Mediation MR analysis reveals that gut microbiota genus Ruminococcaceae UCG-002 may increase the risk of DLBCL by elevating serum MIG levels.
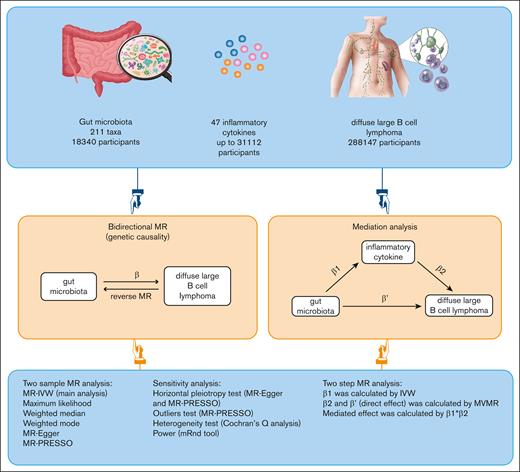
Visual Abstract
Causal relationships between gut microbiota, inflammatory cytokines, and diffuse large B-cell lymphoma (DLBCL) remain elusive. In addressing this gap, our Mendelian randomization (MR) study used data from the MiBioGen consortium encompassing 211 microbiota taxa (n = 18 340), genome-wide association study meta-analyses of 47 inflammatory cytokines, and DLBCL cases and controls from the FinnGen consortium (cases, n = 1010; controls, n = 287 137). Through bidirectional MR analyses, we examined the causal links between gut microbiota and DLBCL and used mediation analyses, including 2-step MR and multivariable MR (MVMR), to identify potential mediating inflammatory cytokines. Our findings revealed that 4 microbiota taxa were causally associated with DLBCL, and conversely, DLBCL influenced the abundance of 20 taxa. Specifically, in the 2-step MR analysis, both the genus Ruminococcaceae UCG-002 (odds ratio [OR], 1.427; 95% confidence interval [CI], 1.011-2.015; P = .043) and the inflammatory cytokine monokine induced by gamma (MIG) (OR, 1.244; 95% CI, 1.034-1.487; P = .020) were found to be causally associated with an increased risk of DLBCL. Additionally, a positive association was observed between genus Ruminococcaceae UCG-002 and MIG (OR, 1.275; 95% CI, 1.069-1.520; P = .007). Furthermore, MVMR analysis indicated that the association between genus Ruminococcaceae UCG-002 and DLBCL was mediated by MIG, contributing to 14.9% of the effect (P = .005). In conclusion, our MR study provides evidence that supports the causal relationship between genus Ruminococcaceae UCG-002 and DLBCL, with a potential mediating role played by the inflammatory cytokine MIG.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most prevalent subtype of non-Hodgkin lymphoma (NHL), accounts for ∼31% of NHL cases in Western countries and ∼37% of all global B-cell tumors.1,2 DLBCL represents a heterogeneous group of biologically distinct entities characterized by the clonal proliferation of malignant B cells from germinal or postgerminal centers. This aggressive disease often results in rapid tumor growth and substantial mass development, infiltrating tissues or obstructing vital organs. Despite extensive research into various risk factors, including demographic, genetic, and tumor-specific characteristics, the exact etiology of DLBCL remains elusive.3-5 Recent studies have emphasized the influence of gut microbiota on hematologic malignancy progression and patient prognosis, suggesting a potential connection between gut microecology and immune modulation in DLBCL development.6,7 Disparities in gut microecological structures between patients with untreated DLBCL at onset and healthy populations have been observed,8,9 implying a critical role of the gut microbiota–immune axis in DLBCL development. Nevertheless, the precise mechanisms underlying the influence of gut microbiota on DLBCL remain unclear.
The tumor microenvironment, encompassing elements such as cytokines, tumor-related stromal cells, infiltrating immune cells, and epithelial cells, has emerged as a pivotal factor influencing lymphoma progression and prognosis.10-12 Specific serum circulating inflammatory cytokines, including soluble interleukin-2 receptors (sIL-2R), IL-6, tumor necrosis factor-α, and IL-8, have been linked to lymphoid cells and associated with the development and poor prognosis of DLBCL.13-18 However, the precise roles of these cytokines in DLBCL remain elusive. Previous research has established connections between gut microbiota and inflammatory cytokines. Alterations in gut microbiome composition are frequently linked to abnormal immune responses and the dysregulated production of inflammatory cytokines, promoting a proinflammatory environment conducive to various diseases.19-21 For instance, Lin et al found that changes in gut microbiota were associated with increased plasma levels of interferon gamma and tumor necrosis factor, contributing to the inflammatory processes in Parkinson disease.22 Therefore, it is reasonable to speculate that causal relationships may exist between gut microbiota, inflammatory cytokines, and DLBCL. Our study aims to elucidate these potential associations and identify specific inflammatory cytokines that could serve as valuable tools for early diagnosis and potential clinical treatment.
Mendelian randomization (MR) is an approach using genetic variants as instrumental variables (IVs) to establish causal relationships between exposures and clinical outcomes while controlling for confounders and reducing reverse causation bias.23 Furthermore, increasing evidence supports the utility of human genetic data related to gut microbial characteristics in clinical investigations, allowing for us to use MR as a powerful tool to infer causal links between the gut microbiota and DLBCL.24 In this study, we conducted a bidirectional MR analysis and mediation analyses using summary statistics from the most extensive and current genome-wide association study (GWAS) of the gut microbiota, inflammatory cytokines, and DLBCL. These analyses aimed to dissect the intricate associations between these variables, offering valuable insights into their causal relationships.
