Monosomy 7 and del(7q) result in a 1-copy deletion of the NAMPT gene at 7q22.3.
NAMPT haploinsufficiency causes a vulnerability to the inhibition of NAMPT.
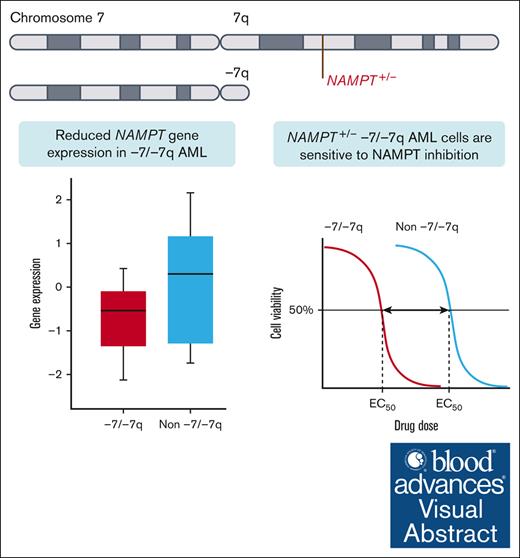
Visual Abstract
Monosomy 7 and del(7q) (-7/-7q) are frequent chromosomal abnormalities detected in up to 10% of patients with acute myeloid leukemia (AML). Despite unfavorable treatment outcomes, no approved targeted therapies exist for patients with -7/-7q. Therefore, we aimed to identify novel vulnerabilities. Through an analysis of data from ex vivo drug screens of 114 primary AML samples, we discovered that -7/-7q AML cells are highly sensitive to the inhibition of nicotinamide phosphoribosyltransferase (NAMPT). NAMPT is the rate-limiting enzyme in the nicotinamide adenine dinucleotide salvage pathway. Mechanistically, the NAMPT gene is located at 7q22.3, and deletion of 1 copy due to -7/-7q results in NAMPT haploinsufficiency, leading to reduced expression and a therapeutically targetable vulnerability to the inhibition of NAMPT. Our results show that in -7/-7q AML, differentiated CD34+CD38+ myeloblasts are more sensitive to the inhibition of NAMPT than less differentiated CD34+CD38– myeloblasts. Furthermore, the combination of the BCL2 inhibitor venetoclax and the NAMPT inhibitor KPT-9274 resulted in the death of significantly more leukemic blasts in AML samples with -7/-7q than the NAMPT inhibitor alone. In conclusion, our findings demonstrate that AML with -7/-7q is highly sensitive to NAMPT inhibition, suggesting that NAMPT inhibitors have the potential to be an effective targeted therapy for patients with monosomy 7 or del(7q).
Introduction
Monosomy 7 and deletion of chromosome arm 7q (-7/-7q) are frequent chromosomal abnormalities in up to 10% of patients with acute myeloid leukemia (AML).1 The alterations are recurrent in myeloid malignancies in both adults and children, including juvenile myelomonocytic leukemia,2,3 myelodysplastic syndromes (MDS),4 and chronic myeloid leukemia.5 In AML, monosomy 7 is correlated with poor prognosis and resistance to chemotherapy, whereas the prognostic significance of deletion 7q (del(7q)) is less clear. This difference can be due to variation in the size of the deletion or co-occurring abnormalities.6 Monosomy 7 and del(7q) are among the most common chromosomal abnormalities in patients with high-risk AML. Monosomy 7 often co-occurs with other markers of adverse prognosis, such as TP53 mutations and inv(3), worsening clinical outcomes.1 A higher frequency of -7/-7q is observed in therapy-related AML and MDS cases. As many as 50% of therapy-related cases have -7/-7q, suggesting that chemotherapy and radiation select cells with these abnormalities.7,8
Monosomy 7 and del(7q) result in the loss of 1 copy of chromosome arm 7q. The homozygous loss of both copies of 7q has not been observed, implying that the deletion of 2 copies of genes in 7q reduces tumor cell viability.6 Monosomy 7 and del(7q) lead to the haploinsufficiency of hundreds of genes. Tumor suppressor genes located on chromosome arm 7q, which have been shown to contribute to the development of myeloid malignancies, include SAMD9, SAMD9L,9KMT2C (MLL3),10,11EZH2,12,13 and CUX1.14,15 Despite unfavorable treatment outcomes for patients with AML with -7/-7q, no targeted therapies for this subgroup of patients have been approved.
Ex vivo drug sensitivity testing has emerged as a powerful tool for identifying therapeutically targetable vulnerabilities in several hematological malignancies.16-18 Results from ex vivo drug sensitivity testing have been demonstrated to correlate with clinical responses for targeted therapies, particularly in relation to the BCL2 inhibitor venetoclax.19 In this study, we aimed to identify novel therapeutic vulnerabilities in AML with -7/-7q. We analyzed ex vivo drug sensitivities of 114 AML samples using a drug library with up to 510 approved and investigational oncology compounds. We performed validation of drug sensitivities using orthogonal assays, including multiparametric flow cytometry, which enables the determination of the sensitivity of distinct cell populations in bone marrow (BM) samples.
We hypothesized that -7/-7q would render AML cells dependent on distinct survival mechanisms that could be exploited to provide novel and efficacious therapeutic strategies for patients with -7/-7q. By analyzing data from ex vivo drug screens in primary AML samples, we discovered that -7/-7q AML cells are highly sensitive to inhibition of nicotinamide phosphoribosyltransferase (NAMPT). The NAMPT gene is located at 7q22.3, and deletion of 1 copy due to -7/-7q results in haploinsufficiency. This leads to a therapeutically targetable vulnerability to the inhibition of NAMPT by reducing NAMPT gene expression in leukemia cells.
