| Proportion of tumoral infiltration (histology) | 74 | 1 (0.93-1.07) | .9 | |||
HR was infinite when no deaths occurred in the selected subgroup. Multivariate analysis was performed with selected relevant variables that were most significantly associated with PFS or OS in the univariate analysis. Bold values indicate that statistical significance was reached (P < .05).
IPI, International Prognostic Index; LDH, lactate dehydrogenase; ref, reference; ULN, upper limit of normal laboratory value.
Based on the univariate analysis, HCD (HR, 21; 95% CI, 2.12-210; P = .009) and AID expression (HR, 1.3; 95% CI, 1.1-1.5; P = .007) were associated with a shorter OS. The multivariate analysis for OS that included HCD and AID expression showed that HCD was an independent prognostic factor for reduced OS (HR, 11.4; 95% CI, 1-128.8; P = .0496; Table 4).
Validation cohort
Within the independent CHB cohort (n = 40; patient characteristics are summarized in supplemental Table 12), we obtained 5′RACE results for 37 patients (92.5%; supplemental Table 13). Within this validation cohort, we identified a subset of 6 (16.2%) patients with HCD using the same HCD cutoffs that were used for the 5ʹRACE set. Despite the small size of the validation cohort, we again observed a significant association between HCD and inferior PFS and OS in the validation cohort (PFS: HR, 12; 95% CI, 3-46; P < .001 and OS: HR, 17; 95% CI, 1.8-170; P < .001; Figure 6). We also detected fusion transcripts involving immunoglobulin genes in 2 patients (supplemental Table 8; supplemental Figure 3).
HCD status correlated with poorer outcomes in the independent validation cohort. (A) PFS and (B) OS according to clonal dominance status (HCD vs others, as assessed by 5′RACE) in the independent validation cohort with available 5′RACE results (n = 37/40). We identified a subset of 6 of 37 patients (16.2%) with the HCD (cutoffs: dominant clonotype representing >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively).
HCD status correlated with poorer outcomes in the independent validation cohort. (A) PFS and (B) OS according to clonal dominance status (HCD vs others, as assessed by 5′RACE) in the independent validation cohort with available 5′RACE results (n = 37/40). We identified a subset of 6 of 37 patients (16.2%) with the HCD (cutoffs: dominant clonotype representing >81.1% and >78.6% of all CDR3 sequences for IgVH and IgVL, respectively).
Discussion
In this study, we deciphered the BCR and TCR repertoires of a large multicenter cohort of patients with PMBL using a 5ʹRACE assay to analyze routine formalin-fixed paraffin-embedded samples. Our results provide a representative description of the immunoglobulin repertoires of the tumoral compartment in PMBL. We confirmed the existence of a high level of somatic mutations in the V(D)J segments, as previously reported by Leithäuser et al,21 and suggested a key role for AID in lymphomagenesis. In addition, our 5ʹRACE analyses also revealed that ∼15% of patients with PMBL carried fusion transcripts involving immunoglobulin loci, involving genes targeted by somatic mutations in PMBL (CIITA, SOCS, IgVH, and IgVL). These findings are consistent with previous reports that showed that chromosomal translocations that involve immunoglobulin loci play a decisive role in B-cell lymphomagenesis.39,40 Notably, translocations between IgVH/IgVL genes are quite common in B-cell lymphomas but do not produce fusion transcripts, do not cause gene expression deregulation, and probably do not have the same value as other translocations.41-43 These translocations may lead to the upregulation of key genes, such as PDL1, which play a role in determining patient prognosis and might be targeted by checkpoint inhibitors.24,44 Our 5ʹRACE assay also revealed an uncommon case of a fusion transcript involving the AID and IgVL genes, reinforcing the hypothesis of the involvement of AID in PMBL oncogenesis. To our knowledge, there is only 1 previously reported case of a similar fusion transcript involving AID and an immunoglobulin gene in a pediatric patient with cHL, but the partner was IgVH.45 However, without DNA data for translocation breakpoints and with our validation based on RNA templates, we cannot exclude trans-splicing, an uncommon process in which 2 independent pre-mRNAs are joined during splicing to form a fusion transcript.46
Nevertheless, the COO of PMBL remains uncertain. Our results suggest a transition through the GC structure, followed by thymic homing. Our assessment of B-cell clonality in PMBLs by the 5′RACE assay and the almost exclusive identification of non-IgM isotypes strongly influenced by AID, which is responsible for CSR, and SHM, reinforce the hypothesis that GC-like activity occurs within the thymus.21 The predominance of IgG rather than IgM isotypes contrasts with that of follicular lymphomas and activated B-cell like (ABC) diffuse LBCL (DLBCL).47 Indeed, ABC DLBCLs express an IgM-BCR in ∼80% of cases as opposed to only ∼34% in germinal center B-cell like DLBCLs48 because of defects in the CSR mechanisms in ABC DLBCLs.48,49 In addition, the frequent involvement of SHM and CSR in the generation of translocations with genes targeted by AID involved in the fusion transcripts described in this study points to the GC as the microenvironment in which many B-cell lymphomas develop.50 AID introduces SHM by converting cytosine to uracil51 and is also responsible for initiating diversity in the V(D)J repertoire and genomic instability in balance with BCL6.52,53 In our cohort, almost all the PMBLs underwent CSR from IgM to IgG, as previously reported.21 Popov et al54 previously observed that secondary CSR from IgG to IgA (Iα-Cγ) or from IgG to IgE (Iε-Cγ) does not occur in most patients with PMBL, although AID may be expressed at a high level. However, this was not analyzed in this study. Furthermore, each patient with PMBL had a different IgVH/IgVL CDR3 sequence without any recurrence/canonic sequence and without any common superantigen or dominant VH family as reported in chronic lymphocytic leukemia or nodular lymphocyte-predominant Hodgkin lymphoma, which counters the hypothesis that PMBL may be promoted by a similar antigen.55,56 BCRs are usually key elements in B-cell oncogenic maintenance.57 As in cHL, multiple components of the BCR signaling cascade are typically expressed at low or undetectable levels in PMBL.3 In cHL, Epstein-Barr Virus+ Epstein-Barr Virus nuclear antigen 2/latent membrane protein 1 can act as a surrogate BCR.58 We hypothesize that other key genetic events, such as XPO1, STAT6, and immune escape drivers (PD1/PDL1/PDL2 axis), may be able to compensate for the lack of the BCR in PMBL, but these events remain to be determined.