Materials and methods
Study design
In our study, we used publicly available GWAS data to investigate the genetic association between gut microbiota and DLBCL. We decomposed the overall effect of gut microbiota on DLBCL into 2 components: direct effects and indirect (mediated) effects. The direct effects of gut microbiota on DLBCL were determined through a MVMR approach, adjusting for the influence of inflammatory cytokines. Additionally, we assessed the indirect effects mediated by inflammatory cytokines.
To explore the potential mediating role of circulating inflammatory cytokines in the relationship between gut microbiota and DLBCL, we conducted a 2-step MR study. This involved initially assessing the causal effect of gut microbiota on inflammatory cytokines, followed by investigating the causal impact of these cytokines on DLBCL. Subsequently, we used a MVMR method to quantify the direct effect of gut microbiota on DLBCL, accounting for potential mediators. This study adhered to the reporting guidelines of the Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization.23Figure 1 illustrates the schematic diagram of the study design.
Study design and flowchart. ‘β’ denotes the total effect of gut microbiota on DLBCL, ‘β1’ indicates the MR effect of gut microbiota on mediators, and ‘β2’ reflects the MR effect of mediators on DLBCL, taking into account genetically determined gut microbiota. ‘β'’ represents the direct effect of gut microbiota on DLBCL. MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier.
Study design and flowchart. ‘β’ denotes the total effect of gut microbiota on DLBCL, ‘β1’ indicates the MR effect of gut microbiota on mediators, and ‘β2’ reflects the MR effect of mediators on DLBCL, taking into account genetically determined gut microbiota. ‘β'’ represents the direct effect of gut microbiota on DLBCL. MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier.
Data sources
We sourced gut microbiota data from an extensive analysis conducted by the MiBioGen consortium,25 encompassing 18 340 individuals across 11 countries, mainly consisting of adults or adolescents of European descent. This data set amalgamated genetic information from 24 cohorts, encompassing whole-genome genotypes, with fecal microbiome data derived via 16S ribosomal RNA gene sequencing. The microbiome data provided information on 211 gut microbial taxa classified at 5 taxonomic levels, spanning 131 genera, 35 families, 20 orders, 16 classes, and 9 phyla.
We obtained summary-level data on DLBCL from the FinnGen consortium, which comprised 1010 cases and 287 137 controls. Furthermore, we integrated GWAS data on circulating levels of 47 inflammatory cytokines from a diverse European population, including Finnish cohorts from the Northern Finland Birth Cohort 1966 (NFBC1966),26 the “Cardiovascular Risk in Young Finns Study,” and the “FINRISK” studies (FINRISK1997 and FINRISK2002).27 Additional robust estimates were drawn from the SCALLOP consortium and the INTERVAL study,28,29 allowing for us to pool single-nucleotide polymorphism (SNP)–cytokine association data effectively. This comprehensive meta-analysis covered up to 31 112 individuals, providing us with the most accurate effect estimates for each cytokine. Characteristics of corresponding GWAS data sources are described in supplemental Table 1, whereas the sources and sample sizes for the 47 inflammatory cytokines are outlined in supplemental Table 2.
IV selection and data harmonization
In our MR analysis, we used SNPs as IVs to investigate the causal connections between exposures and outcomes at the gene level. Initially, we intended to select SNPs that exhibited significant associations with gut microbiota and inflammatory cytokines (P < 5 × 10−⁸). However, due to the limited availability of genome-wide significant SNPs, we adopted a more relaxed statistical threshold (P < 1 × 10−⁵) in our MR investigations.24,30 This adjustment allowed for us to access a broader set of SNPs related to gut microbiota, inflammatory cytokines, and DLBCL.
We established specific criteria for SNP selection, using a threshold of R2 <0.001 and 10 000 kb to eliminate SNPs located within a 10 Mb region that exhibited linkage disequilibrium with the most significant SNP (R2 > 0.001).31 This strategy enabled the exclusion of SNPs susceptible to linkage disequilibrium effects. Moreover, SNPs exhibiting palindromic or ambiguous characteristics were systematically excluded from the IVs used in the MR analysis. To prevent weak instrument bias, we also calculated F statistics (F for a single SNP is calculated as β2/standard error2).32
MR analysis and mediation analysis
We performed a comprehensive 2-sample bidirectional MR analysis to assess the reciprocal causality between gut microbiota and DLBCL, termed “total effect.” After this, a mediation analysis was performed using a 2-step MR approach to identify inflammatory cytokines that mediate the relationship between gut microbiota and DLBCL. Firstly, we determined the direct effects of gut microbiota on DLBCL through MVMR, accounting for the role of inflammatory cytokines. Next, we calculated the indirect effects, denoted as “mediated effect,” by multiplying β1 by β2. Here, β1 represents the MR effect of gut microbiota on mediators, whereas β2 signifies the MR effect of mediators on DLBCL, considering genetically determined gut microbiota.33 To quantify the extent of mediation, we divided the indirect effect by the total effect of gut microbiota on DLBCL. This analysis provided insights into the proportion of the gut microbiota's influence on DLBCL that is mediated by inflammatory cytokines in the causal pathway.