Materials and methods
Patient samples
BM or peripheral blood (PB) samples were collected after informed consent from patients with AML using protocols approved by the Institutional Review Board at the Helsinki University Hospital (permit numbers 239/13/03/00/2010 and 303/13/03/01/2011, Helsinki University Hospital Ethics Committee) in compliance with the Declaration of Helsinki. Mononuclear cells (MNCs) were isolated by Ficoll density gradient and suspended in conditioned medium.
Ex vivo drug sensitivity testing
We evaluated the drug sensitivity of freshly isolated MNCs from patients with AML to 510 small molecule inhibitors, including the NAMPT inhibitor daporinad, using CellTiter-Glo (CTG) and CellTox Green (CTxG) assays (Promega). Multiple NAMPT inhibitors were then tested at increasing concentrations using viably cryopreserved MNCs from AML samples that were left to recover in conditioned medium before being plated onto predrugged plates. After 72-hour incubation with the drugs, viability readouts from the CTG assay were normalized to negative and positive controls, and a dose-response curve was fitted to the drug sensitivity data. For each dose-response curve, a drug sensitivity score was calculated as previously described.20 Detailed quality metrics used to evaluate the ex vivo screening results, and their reproducibility were previously described 21 and are summarized for this study cohort in supplemental Figures 1 and 2.
Ex vivo drug sensitivity testing by multiparametric flow cytometry
Four NAMPT inhibitors (daporinad, GMX1778, KPT-9274, and LSN3154567) were preplated on 96-well plates, and thawed cryopreserved MNCs were added to the plates. After a 72-hour incubation, the cells were stained using the antibody panel shown in supplemental Table 1 and analyzed using flow cytometry to determine cell viability. The gating strategy is illustrated in supplemental Figure 3. The experiment was also performed to determine the sensitivity of -7/-7q AML to combinations of KPT-9274 and venetoclax in samples from 3 patients with AML with an immature blast population. The viability of the total leukemic blast population was determined after incubating the cells with either a single dose of KPT-9274, a single dose of venetoclax, or both.
Identification of monosomy 7 and del(7q)
We retrieved clinical karyotype data for patients with AML samples from the Finnish Hematology Registry and Biobank, the Hospital District of Helsinki, and Uusimaa Hematological Datalake. For AML samples without karyotype data, chromosomal copy number aberrations were identified through exome sequencing of BM or PB and a matched skin biopsy sample. Within the Cancer Genome Atlas (TCGA) AML cohort, the -7/-7q samples were identified based on chromosomal aberration annotations in the clinical data retrieved from https://gdc.cancer.gov/about-data/publications/laml_2012.
RNA sequencing and gene expression analysis
We retrieved bulk RNA sequencing data for AML samples sequenced in-house and for TCGA AML samples from the TCGA data portal.22 Raw gene expression counts were normalized and converted to log2–transformed counts per million values with edgeR v3.40.2 package functions in R.23 Single-cell gene expression profiles were determined using the 10X Genomics Chromium Single Cell 3'RNAseq platform. The data were processed using the Cell Ranger mfastq and count pipelines, aligned to the reference genome GRCh38, and analyzed using Seurat v4.3.024 and R 4.2.1. Human Cell Atlas healthy BM data from 8 different donors25 were downloaded through SeuratData R package v0.2.2 and analyzed similarly to the AML single cell data from the Institute for Molecular Medicine Finland (FIMM AML).
The supplemental Materials accompanying this article provide a complete description of the methods used in this study.
Results
In total, 180 patients with AML and 21 healthy donors were included in the study. Patients with acute promyelocytic leukemia were excluded. Chromosomal abnormalities were identified for all patients with AML, of whom 18 (10%) had -7 or del(7q). Biopsy samples taken at multiple time points (ie, at diagnosis and relapse) were obtained from 31 patients, however, only a single sample per patient was included in a given analysis. For patients from whom multiple samples were available, the sample taken at the time of AML diagnosis was selected if available. If no diagnosis sample was available, the relapse sample with the highest blast count was selected, because samples with high blast count provide the best readout of drug sensitivity of the leukemia cells. The clinical characteristics of the study cohort (FIMM AML) are shown in Table 1. The number of samples used in the experiments is listed in supplemental Table 2. Overall, patients with -7/-7q were significantly more likely to have a secondary or therapy-related AML than those without -7/-7q; otherwise, the clinical characteristics of patients in both groups were comparable. Cytogenetic aberrations and gene mutations in -7/-7q cases, for which exome-seq data are available, are illustrated in supplemental Figure 4.