Finally, by correlating the 5ʹRACE results of PMBLs with outcomes, we identified a subset of patients with HCD that represented ∼16% of our patients. These cases were isolated based on thresholds of CDR3 sequence predominance in each sample as determined by the receiver operating characteristic analysis of PFS. To ensure that the apparent low clonal dominance of other cases was not a reflection of ongoing SHM, we used only the CDR3 to identify clones and we aggregated all CDR3 cases with a Levenstein distance of <0.2 from the dominant CDR3 as previously described.25 Based on the multivariate analyses, HCD was an independent prognostic factor for the outcomes of both a shorter PFS and shorter OS. This finding was confirmed in the validation cohort using the same cutoffs for HCD, which emphasizes the robustness of our results. Although PMBL is a disease with an overall good prognosis, we observed, to our knowledge for the first time, that a subset of PMBL tumors may present with enrichment of tumor cells (markedly augmented expression of tumor driver genes and median VAF), expansion of a strongly immunodominant clonotype, and a reduction in the reactive microenvironment cells, namely, a decrease in normal B- and T-cell infiltration. This result is consistent with other data from the literature on DLBCL, peripheral T-cell lymphomas, and solid tumors and contributes to the increasing evidence that tumor-infiltrating lymphocytes are associated with improved long-term survival across malignancies.59-65 Recently, the positive association between improved survival and tumor-infiltrating B lymphocyte abundance in DLBCL was demonstrated by Xu-Monette et al who performed ultradeep sequencing of immunoglobulin genes to determine the B-cell clonotypes of 269 patients with de novo DLBCL.59 In our study, the normal B-cell component was markedly lower in HCD cases than in other cases, as shown by the lower entropy of the BCR repertoire (ie, higher Simpson index among BCR sequences), thereby demonstrating a reduction in the heterogeneity of the overall B-cell population in the tumor. Notably, we did not observe a difference in the MTV or maximum standardized uptake values between the HCD and non-HCD groups, suggesting no relationship between HCD and tumor burden.
A majority of the patients with PMBL in the HCD group showed augmented PDL1 and PDL2 gene expression; thus, high PDL1/PDL2 gene expression, as determined by the cutoffs defined in our previous GEP work on this cohort,24 seems to be linked with HCD. Considering that these patients also strongly express AID and the main B driver genes with a weak tumor microenvironment (lower number of polyclonal BCR and TCR CDR3 sequences), we hypothesized that these cases may represent PMBLs with higher tumoral infiltration.