Sensitivity analysis
In our univariable MR analysis, the inverse-variance weighted (IVW) method served as the primary approach due to its robustness in integrating the effect estimates from individual SNPs into a single aggregated effect size. This method is particularly advantageous when horizontal pleiotropy is absent, because it uses a random-effects meta-analysis framework to effectively accommodate the variability across different SNP effects. Consequently, unless there is evidence suggesting the presence of horizontal pleiotropy, the results derived from the IVW method are considered the main findings of our study. To support the robustness of our IVW-based results and to provide a comprehensive analysis, we included several complementary MR methods. The maximum likelihood method complements the IVW by optimizing the fit of the model to the observed data, making it particularly useful in situations with normally distributed SNP effects. The weighted median method adds an extra layer of robustness, yielding valid estimates even when up to 50% of the instrumental variables are invalid due to pleiotropy or other biases.34 Furthermore, we used MR-Egger regression, which not only offers MR estimates but also enables the detection of directional pleiotropy through its intercept term. This method provides insights into the average pleiotropic effect of the genetic variants used as instruments.35 However, due to its larger standard errors, the MR-Egger method is usually considered secondary to the IVW when there is no evidence of pleiotropy. The Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) method was also used for its ability to detect and adjust for outliers that could be influencing the results due to pleiotropic effects not accounted for by the other methods. Lastly, Cochran Q analysis was conducted to evaluate the heterogeneity among the individual causal effects estimated by each SNP. A P value >.05 in this analysis suggests that the heterogeneity is not statistically significant, supporting the consistency of the findings across the different genetic variants.
In our analysis, we assessed the power for detecting true causal relationships using the mRnd tool, which applies a noncentrality parameter for a 2-sample MR approach to binary outcomes. This advanced calculation considers the variance explained by the IVs, the odds ratio (OR) for exposure-effect on outcome, the proportion of cases in the outcome, and the overall sample size, providing a rigorous estimate of our study's ability to identify significant results.36 In addition, multiple testing correction was addressed through the application of the Bonferroni correction procedure.37 A 2-sided P value that passed the correction was defined as statistically significant, which is .00024 (0.05/211) for gut microbiota and .001 (0.05/47) for inflammatory cytokines, and P value <.05 was considered to have a suggestively significant association. All statistical analyses were performed using R version 4.2.2 (R Foundation for Statistical Computing, Vienna, Austria). MR analyses were performed using the TwosampleMR (version 0.5.7), MR-PRESSO (version 1.0), and MVMR (version 0.4) R packages.
Results
Genetic instruments for exposures
The number of SNPs serving as IVs varied among data sets. For the 211 gut microbiota taxa, SNPs ranged from 3 to 24 (median, 12). In the case of 47 inflammatory cytokines, the range was 10 to 101 (median, 21). In the reverse MR analysis of DLBCL on gut microbiota, the range for DLBCL was 1 to 8 (median, 8). The median F statistic was 21.0 (range, 14.6-88.4) for gut microbiota, 22 (range, 19.4-4133.5) for inflammatory cytokines, and 21.4 (range, 20.0-61.4) for DLBCL (supplemental Tables 3-5). An F statistic >10 is typically informative for MR analyses, and all IVs had F statistics >10, indicating no bias due to weak IVs. These results confirmed the robustness of IVs, making the data suitable for MR investigations.
Associations of gut microbiota and DLBCL
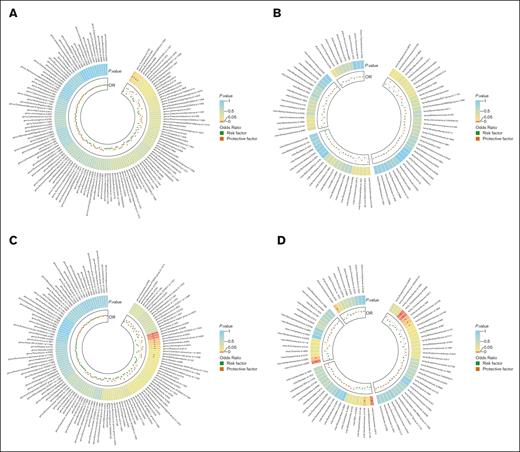
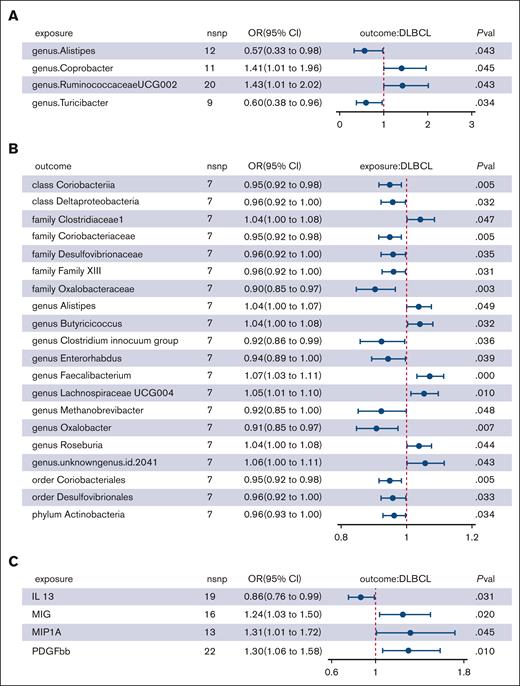
In our genetic analysis of bacterial taxa associated with DLBCL, we used 2676 SNPs as IVs. Multiple MR analysis methods provided evidence of potential causal links between 4 genera and DLBCL risk (supplemental Table 6; Figure 2A-B). IVW analysis indicated that genus Coprobacter (OR, 1.407; 95% confidence interval [CI], 1.008-1.963; P = .045) and genus Ruminococcaceae UCG-002 (OR, 1.427; 95% CI, 1.011-2.015; P = .043) were risk factors for DLBCL, whereas genus Alistipes (OR, 0.568; 95% CI, 0.328-0.982; P = .043) and genus Turicibacter (OR, 0.604; 95% CI, 0.379-0.964; P = .034) were protective factors (Table 1; Figure 3A). However, after applying the Bonferroni correction for multiple comparisons, these associations were not statistically significant.