Clinical characteristics of the FIMM AML patient cohort
| . | . | Total (N = 180) . | Patients with -7/-7q (n = 18; 10%) . | Patients without -7/-7q (n = 162; 90%) . | P value . |
|---|---|---|---|---|---|
| Median age (range), y | 6 (3.3%) NA | 63 (19-81) | 62.5 (22-76) | 63 (19-81) | NS (.66) |
| AML type | De novo, n (%) | 125 (69.4%) | 8 (44.4%) | 117 (72.2%) | .03 |
| sAML/tAML, n (%) | 55 (30.6%) | 10 (55.6%) | 45 (27.8%) | ||
| Disease stage | Diagnosis, n (%) | 115 (63.9%) | 10 (55.6%) | 105 (64.8%) | NS (.08) |
| Relapse, n (%) | 45 (25.0%) | 3 (16.7%) | 42 (25.9%) | ||
| Refractory, n (%) | 20 (11.1%) | 5 (27.8%) | 15 (9.3%) | ||
| Blast percentage (range) | 6 (3.3%) NA | 52.5 (3-95) | 65 (14-92) | 51 (3-95) | NS (.33) |
| FAB | AML M0, n (%) | 8 (4.4%) | 1 (5.6%) | 7 (4.3%) | NS (.19) |
| AML M1, n (%) | 25 (13.9%) | 1 (5.6%) | 24 (14.8%) | ||
| AML M2, n (%) | 42 (23.3%) | 2 (11.1%) | 40 (24.7%) | ||
| AML M4, n (%) | 14 (7.8%) | 1 (5.6%) | 13 (8.0%) | ||
| AML M5, n (%) | 25 (13.9%) | 1 (5.6%) | 24 (14.8%) | ||
| AML M6, n (%) | 1 (0.6) | 0 (0.0%) | 1 (0.6%) | ||
| NA, n (%) | 65 (36.1%) | 12 (66.7%) | 53 (32.7%) |
| . | . | Total (N = 180) . | Patients with -7/-7q (n = 18; 10%) . | Patients without -7/-7q (n = 162; 90%) . | P value . |
|---|---|---|---|---|---|
| Median age (range), y | 6 (3.3%) NA | 63 (19-81) | 62.5 (22-76) | 63 (19-81) | NS (.66) |
| AML type | De novo, n (%) | 125 (69.4%) | 8 (44.4%) | 117 (72.2%) | .03 |
| sAML/tAML, n (%) | 55 (30.6%) | 10 (55.6%) | 45 (27.8%) | ||
| Disease stage | Diagnosis, n (%) | 115 (63.9%) | 10 (55.6%) | 105 (64.8%) | NS (.08) |
| Relapse, n (%) | 45 (25.0%) | 3 (16.7%) | 42 (25.9%) | ||
| Refractory, n (%) | 20 (11.1%) | 5 (27.8%) | 15 (9.3%) | ||
| Blast percentage (range) | 6 (3.3%) NA | 52.5 (3-95) | 65 (14-92) | 51 (3-95) | NS (.33) |
| FAB | AML M0, n (%) | 8 (4.4%) | 1 (5.6%) | 7 (4.3%) | NS (.19) |
| AML M1, n (%) | 25 (13.9%) | 1 (5.6%) | 24 (14.8%) | ||
| AML M2, n (%) | 42 (23.3%) | 2 (11.1%) | 40 (24.7%) | ||
| AML M4, n (%) | 14 (7.8%) | 1 (5.6%) | 13 (8.0%) | ||
| AML M5, n (%) | 25 (13.9%) | 1 (5.6%) | 24 (14.8%) | ||
| AML M6, n (%) | 1 (0.6) | 0 (0.0%) | 1 (0.6%) | ||
| NA, n (%) | 65 (36.1%) | 12 (66.7%) | 53 (32.7%) |
FAB, French-American-British classification; NA, data not available; NS, not significant; sAML, secondary AML; tAML, therapy-related AML.
AML samples with -7 or del(7q) are sensitive to the inhibition of NAMPT
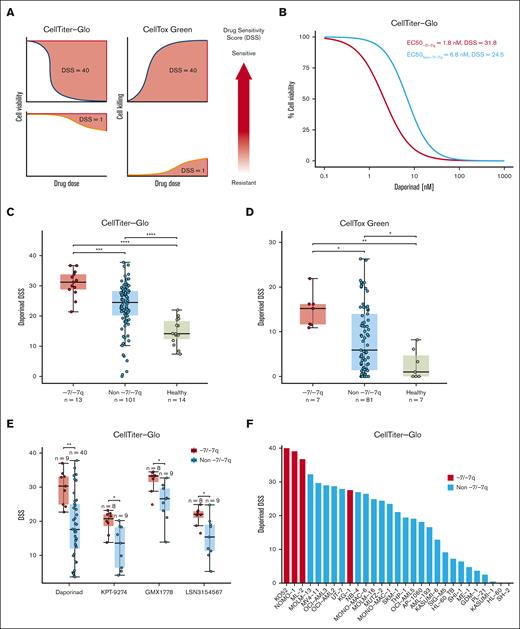
To identify therapeutic vulnerabilities in AML with -7/-7q, we performed ex vivo drug sensitivity testing in AML samples with -7/-7q and samples with diploid chromosome 7. To facilitate the statistical analysis of drug sensitivities between the sample groups, we computed drug sensitivity scores from the dose-response curves (Figure 1A). By analyzing results from 510 tested drugs, we found that AML samples with -7/-7q were most sensitive to the NAMPT inhibitor daporinad, showing the highest drug sensitivity score mean difference compared with samples without -7/-7q and the smallest P value (supplemental Table 3). A comparison of daporinad dose-response curves of the representative samples with a median half-maximal effective concentration (EC50) in the 2 groups showed that half-maximal inhibition was achieved with a 5-nM lower daporinad concentration in the AML sample with -7/-7q than with the AML sample with diploid chromosome 7 (Figure 1B).
AML cells with -7/-7q are sensitive to the inhibition of NAMPT. (A) Schematic diagram illustrating the distinct relationships of dose-response curves and drug sensitivity scores (DSS) for 2 different assays. In the CTG viability assay, the area over the dose-response curve is transformed into a DSS. Conversely, in the CTxG apoptosis assay, the area under the dose-response curve is used to calculate the DSS. Larger DSS values indicate increased drug sensitivity. (B) Comparison of dose-response to NAMPT inhibitor daporinad in cells from an AML harboring del(7q) with an AML with diploid chromosome 7 (non -7/-7q). Representative AML samples from -7/-7q and non -7/-7q cohorts with median daporinad EC50 are shown. BM MNCs obtained from fresh biopsy samples were incubated with the drug for 72 hours in 5 concentrations, and viability was determined using the CTG assay. (C) Comparison of daporinad sensitivity of BM or PB MNCs from a cohort of patients with AML and healthy individuals. AML with -7/-7q is compared with AML without -7/-7q and healthy individuals. Cell viability was measured using CTG. (D) Sensitivity to daporinad-induced cell death was measured using CTxG. (E) Ex vivo sensitivity to NAMPT inhibitors in viably frozen cells from patients with and without -7/-7q AML. (F) Sensitivity to daporinad in AML cell lines. Cell lines with del(7q) are shown in red and are significantly more sensitive than cell lines without -7/-7q cell lines by the t test (P ∼ .00094). Cells were incubated with daporinad for 72 hours, and viability was measured with CTG. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001, by Mann-Whitney U test.