We would like to highlight several limitations of our work. Regarding the technical aspects of the 5ʹRACE assay, the high failure rate (45%) observed in the multicenter cohort was probably related to poor RNA quality (reverse transcription multiplex ligation-dependent probe amplification quality score low, fragmented RNAs) and could be attributed to the age of the collected samples (between 2007 and 2017) and the multicentric and heterogeneous nature of the sampling and fixation methods. In contrast, the monocentric validation cohort (collected between 2007 and 2022) demonstrated a lower failure rate of 7.5%. This discrepancy may be a consequence of the more consistent and controlled conditions of the monocentric cohort. Alternatively, 5ʹRACE may fail if there are many somatic mutations that affect the IgVH/IgVL genes that could prevent primer hybridization, but our primers targeted constant regions that typically exhibited few or no mutations. In addition, tumors for which the 5ʹRACE assay did not fail but clonality was not established (polyclonal profiles, ∼20% of cases) were associated with lower tumoral infiltration. Our results are consistent with those reported by Leithäuser et al21 who detected clonal IgVH in only 13 of 20 (65%) patients (using DNA as template and PCR amplification), and immunoglobulin transcripts in only 61.5% of patients. However, our RNA-based approach that relied on whole tissue sections overlooked the variable levels of immunoglobulin transcripts in B cells, which is particularly concerning for PMBL that may exhibit low immunoglobulin transcription in tumor cells. Consequently, our immunoglobulin transcript analysis may underestimate the fraction of lymphoma cells, potentially explaining why clonal dominance does not correlate well with the actual lymphoma clone size. Notably, the 5ʹRACE failure seems to be more common in the subset of patients with PMBL within the LYSA cohort with a PDL1high/PDL2high profile. Therefore, we were not able to determine the synergistic prognostic effect of the combination of a PDL1high/PDL2high status and HCD. We previously demonstrated that patients with PDL1high/PDL2high have a poor prognosis.24 The hypothesis that these 2 variables define the same population with markedly poorer outcomes that are related to more aggressive biology should be assessed in another cohort of patients. It would be intriguing to investigate the potential use of anti-PD1 antibodies in patients with HCD. However, the depleted tumor microenvironment could contribute to a lack of response to anti-PD1 therapy.66 To date, we lack data that correlate 5ʹRACE results with the response to anti-PD1 treatment in PMBLs. To date, 5ʹRACE has not been used to assess the clonality profile of other LBCLs. Therefore, we lack additional data sets to compare with ours or to evaluate the prognostic impact of HCD in other LBCLs. This highlights an area of potential future research to further our understanding of LBCLs and the role of HCD as a prognostic biomarker. Given that 5ʹRACE is unlikely to be routinely adopted, we suggest considering alternative commercially available assays that use DNA as a template to determine if they can provide similar insights into the dominant clonotypes and the BCR repertoire. Finally, our 5ʹRACE assay cannot detect fusion transcripts that do not involve immunoglobulin loci that have been described using fluorescent in situ hybridization experiments, including translocations of CIITA (38%)67 and PDL1/PDL2 (20%)68 when the partner is not an immunoglobulin locus or when the translocation does not produce a fusion transcript.
In conclusion, analysis of the BCR repertoire by 5ʹRACE highlighted that most PMBL tumors are strongly subject to SHM and CSR, suggesting strong involvement of AID in oncogenesis. This work suggests that there exists a rare subgroup of PMBLs with HCD and poorer outcomes. HCD may reflect greater tumor cell infiltration in these tumors and contribute to a better understanding of the biology of this lymphoma and improve patient management.
Acknowledgments
The authors thank the patients and their families, Marie-Hélène Delfau-Larue who reviewed this manuscript for the Lymphoma Study Association (LYSA) scientific committee, and all of the investigators at the LYSA centers. The authors thank the Biological Resource Center of the Centre Henri Becquerel for tumor sample banking and nucleic acid extractions. The authors thank the LYSA Pathology Group and platform for project management, especially Maryse BAIA. The authors thank the Centre de Traitement des Données du Cancéropôle Nord-Ouest and Julie Libraire, clinical data manager at the Clinical Research Unit, for their data management support. The authors thank the Lymphopath consortium and pathologists at the LYSA centers for sending their samples for this study.
The PMBL LYSA study was supported by grants from the Ligue Contre le Cancer (Comité de Seine-Maritime, AO_2020), the groupement d'intérêt public Cancéropôle Nord-Ouest (N°2021/01 and N°2021/13), Force Hémato (N°02-2020), and Institut Carnot Consortium for Lymphoma Research (ANR-2020).
Authorship
Contribution: V.C. designed and supervised the study, collected, analyzed, and interpreted the data, and wrote the manuscript; M.V. performed and interpreted the 5′ rapid amplification of cDNA ends (5′RACE), reverse transcriptase polymerase chain reaction, and Sanger experiments, conducted the statistical analysis, and created the tables and figures; P.-J.V. performed the bioinformatics analysis; F.D., E.-L.V., V.B., V.R., E.B., P.S., C.H., E.D., M.B., C.R., L.M., D.P., S.K., J.B., J.P., O.T., N.G., A.W., C.A., L.R., E.L., P.G., H.T., T.J.M., A.T.-G., M.D., P.R., and F.J. collected and interpreted the data; E.B. performed and interpreted the next-generation sequencing experiments; V.R. and P.R. performed and interpreted the gene expression profile experiments; P.R. designed and interpreted the 5ʹRACE experiments; P.D., S.B., and D.T. analyzed the metabolic imaging data; F.J. designed and supervised the study, analyzed and interpreted the data, and edited the manuscript; and all authors interpreted the data, edited the manuscript, and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vincent Camus, Department of Hematology, Centre Henri Becquerel, 1 Rue d’Amiens, 76038 Rouen Cedex, France; email: vincent.camus@chb.unicancer.fr.
References
Author notes
The sequencing data are publicly available on the National Center for Biotechnology Information (reference PRJNA1139660).
Data are available on request from the corresponding author, Vincent Camus (vincent.camus@chb.unicancer.fr).
The full-text version of this article contains a data supplement.