Heat map showing bidirectional causality between gut microbiota and DLBCL. (A) Causality of 131 genera on DLBCL. (B) Causality of other 4 microbiota taxonomic levels on DLBCL. (C) Causality of DLBCL on 131 genera. (D) Causality of DLBCL on other microbiota taxonomic levels. Results that reached statistical significance in the IVW analysis are marked with an asterisk (∗) and represent a P value of <.05.
Heat map showing bidirectional causality between gut microbiota and DLBCL. (A) Causality of 131 genera on DLBCL. (B) Causality of other 4 microbiota taxonomic levels on DLBCL. (C) Causality of DLBCL on 131 genera. (D) Causality of DLBCL on other microbiota taxonomic levels. Results that reached statistical significance in the IVW analysis are marked with an asterisk (∗) and represent a P value of <.05.
MR analyses of causal effects of 4 gut microbial genera on DLBCL
| Exposure . | Method . | SNP . | β (95% CI) . | P value . | OR (95% CI) . | Power . |
|---|---|---|---|---|---|---|
| Genus Alistipes | IVW | 12 | −0.566 (−1.115 to −0.017) | .043 | 0.568 (0.328-0.982) | 0.4 |
| Maximum likelihood | 12 | −0.570 (−1.128 to −0.012) | .045 | 0.565 (0.324-0.988) | ||
| MR-PRESSO | 12 | −0.441 (−0.878 to −0.004) | .071 | 0.643 (0.415-0.996) | ||
| WM | 12 | −0.545 (−1.247 to 0.157) | .128 | 0.58 (0.287-1.170) | ||
| MR-Egger | 12 | 1.235 (−1.376 to 3.846) | .376 | 3.439 (0.253-46.812) | ||
| Genus Coprobacter | IVW | 11 | 0.341 (0.008-0.674) | .045 | 1.407 (1.008-1.963) | 0.82 |
| Maximum likelihood | 11 | 0.355 (0.018-0.692) | .039 | 1.426 (1.018-1.997) | ||
| MR-PRESSO | 11 | 0.346 (0.062-0.63) | .033 | 1.426 (1.063-1.878) | ||
| WM | 11 | 0.388 (−0.061 to 0.837) | .091 | 1.474 (0.941-2.309) | ||
| MR-Egger | 11 | 0.62 (−0.734 to 1.974) | .393 | 1.859 (0.48-7.205) | ||
| Genus Ruminococcaceae UCG-002 | IVW | 20 | 0.356 (0.011-0.701) | .043 | 1.427 (1.011-2.015) | 0.75 |
| Maximum likelihood | 20 | 0.367 (0.016-0.718) | .040 | 1.443 (1.016-2.050) | ||
| MR-PRESSO | 20 | 0.298 (0.078-0.518) | .014 | 1.443 (1.083-1.677) | ||
| WM | 20 | 0.358 (−0.132 to 0.848) | .152 | 1.43 (0.876-2.335) | ||
| MR-Egger | 20 | 0.503 (−0.391 to 1.397) | .285 | 1.653 (0.676-4.044) | ||
| Genus Turicibacter | IVW | 9 | −0.504 (−0.97 to −0.038) | .034 | 0.604 (0.379-0.964) | 0.66 |
| Maximum likelihood | 9 | −0.532 (−0.964 to −0.100) | .016 | 0.587 (0.381-0.905) | ||
| MR-PRESSO | 9 | −0.226 (−0.692 to 0.24) | .36 | 0.587 (0.501-1.271) | ||
| WM | 9 | −0.516 (−1.088 to 0.056) | .078 | 0.597 (0.337-1.059) | ||
| MR-Egger | 9 | −0.359 (−2.38 to 1.662) | .738 | 0.698 (0.093-5.264) |
| Exposure . | Method . | SNP . | β (95% CI) . | P value . | OR (95% CI) . | Power . |
|---|---|---|---|---|---|---|
| Genus Alistipes | IVW | 12 | −0.566 (−1.115 to −0.017) | .043 | 0.568 (0.328-0.982) | 0.4 |
| Maximum likelihood | 12 | −0.570 (−1.128 to −0.012) | .045 | 0.565 (0.324-0.988) | ||
| MR-PRESSO | 12 | −0.441 (−0.878 to −0.004) | .071 | 0.643 (0.415-0.996) | ||
| WM | 12 | −0.545 (−1.247 to 0.157) | .128 | 0.58 (0.287-1.170) | ||
| MR-Egger | 12 | 1.235 (−1.376 to 3.846) | .376 | 3.439 (0.253-46.812) | ||
| Genus Coprobacter | IVW | 11 | 0.341 (0.008-0.674) | .045 | 1.407 (1.008-1.963) | 0.82 |
| Maximum likelihood | 11 | 0.355 (0.018-0.692) | .039 | 1.426 (1.018-1.997) | ||
| MR-PRESSO | 11 | 0.346 (0.062-0.63) | .033 | 1.426 (1.063-1.878) | ||
| WM | 11 | 0.388 (−0.061 to 0.837) | .091 | 1.474 (0.941-2.309) | ||
| MR-Egger | 11 | 0.62 (−0.734 to 1.974) | .393 | 1.859 (0.48-7.205) | ||
| Genus Ruminococcaceae UCG-002 | IVW | 20 | 0.356 (0.011-0.701) | .043 | 1.427 (1.011-2.015) | 0.75 |
| Maximum likelihood | 20 | 0.367 (0.016-0.718) | .040 | 1.443 (1.016-2.050) | ||
| MR-PRESSO | 20 | 0.298 (0.078-0.518) | .014 | 1.443 (1.083-1.677) | ||