AML cells with -7/-7q are sensitive to the inhibition of NAMPT. (A) Schematic diagram illustrating the distinct relationships of dose-response curves and drug sensitivity scores (DSS) for 2 different assays. In the CTG viability assay, the area over the dose-response curve is transformed into a DSS. Conversely, in the CTxG apoptosis assay, the area under the dose-response curve is used to calculate the DSS. Larger DSS values indicate increased drug sensitivity. (B) Comparison of dose-response to NAMPT inhibitor daporinad in cells from an AML harboring del(7q) with an AML with diploid chromosome 7 (non -7/-7q). Representative AML samples from -7/-7q and non -7/-7q cohorts with median daporinad EC50 are shown. BM MNCs obtained from fresh biopsy samples were incubated with the drug for 72 hours in 5 concentrations, and viability was determined using the CTG assay. (C) Comparison of daporinad sensitivity of BM or PB MNCs from a cohort of patients with AML and healthy individuals. AML with -7/-7q is compared with AML without -7/-7q and healthy individuals. Cell viability was measured using CTG. (D) Sensitivity to daporinad-induced cell death was measured using CTxG. (E) Ex vivo sensitivity to NAMPT inhibitors in viably frozen cells from patients with and without -7/-7q AML. (F) Sensitivity to daporinad in AML cell lines. Cell lines with del(7q) are shown in red and are significantly more sensitive than cell lines without -7/-7q cell lines by the t test (P ∼ .00094). Cells were incubated with daporinad for 72 hours, and viability was measured with CTG. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001, by Mann-Whitney U test.
A comparison of daporinad sensitivities in a cohort consisting of -7/-7q AML samples (n = 13), AML samples with diploid chromosome 7 (n = 101), and healthy controls (n=14) showed that the viability of -7/-7q AML samples was significantly lower after incubation with daporinad than in samples without -7/-7q AML samples and samples from healthy donors (Figure 1C). BM MNCs from healthy individuals were significantly more resistant to daporinad than cells from patients with AML.
To determine whether AML cells with -7/-7q were more efficiently killed by daporinad, we compared daporinad-induced cell death in -7/-7q AML samples (n = 7) with samples without -7/-7q AML (n = 81) and healthy samples (n = 7) using the CTxG cytotoxicity assay (Figure 1D; supplemental Table 4). We found that daporinad killed significantly more cells in the -7/-7q AML samples than in AML samples with diploid chromosome 7 or samples from healthy controls.
To determine whether the reduced viability induced by NAMPT inhibitors in the -7/-7q AML samples was caused by on-target inhibition of NAMPT, we tested the sensitivity of a cohort of AML samples to 4 NAMPT inhibitors with distinct chemical structures (daporinad, KPT-9274, GMX1778, and LSN3154567). We used viably frozen BM or PB MNCs from up to 49 patients with AML and determined the viability of the cells after incubation with each of the inhibitors using the CTG assay (supplemental Table 5). The rationale for this experiment was that because structurally distinct NAMPT inhibitors have unique off-target activity profiles, they may exhibit activity against distinct groups of samples if the off-target effects are the dominant cause of cell killing. We found that -7/-7q AML samples were significantly more sensitive than AML samples without -7/-7q to all 4 NAMPT inhibitors tested, indicating that the preferential inhibition of -7/-7q AML was due to the on-target inhibition of NAMPT (Figure 1E).
Next, we examined the sensitivity of immortalized AML cell lines to NAMPT inhibition. The sensitivity of 29 AML cell lines was tested by exposing cells to increasing concentrations of daporinad for 72 hours, followed by viability assessment using the CTG assay. We found that the AML cell lines most sensitive to daporinad carried -7 or del(7q), and they were significantly more sensitive than AML cell lines without -7/-7q (Figure 1F).
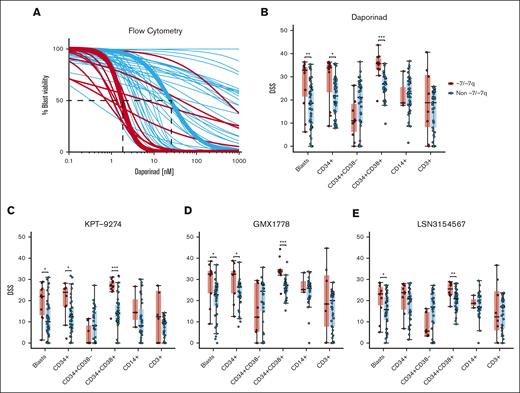
Next, we aimed to determine whether our initial findings could be demonstrated using a different orthogonal assay. For this purpose, we performed a dose-response evaluation using 4 NAMPT inhibitors (daporinad, KPT-9274, GMX1778, and LSN3154567) with a multiparametric flow cytometry readout in viably frozen BM or PB MNCs from up to 62 cryopreserved primary AML samples with (n = 11) and without (n = 51) -7/-7q (supplemental Table 6). In the flow cytometry-based drug sensitivity testing, live cells were quantified after incubation with the drugs. Dimly positive CD45 expression and low side scatter were used to identify leukemic blasts. Analysis of the representative samples with median daporinad EC50 showed that AML blast cells in samples with -7/-7q had an EC50 value of 1.5 nM, which was 16 times lower than the 24.6 nM EC50 value observed in the AML blast cells without -7/-7q (Figure 2A). We found that the blast cells in -7/-7q AML samples were significantly more sensitive to all 4 NAMPT inhibitors tested than blasts from AML samples without -7/-7q (Figures 2B-E). These results provided an orthogonal validation of our prior findings. Together, our results show that -7/-7q is associated with sensitivity to NAMPT inhibition in AML.
NAMPT inhibitor sensitivity of BM cell populations in AML with -7/-7q. (A) Daporinad dose response of leukemic blast cells from the BM or PB of patients with AML, measured by multiparametric flow cytometry. Samples were incubated with daporinad for 72 hours, followed by high-throughput flow cytometry–based cell viability readout. The curves highlighted in bold show patient samples with median EC50 for AML with -7/-7q (red) and AML with diploid chromosome 7 (blue). (B) Daporinad sensitivity of cell populations in AML with -7/-7q (n = 11) compared with AML without the deletion (n = 51). (C) KPT-9274 sensitivity in AML with -7/-7q (n = 10) vs without -7/-7q (n = 40). (D) GMX1778 sensitivity -7/-7q (n = 9) vs non -7/-7q (n = 40). (E) LSN3154567 sensitivity in -7/-7q (n = 10) vs non -7/-7q (n = 40). Groups were compared using Mann-Whitney U test; ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001.
NAMPT inhibitor sensitivity of BM cell populations in AML with -7/-7q. (A) Daporinad dose response of leukemic blast cells from the BM or PB of patients with AML, measured by multiparametric flow cytometry. Samples were incubated with daporinad for 72 hours, followed by high-throughput flow cytometry–based cell viability readout. The curves highlighted in bold show patient samples with median EC50 for AML with -7/-7q (red) and AML with diploid chromosome 7 (blue). (B) Daporinad sensitivity of cell populations in AML with -7/-7q (n = 11) compared with AML without the deletion (n = 51). (C) KPT-9274 sensitivity in AML with -7/-7q (n = 10) vs without -7/-7q (n = 40). (D) GMX1778 sensitivity -7/-7q (n = 9) vs non -7/-7q (n = 40). (E) LSN3154567 sensitivity in -7/-7q (n = 10) vs non -7/-7q (n = 40). Groups were compared using Mann-Whitney U test; ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001.
Sensitivity of BM cell populations to NAMPT inhibition in -7/-7q AML
To determine the effect of the inhibition of NAMPT on blast, progenitor, and lymphocyte cell populations from the BM of patients with -7/-7q AML, we analyzed the sensitivity of cryopreserved patient cells against daporinad, KPT-9274, GMX1778, and LSN3154567 using multiparametric flow cytometry. The flow antibody panel (supplemental Table 1) consisted of viability and cell surface markers that allowed for us to distinguish the major viable cell populations. These included CD34 for immature stem and progenitor cells, CD38 for more-differentiated myeloid cells, CD14 for (pro-)monocytes, and CD3 for lymphocytes. Dimly positive CD45 expression and low side scatter were used to identify leukemic blasts. We found that more-differentiated myeloblasts identified by CD34 and CD38 expression were significantly more sensitive to NAMPT inhibition in AML samples with -7/-7q than in those without -7/-7q. In contrast, less-differentiated CD34+CD38– myeloblasts from -7/-7q AML did not exhibit increased sensitivity to NAMPT inhibition. We did not observe a significant difference between the sensitivity of differentiated monocytes or lymphocytes in -7/-7q AML and AML lacking the deletion. Lymphocytes from -7/-7q AML showed notably large variability in responses to NAMPT inhibition (Figures 2B-E). Overall, our results show that in -7/-7q AML, differentiated myeloblasts are the most sensitive cell population to the inhibition of NAMPT.
AML with del(7q) that does not encompass the NAMPT gene locus lack sensitivity to NAMPT inhibition
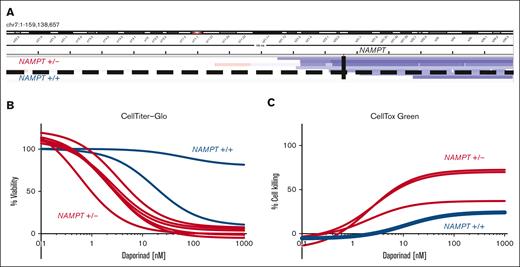
The NAMPT locus remains diploid in cases in which the del(7q) breakpoint occurs on the telomeric side of the gene. We sought to determine whether the location of the del(7q) breakpoint influences drug sensitivity. To identify breakpoint locations in the AML samples with del(7q), we analyzed copy number profiles defined by exome sequencing del(7q) AML. We quantified the NAMPT gene copy number based on exome sequencing (supplemental Table 7). We found that in 2 patients the breakpoint was on the telomeric side of the NAMPT locus, maintaining diploid NAMPT (Figure 3A). We evaluated the effect of daporinad on viability by CTG (Figure 3B) and cell killing using CTxG assays (Figure 3C) in del(7q) AML with diploid (NAMPT+/+) and -7/-7q AML with haploid NAMPT (NAMPT+/−). This revealed that NAMPT+/+ AML samples lacked sensitivity to daporinad, whereas NAMPT+/− samples were highly sensitive. These results show that del(7q) AML are sensitive to the inhibition of NAMPT when the deletion breakpoint is centromeric to the NAMPT locus at 7q22.3, leading to NAMPT haploinsufficiency. In comparison, AML with the del(7q) breakpoint on the telomeric side of the NAMPT locus do not exhibit increased sensitivity to NAMPT inhibitors.