| WM | 20 | 0.358 (−0.132 to 0.848) | .152 | 1.43 (0.876-2.335) | ||
| MR-Egger | 20 | 0.503 (−0.391 to 1.397) | .285 | 1.653 (0.676-4.044) | ||
| Genus Turicibacter | IVW | 9 | −0.504 (−0.97 to −0.038) | .034 | 0.604 (0.379-0.964) | 0.66 |
| Maximum likelihood | 9 | −0.532 (−0.964 to −0.100) | .016 | 0.587 (0.381-0.905) | ||
| MR-PRESSO | 9 | −0.226 (−0.692 to 0.24) | .36 | 0.587 (0.501-1.271) | ||
| WM | 9 | −0.516 (−1.088 to 0.056) | .078 | 0.597 (0.337-1.059) | ||
| MR-Egger | 9 | −0.359 (−2.38 to 1.662) | .738 | 0.698 (0.093-5.264) |
IVW was used as the primary method, synthesizing SNP effects by prioritizing those with higher precision under the assumption of no horizontal pleiotropy. Complementing this, the maximum likelihood method was used to optimize model fit with normally distributed effects. MR-PRESSO was applied to detect and correct for pleiotropic biases from SNPs. The WM provided robust estimates against invalid instrumental variables, focusing on the median of their weighted effects. Lastly, MR-Egger was used to identify directional pleiotropy, essential for unbiased estimation under the Instrument Strength Independent of Direct Effect (InSIDE) assumption.
MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; WM, weighted median.
Forest plot showing significant causal effects between gut microbiota, inflammatory cytokines, and DLBCL from the IVW analysis. (A) Causal effects of 4 gut microbial genera on DLBCL. (B) Causal effects of DLBCL on 20 gut microbial taxa. (C) Causal effects of 4 inflammatory cytokines on DLBCL. nsnp, number of single nucleotide polymorphism; pval, P value.
Forest plot showing significant causal effects between gut microbiota, inflammatory cytokines, and DLBCL from the IVW analysis. (A) Causal effects of 4 gut microbial genera on DLBCL. (B) Causal effects of DLBCL on 20 gut microbial taxa. (C) Causal effects of 4 inflammatory cytokines on DLBCL. nsnp, number of single nucleotide polymorphism; pval, P value.
When assessing the causal effects of DLBCL on gut microbiota, we found evidence suggesting a genetic predisposition to DLBCL was linked to specific bacterial groups, with lower abundance in 1 phylum, 2 classes, 2 orders, 4 families, and 4 genera and higher abundance in 1 family and 6 genera (supplemental Tables 7-8; Figure 2C-D). The most significant result was for the genus Faecalibacterium, with an OR of 1.071 (95% CI, 1.031-1.113; P = .0004).
Notably, DLBCL was positively associated with genus Alistipes (OR, 1.037; 95% CI, 1.000-1.075; P = .049), whereas genus Alistipes had a protective effect against DLBCL when considered as an exposure (Figure 3B). Nevertheless, no bacterial taxa remained significantly associated after Bonferroni correction.
Heterogeneity tests for the bacterial taxa and DLBCL showed all P values >.05, and no evidence of horizontal pleiotropy was found using the MR-Egger method. MR-PRESSO analysis confirmed the absence of outliers, enhancing the reliability of our IVW analysis results (supplemental Tables 9-10).
Mediation analysis of gut microbiota, inflammatory cytokines, and DLBCL
In the 2-step MR analysis, only 4 inflammatory cytokines were causally associated with DLBCL (supplemental Table 10; supplemental Figure 1). IVW analysis revealed that genetically predicted monokine induced by gamma (MIG; OR, 1.244; 95% CI, 1.034-1.487; P = .020), macrophage inflammatory protein-1 alpha (MIP-1α; OR, 1.314; 95% CI, 1.006-1.718; P = .045), and platelet-derived growth factor BB (PDGFbb; OR, 1.298; 95% CI, 1.063-1.585; P = .010) were risk factors for DLBCL. In contrast, IL-13 (OR, 0.864; 95% CI, 0.756-0.987; P = .031) was identified as a protector for DLBCL (Table 2; Figure 3C). However, after Bonferroni correction, these associations did not reach statistical significance.