AML with 7q deletion that does not encompass NAMPT lacks sensitivity to NAMPT inhibition. (A) Genomic coverage plot showing the location of the deletion breakpoint in del(7q) AML samples with respect to the NAMPT gene locus. In 2 AML cases, a del(7q) breakpoint was on the telomeric side of NAMPT, maintaining the locus diploid (NAMPT+/+). The deletion breakpoint locations were determined based on exome sequencing. (B) Ex vivo daporinad dose response of 2 NAMPT+/+ AML samples with del(7q) compared with AML samples with NAMPT haploinsufficiency (NAMPT+/−). Cell viability was measured by CTG (B), and cell death by CTxG (C).
AML with 7q deletion that does not encompass NAMPT lacks sensitivity to NAMPT inhibition. (A) Genomic coverage plot showing the location of the deletion breakpoint in del(7q) AML samples with respect to the NAMPT gene locus. In 2 AML cases, a del(7q) breakpoint was on the telomeric side of NAMPT, maintaining the locus diploid (NAMPT+/+). The deletion breakpoint locations were determined based on exome sequencing. (B) Ex vivo daporinad dose response of 2 NAMPT+/+ AML samples with del(7q) compared with AML samples with NAMPT haploinsufficiency (NAMPT+/−). Cell viability was measured by CTG (B), and cell death by CTxG (C).
Influence of leukemic clone size on NAMPT inhibitor sensitivity in AML samples with -7/-7q
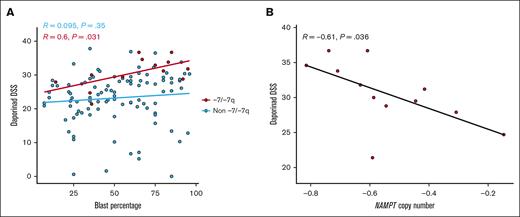
Next, we assessed the influence of leukemic clone size on the sensitivity to the NAMPT inhibitor daporinad in -7/-7q samples (n = 13). We performed a correlation test to evaluate the relationship between the proportion of blast cells in the sample and daporinad response. A positive and significant relationship was observed between the percentage of blast cells and sensitivity to daporinad (Figure 4A).
Impact of blast percentage and NAMPT copy number on daporinad sensitivity in AML samples with -7/-7q. (A) Scatterplot showing the relationship between daporinad sensitivity and blast percentage in AML samples with -7/-7q (n = 13). (B) Scatterplot showing the relationship between daporinad sensitivity and NAMPT copy number in AML samples with -7/-7q (n = 12). Analyzed using Pearson correlation test.
Impact of blast percentage and NAMPT copy number on daporinad sensitivity in AML samples with -7/-7q. (A) Scatterplot showing the relationship between daporinad sensitivity and blast percentage in AML samples with -7/-7q (n = 13). (B) Scatterplot showing the relationship between daporinad sensitivity and NAMPT copy number in AML samples with -7/-7q (n = 12). Analyzed using Pearson correlation test.
Due to 1 copy deletion of the NAMPT gene locus, the size of the -7/-7q clone can be expected to be inversely related to the NAMPT copy number in the sample. We performed a correlation test to evaluate the relationship between the NAMPT gene copy number and daporinad sensitivity in -7/-7q samples (n = 12). This analysis revealed a significant, inverse correlation, demonstrating that a decrease in NAMPT copy number is associated with increased daporinad sensitivity in -7/-7q AML samples (Figure 4B). Together, these results suggest that the size of the -7/-7q clone is a significant factor influencing the sensitivity of AML samples to NAMPT inhibition.
AML samples with -7/-7q have low NAMPT expression levels
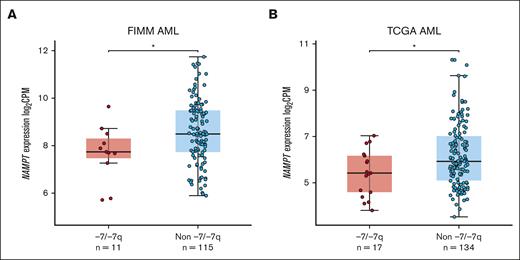
Monosomy 7 and del(7q) lead to the deletion of 1 copy of the NAMPT gene in most cases. Haploinsufficient gene expression is characterized by the reduced ability of cells to upregulate RNA expression. To determine whether NAMPT exhibits haploinsufficient gene expression in AML with -7/-7q, we analyzed NAMPT gene expression data from 2 AML cohorts. We found that -7/-7q AML had significantly lower NAMPT gene expression in our in-house cohort of AML samples (Figure 5A). A similar analysis in the TCGA AML cohort also showed significantly lower NAMPT gene expression in samples with -7/-7q (Figure 5B). These results suggest that NAMPT exhibits haploinsufficient gene expression in AML with -7/-7q.
AML samples with -7/-7q have reduced NAMPT expression. Comparison of the level of NAMPT gene expression in the BM or PB MNCs from AML with -7/-7q compared with AML lacking the deletion (non -7/-7q). Gene expression was determined by whole-transcriptome RNA sequencing. NAMPT gene expression analysis was performed on independent cohorts of patients with AML from the Institute for Molecular Medicine Finland (FIMM) (A) and TCGA (B). Dots represent gene expression values in individual patient samples as log2–transformed counts per million (CPM) of RNA sequencing reads. ∗ P < .05 by Mann-Whitney U test.
AML samples with -7/-7q have reduced NAMPT expression. Comparison of the level of NAMPT gene expression in the BM or PB MNCs from AML with -7/-7q compared with AML lacking the deletion (non -7/-7q). Gene expression was determined by whole-transcriptome RNA sequencing. NAMPT gene expression analysis was performed on independent cohorts of patients with AML from the Institute for Molecular Medicine Finland (FIMM) (A) and TCGA (B). Dots represent gene expression values in individual patient samples as log2–transformed counts per million (CPM) of RNA sequencing reads. ∗ P < .05 by Mann-Whitney U test.
Differentiated CD34+CD38+ AML myeloblasts have high NAMPT expression
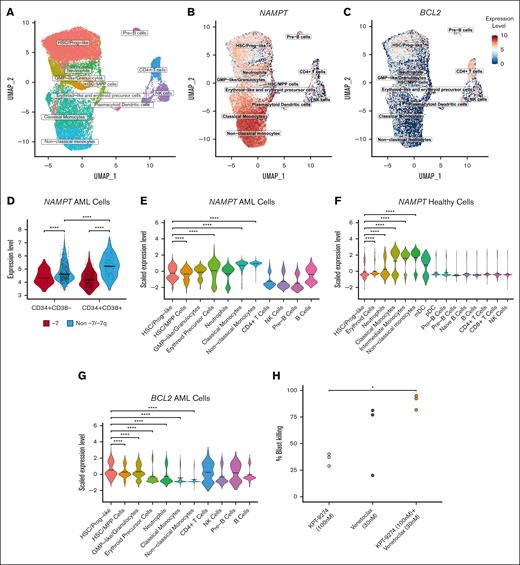
Because we found that more differentiated CD34+CD38+ BM myeloblasts from patients with AML were more sensitive to the inhibition of NAMPT than their less differentiated counterparts, we sought to determine whether these cells expressed higher levels of NAMPT. To determine NAMPT expression levels in myeloid cell populations in the BM of patients with AML, we analyzed single-cell RNA sequence data of viably frozen MNCs from 7 patients, of whom 1 had -7 (Figure 6A). We found that more differentiated myeloblasts had significantly higher levels of NAMPT expression than primitive myeloid cells and hematopoietic stem cells (HSCs) (Figure 6B,D,E), with some patient-to-patient variation (supplemental Figure 5A). Furthermore, when analyzing NAMPT expression in BM MNCs of healthy individuals, we observed higher expression in mature myeloid cells (Figure 6F), indicating a similar trend to that seen in AML. We examined similar cell populations as we did by flow cytometry–based drug sensitivity testing by selecting cells based on their CD34 and CD38 gene expression. In these cells, we found that NAMPT gene expression was significantly lower in AML cells with -7 than in cells without -7/-7q cells (Figure 6D), supporting our findings from the bulk RNA sequencing analyses. In addition, CD34+CD38+ cells in samples without -7/-7q also had significantly higher expression of NAMPT than CD34+CD38– cells, indicating that CD34+CD38+ cells are likely to be more reliant on NAMPT for their metabolism and, therefore, more sensitive to NAMPT inhibition.
AML cell populations display distinct gene expression and drug sensitivity profiles at the single-cell level. (A) Uniform Manifold Approximation and Projection (UMAP) representation of the cell types identified from single-cell RNA sequencing data from samples from 7 patients with AML. The group consisted of 1 patient with monosomy 7 and 6 patients with AML without -7/-7q. Cells are colored based on clusters identified using ScType and in-house gene sets. (B) UMAP representation of log normalized NAMPT gene expression level across the total cell cohort. NAMPT expression increases toward the differentiated myeloid cell types. (C) UMAP representation of log normalized BCL2 gene expression. Cells that belong to the HSC/progenitor-like cell cluster show the most expression in the myeloid cells. (D) Violin plot showing log normalized NAMPT gene expression in CD34+CD38– and CD34+CD38+ cells. Cells were selected from the total cell cohort based on their CD34 and CD38 expression levels. NAMPT expression is significantly lower in cells from the patient with monosomy 7 than cells from the patients without -7/-7q. In the samples without -7/-7q, NAMPT expression is significantly higher in CD34+CD38+ cells than in CD34+CD38– cells. Comparisons were done using the Mann-Whitney U test. (E) Violin plot showing scaled NAMPT gene expression in the 11 different cell clusters. NAMPT expression is highest in the more-differentiated myeloid cells (erythroid-like, neutrophil-like, and monocyte-like cells) compared with myeloid progenitor or lymphoid cells. Scaled expression values were calculated using Seurat functions to bring NAMPT and BCL2 expression to the same level for a more accurate comparison. (F) Violin plot showing scaled NAMPT expression in different myeloid and lymphoid cells in healthy BM cells. Expression data are obtained from 8 different donors from the Human Cell Atlas. (G) Violin plot showing scaled BCL2 gene expression in cell clusters. BCL2 is expressed at a significantly higher level in the HSC/progenitor-like cells than other myeloid cell clusters. Mean gene expression is marked with thick lines in each violin plot. Cell clusters in each violin plot were compared with each other with Dunn test followed by the Kruskal-Wallis H test. (H) Dotplot illustrating the proportion of total blast killing as measured by multiparametric flow cytometry in 3 distinct samples from patients with AML with -7/-7q after incubation for 72 hours with NAMPT inhibitor KPT-9274 at 100 nM (light gray), BCL2 inhibitor venetoclax at 30 nM (dark gray), or a combination of both agents (orange). The combination leads to the death of significantly more AML blasts than the NAMPT inhibitor alone due to an additive effect. Statistical comparisons by the Tukey test subsequent to an ANOVA. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001. ANOVA, analysis of variance; mDC, myeloid dendritic cells; NK cells, natural killer cells; pDC, plasmacytoid dendritic cells.