MR analyses of causal effects of 4 inflammatory cytokines on DLBCL
| Exposure . | Method . | SNP . | β (95% CI) . | P value . | OR (95% CI) . | Power . |
|---|---|---|---|---|---|---|
| IL13 | IVW | 19 | −0.146 (−0.279 to −0.013) | .031 | 0.864 (0.756-0.987) | 0.58 |
| Maximum likelihood | 19 | −0.147 (−0.282 to −0.011) | .034 | 0.864 (0.754-0.989) | ||
| MR-PRESSO | 19 | −0.033 (−0.138 to 0.073) | .55 | 0.968 (0.871-1.075) | ||
| WM | 19 | −0.062 (−0.254 to 0.13) | .527 | 0.94 (0.776-1.139) | ||
| MR-Egger | 19 | 0.021 (−0.238 to 0.28) | .875 | 1.021 (0.788-1.323) | ||
| MIG | IVW | 16 | 0.219 (0.035-0.403) | .02 | 1.244 (1.034-1.497) | 0.79 |
| Maximum likelihood | 16 | 0.229 (0.039-0.418) | .018 | 1.257 (1.040-1.519) | ||
| MR-PRESSO | 16 | 0.219 (0.050-0.388) | .023 | 1.244 (1.051-1.474) | ||
| WM | 16 | 0.503 (0.117-0.889) | .023 | 1.654 (1.125-2.432) | ||
| MR-Egger | 16 | 0.209 (−0.044 to 0.462) | .105 | 1.233 (0.957-1.589) | ||
| MIP1α | IVW | 13 | 0.274 (0.005-0.543) | .045 | 1.315 (1.006-1.718) | 0.89 |
| Maximum likelihood | 13 | 0.291 (0.063-0.519) | .012 | 1.337 (1.065-1.680) | ||
| MR-PRESSO | 13 | 0.274 (0.005-0.543) | .068 | 1.315 (1.006-1.718) | ||
| WM | 13 | 0.28 (−0.359 to 0.919) | .409 | 1.323 (0.698-2.507) | ||
| MR-Egger | 13 | 0.15 (−0.171 to 0.471) | .359 | 1.162 (0.843-1.601) | ||
| PDGFbb | IVW | 22 | 0.261 (0.062-0.461) | .01 | 1.298 (1.063-1.585) | 0.95 |
| Maximum likelihood | 22 | 0.267 (0.081-0.454) | .005 | 1.307 (1.084-1.575) | ||
| MR-PRESSO | 22 | 0.29 (0.113-0.467) | .004 | 1.336 (1.119-1.595) | ||
| WM | 22 | 0.13 (−0.146 to 0.407) | .356 | 1.139 (0.864-1.503) | ||
| MR-Egger | 22 | 0.17 (−0.253 to 0.594) | .44 | 1.186 (0.776-1.811) |
| Exposure . | Method . | SNP . | β (95% CI) . | P value . | OR (95% CI) . | Power . |
|---|---|---|---|---|---|---|
| IL13 | IVW | 19 | −0.146 (−0.279 to −0.013) | .031 | 0.864 (0.756-0.987) | 0.58 |
| Maximum likelihood | 19 | −0.147 (−0.282 to −0.011) | .034 | 0.864 (0.754-0.989) | ||
| MR-PRESSO | 19 | −0.033 (−0.138 to 0.073) | .55 | 0.968 (0.871-1.075) | ||
| WM | 19 | −0.062 (−0.254 to 0.13) | .527 | 0.94 (0.776-1.139) | ||
| MR-Egger | 19 | 0.021 (−0.238 to 0.28) | .875 | 1.021 (0.788-1.323) | ||
| MIG | IVW | 16 | 0.219 (0.035-0.403) | .02 | 1.244 (1.034-1.497) | 0.79 |
| Maximum likelihood | 16 | 0.229 (0.039-0.418) | .018 | 1.257 (1.040-1.519) | ||
| MR-PRESSO | 16 | 0.219 (0.050-0.388) | .023 | 1.244 (1.051-1.474) | ||
| WM | 16 | 0.503 (0.117-0.889) | .023 | 1.654 (1.125-2.432) | ||
| MR-Egger | 16 | 0.209 (−0.044 to 0.462) | .105 | 1.233 (0.957-1.589) | ||
| MIP1α | IVW | 13 | 0.274 (0.005-0.543) | .045 | 1.315 (1.006-1.718) | 0.89 |
| Maximum likelihood | 13 | 0.291 (0.063-0.519) | .012 | 1.337 (1.065-1.680) | ||
| MR-PRESSO | 13 | 0.274 (0.005-0.543) | .068 | 1.315 (1.006-1.718) | ||
| WM | 13 | 0.28 (−0.359 to 0.919) | .409 | 1.323 (0.698-2.507) | ||
| MR-Egger | 13 | 0.15 (−0.171 to 0.471) | .359 | 1.162 (0.843-1.601) | ||
| PDGFbb | IVW | 22 | 0.261 (0.062-0.461) | .01 | 1.298 (1.063-1.585) | 0.95 |
| Maximum likelihood | 22 | 0.267 (0.081-0.454) | .005 | 1.307 (1.084-1.575) | ||
| MR-PRESSO | 22 | 0.29 (0.113-0.467) | .004 | 1.336 (1.119-1.595) | ||
| WM | 22 | 0.13 (−0.146 to 0.407) | .356 | 1.139 (0.864-1.503) | ||
| MR-Egger | 22 | 0.17 (−0.253 to 0.594) | .44 | 1.186 (0.776-1.811) |
MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; WM, weighted median.
Among the 4 taxa that were causally associated with DLBCL, we observed that genus Ruminococcaceae UCG-002, a risk taxon against DLBCL, was positively associated with MIG (OR, 1.275; 95% CI, 1.069-1.520; P = .007) by IVW analysis, which in turn was associated with an increased risk of DLBCL (supplemental Tables 12-19). We further performed MVMR analysis of Ruminococcaceae UCG-002–MIG-DLBCL; the direct effect of genus Ruminococcaceae UCG-002 on DLBCL after taking MIG into account was an OR of 1.348 (95% CI, 1.049-1.732; P = .025). The indirect effect of genus Ruminococcaceae UCG-002 on DLBCL via MIG was β of 0.053 (95% CI, 0.0009-0.105) and the proportion mediated by MIG was 14.9% (95% CI, 0.3-29.6; P = .005). Similarly, there was no evidence of heterogeneity or horizontal pleiotropy, and no outliers were identified by MR-PRESSO analysis (supplemental Tables 9 and 10).
Discussion
In this comprehensive MR investigation, we established a suggestive causal link between 4 gut microbial taxa and DLBCL. Additionally, DLBCL was found to alter the relative abundances of 20 gut microbial taxa. Through advanced analytical approaches, including 2-step MR and MVMR, we identified a potential mechanistic pathway implicating an inflammatory cytokine, MIG, in the relationship between the gut microbiota genus Ruminococcaceae UCG-002 and DLBCL. We propose that the genus Ruminococcaceae UCG-002 may increase the risk of DLBCL by elevating serum MIG levels. This study is pioneering in its thorough examination of the genetic ties between the gut microbiota and DLBCL, highlighting the essential role of gut microorganisms in the host-microbiota interaction pertinent to DLBCL. Given these findings, the modulation of this balance is crucial for both prevention and treatment strategies.
The gut microbiome's interactions with the host are vital for immune development, equilibrium, and the maintenance of the intestinal barrier's integrity. There is a growing body of evidence indicating that the gut microbiota can affect a range of outcomes in patients with hematologic cancers, influencing disease pathogenesis, blood cell formation, the metabolism and efficacy of chemotherapy, and overall survival.38-43 Specifically, the connection between gut microbiota and lymphoma development has been explored, with studies demonstrating the influence of gut microbes on disease penetrance, latency, oxidative stress, and the genotoxicity of certain pathogens, pointing to a potentially modifiable aspect of lymphomagenesis.44,45 In humans, research by Asao et al indicated a higher prevalence of the genus Delftia and reduced levels of Bacteroides and Clostridium in the conjunctival microbiome of patients with mucosa-associated lymphoid tissue lymphoma, suggesting a pathogenetic role for genus Delftia.46 Yet, the causative relationship between gut microbiota and DLBCL remained underexplored until now.
Our findings implicate the genera Coprobacter and Ruminococcaceae UCG-002 as risk factors for DLBCL. The genus Coprobacter, part of the Barnesiellaceae family, has previously been implicated in neurological conditions,47,48 whereas the Ruminococcaceae UCG-002 group has been linked to immune thrombocytopenia.49 Conversely, the genera Alistipes and Turicibacter appear to offer protection against DLBCL. The genus Alistipes, from the Rikenellaceae family, has been associated with varying conditions. Although some studies suggest its protective role against diseases such as liver fibrosis, colitis, and cardiovascular diseases,50-54 others indicate its pathogenic potential in colorectal cancer and its association with depression.55,56 Zhou et al reported an increased abundance of genus Alistipes and genus Roseburia in patients with DLBCL, aligning with our observations.9 Our reverse MR analysis identified that DLBCL could increase the abundance of genus Alistipes, indicating a strong association between genus Alistipes and DLBCL. Furthermore, these data suggest potential adaptive feedback by genus Alistipes in response to the inflammatory milieu of DLBCL. This adaptive mechanism warrants further investigation.
Our MR study presents compelling genetic evidence that implicates 4 inflammatory cytokines, IL-13, MIP-1α, MIG, and PDGF, in the etiology of DLBCL. Notably, IL-13 was identified as a protective factor against DLBCL, a finding that is in line with the research conducted by Yian et al. In their prospective study of 92 B-cell NHL cases with 2 matched controls each, Yian et al identified IL-13 as a protective factor and sIL-2R as a risk factor for B-cell NHL.18 MIP-1α, also recognized as C-C motif chemokine ligand 3 (CCL3), is a chemokine pivotal in the acute inflammatory response, known for its role in recruiting and activating polymorphonuclear leukocytes.57 Previous research has accentuated CCL3 as a critical gene responsive to B-cell receptor signaling in B cells. The work by Koichi et al further established that elevated serum levels of CCL3 are associated with poorer progression-free and overall survival outcomes in patients with DLBCL, suggesting its utility as a prognostic serum marker for this malignancy.58,59 Our findings suggest MIP-1α as a risk factor for DLBCL, thus reinforcing its linkage to the disease and offering novel insights into its role in DLBCL pathogenesis. Furthermore, our MR analysis also determined a direct association between PDGF and an increased risk of DLBCL, although the specific contributions of PDGF in DLBCL pathogenesis warrant further study.
Our study provides genetic evidence that MIG acts as a mediator in the effect of the bacterial genus Ruminococcaceae UCG-002 on DLBCL. MIG, or C–X–C motif chemokine ligand 9, is a member of the CXC chemokine subfamily and is implicated in leukocyte chemotaxis, differentiation, multiplication, and tissue extravasation.60 The significance of MIG within the lymphoma microenvironment and its role in lymphomagenesis have been previously described. For instance, Cheng et al demonstrated that MIG supports the progression of DLBCL via a β-catenin–dependent pathway,61 and Zhou et al reported significantly elevated C–X–C motif chemokine ligand 9 expression levels in DLBCL tissues compared with normal counterparts.62 These investigations corroborate our findings and highlight the importance of MIG in the pathogenesis and prognosis of DLBCL.
Our mediation analyses have yielded genetic evidence linking gut microbiota with inflammatory cytokines. This study, to our knowledge, appears to be the first to establish a direct connection between the genus Ruminococcaceae UCG-002 and the cytokine MIG. Although previous research has suggested a general association between gut microbiota composition and inflammatory cytokines, none have specifically linked Ruminococcaceae UCG-002 with MIG. Ratnakar et al reported that dysbiosis in gut microbiota may trigger immune activation, leading to intestinal inflammation through the stimulation of inflammatory cytokines, including IL-6, IL-10, CXC chemokine receptor-3, and CXCL-11, in patients with irritable bowel syndrome.63 Furthermore, Yang et al found that gut microbiota could orchestrate monocyte-like macrophage accumulation in a CCL2-dependent manner, stimulate IL-1β production from macrophages, and create a precancerous inflammatory environment conducive to tumorigenesis in a mouse model of colitis.64 These studies, together with our own, elucidate the intricate causal relationship between gut microbiota, inflammatory cytokines, and the pathogenesis of inflammatory diseases and certain cancers.
Our study, exploring the relationship between gut microbiota and DLBCL, encounters several constraints. Primarily, the applicability of our findings is limited due to the sample's significant European ancestry, which may not accurately reflect the genetic and lifestyle diversity influencing gut microbiota in varied populations. Additionally, the use of 16S ribosomal RNA gene sequencing provides only genus- to phylum-level insights into microbial taxonomy, lacking the depth that metagenomic sequencing could offer for detailed species-level analysis. The associations we observed between certain microbiota and DLBCL risk are preliminary and should be interpreted cautiously, considering the complex nature of gut flora interactions. Moreover, our methodology, based on MR, may not adequately capture the intricate, possibly nonlinear relationships between the gut microbiome and DLBCL. Furthermore, the current limitations of GWAS data sets specific to DLBCL hinder our ability to conduct robust replication studies with different GWAS data, which are crucial for establishing definitive causal relationships, and the relatively small sample sizes for DLBCL GWAS currently available have led to lower power in detecting true causal effects for certain exposures, and this could inadvertently increase the risk of false-negative results. Therefore, future research with more extensive GWAS data is imperative to solidify our findings and to further elucidate the complex role of gut microbiota in the pathogenesis of DLBCL. Lastly, it is also crucial to acknowledge that our findings did not withstand the rigorous thresholds of Bonferroni correction. Yet, the application of such stringent corrections, particularly within the realm of exploratory and hypothesis-generating research, risks obscuring discoveries of biological significance. MR's hypothesis-driven nature enables the detection of causally relevant associations with biological underpinnings, independent of multiple comparison adjustments. Our exploratory work thus prioritizes biologically significant associations that merit further research in larger data sets, despite not achieving strict statistical significance after correction.
Conclusion
To our knowledge, this is the first study to comprehensively evaluate the causal links between gut microbiota, inflammatory cytokines, and DLBCL. We have identified a significant role for gut microorganisms and inflammatory cytokines in the pathogenesis and progression of host DLBCL. These results underscore the necessity of exploring the mechanisms underlying the interaction between gut microbiota and DLBCL. Furthermore, our findings provide novel insights that may guide the development of microbiome-based therapies and cytokine-targeted interventions for DLBCL.
Acknowledgments
In this study, the authors used GWAS summary data from the MiBioGen, NFBC1966, YFS, FINRISK, SCALLOP, INTERVAL, and FinnGen studies. The authors extend heartfelt gratitude to all the investigators for their invaluable contributions in providing these data and to the individual patients whose sample donations made this research possible.
This work was supported by grants from National Natural Science Foundation of China (82270226) and Shanghai Shen Kang Hospital Development Center (SHDC2020CR1012B; X.S.).
Authorship
Contribution: P.J. conducted data collection, data analysis, and drafted the manuscript; F.Y. and X.Z. assisted with data analysis and data interpretation; H.S. and Q.H. contributed to manuscript revision; and all authors contributed to the conception and design of the study, and read and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xianmin Song, Department of Hematology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, No. 100 Haining Rd, Building 2, 17th Floor, Shanghai 200080, China; email: shongxm@sjtu.edu.cn.
References
Author notes
P.J., F.Y., and X.Z. are first authors.
The original contributions presented in the study are included in the article and supplemental Table 1. For additional information or inquiries, please contact the corresponding author, Xianmin Song (shongxm@sjtu.edu.cn).
The full-text version of this article contains a data supplement.