AML cell populations display distinct gene expression and drug sensitivity profiles at the single-cell level. (A) Uniform Manifold Approximation and Projection (UMAP) representation of the cell types identified from single-cell RNA sequencing data from samples from 7 patients with AML. The group consisted of 1 patient with monosomy 7 and 6 patients with AML without -7/-7q. Cells are colored based on clusters identified using ScType and in-house gene sets. (B) UMAP representation of log normalized NAMPT gene expression level across the total cell cohort. NAMPT expression increases toward the differentiated myeloid cell types. (C) UMAP representation of log normalized BCL2 gene expression. Cells that belong to the HSC/progenitor-like cell cluster show the most expression in the myeloid cells. (D) Violin plot showing log normalized NAMPT gene expression in CD34+CD38– and CD34+CD38+ cells. Cells were selected from the total cell cohort based on their CD34 and CD38 expression levels. NAMPT expression is significantly lower in cells from the patient with monosomy 7 than cells from the patients without -7/-7q. In the samples without -7/-7q, NAMPT expression is significantly higher in CD34+CD38+ cells than in CD34+CD38– cells. Comparisons were done using the Mann-Whitney U test. (E) Violin plot showing scaled NAMPT gene expression in the 11 different cell clusters. NAMPT expression is highest in the more-differentiated myeloid cells (erythroid-like, neutrophil-like, and monocyte-like cells) compared with myeloid progenitor or lymphoid cells. Scaled expression values were calculated using Seurat functions to bring NAMPT and BCL2 expression to the same level for a more accurate comparison. (F) Violin plot showing scaled NAMPT expression in different myeloid and lymphoid cells in healthy BM cells. Expression data are obtained from 8 different donors from the Human Cell Atlas. (G) Violin plot showing scaled BCL2 gene expression in cell clusters. BCL2 is expressed at a significantly higher level in the HSC/progenitor-like cells than other myeloid cell clusters. Mean gene expression is marked with thick lines in each violin plot. Cell clusters in each violin plot were compared with each other with Dunn test followed by the Kruskal-Wallis H test. (H) Dotplot illustrating the proportion of total blast killing as measured by multiparametric flow cytometry in 3 distinct samples from patients with AML with -7/-7q after incubation for 72 hours with NAMPT inhibitor KPT-9274 at 100 nM (light gray), BCL2 inhibitor venetoclax at 30 nM (dark gray), or a combination of both agents (orange). The combination leads to the death of significantly more AML blasts than the NAMPT inhibitor alone due to an additive effect. Statistical comparisons by the Tukey test subsequent to an ANOVA. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001. ANOVA, analysis of variance; mDC, myeloid dendritic cells; NK cells, natural killer cells; pDC, plasmacytoid dendritic cells.
Because HSC-like blasts (CD34+CD38–) are less sensitive to NAMPT inhibitors, other vulnerabilities must be exploited to inhibit these cells. HSC-like blasts are known to be more reliant on BCL2 for their survival.19,26 We studied the level of BCL2 expression in the myeloid cell clusters because BCL2 can be targeted with BCL2 inhibitors (Figure 6C; supplemental Figure 5B). BCL2 was expressed most in HSC/progenitor-like cells, and the expression decreased significantly toward more-differentiated cell states (Figure 6G), whereas the opposite was true for NAMPT (Figure 6E). These results suggest that a combination with inhibitors of BCL2 could help to eradicate the stem cell–like blasts that are less dependent on NAMPT but depend on BCL2 for survival.
A combination of BCL2 inhibitor venetoclax and a NAMPT inhibitor efficiently eradicated AML blasts with -7/-7q
We hypothesized that less differentiated cells could be inhibited by combining a NAMPT inhibitor with a BCL2-specific inhibitor. To test this, we incubated samples from 3 patients with AML harboring -7/-7q and a quantifiable CD34+CD38– cell population with either the NAMPT inhibitor KPT-9274 at 100 nM alone, the BCL2-specific inhibitor venetoclax at 30 nM alone, or a combination of both agents for 72 hours. Afterward, we quantified the total blast cell killing caused by the treatments in each sample by flow cytometry (Figure 6H). As expected, although KPT-9274 caused partial cell killing of the leukemic blast compartment in these samples, the combination with venetoclax resulted in significantly higher, near-total cell death of the blasts, indicating that the dual inhibition of NAMPT and BCL2 represents an effective strategy for targeting leukemic cells with -7/-7q, as a result of an additive effect of the 2 drugs.
Discussion
In this study, we identified NAMPT inhibition as a novel therapeutically targetable vulnerability in AML cells with -7/-7q. We analyzed ex vivo drug sensitivities of primary AML samples from patients in the FIMM AML cohort18,27 that were based on viability and cytotoxicity assays. As a result, we found that samples with -7/-7q were more sensitive to NAMPT inhibition than AML samples without -7/-7q and healthy samples. Additional experiments using structurally distinct NAMPT inhibitors confirmed that the sensitivity of -7/-7q AML samples to NAMPT inhibitors was due to on-target inhibition. We observed a correlation between blast count and NAMPT inhibitor sensitivity in -7/-7q AML cases. Furthermore, there was an inverse relationship between NAMPT copy number and NAMPT inhibitor sensitivity. These observations provide evidence that the size of the -7/-7q clone plays a role in determining the sensitivity of AML samples to NAMPT inhibitors. These results further support the hypothesis that NAMPT inhibitors preferentially target cells harboring -7/-7q deletions.
Cancer cells are known to have high metabolic activity and increased nicotinamide adenine dinucleotide (NAD+) turnover compared with normal cells because of the activity of enzymes such as sirtuins and poly (adenosine diphosphate ribose) and polymerase 1 (PARP1) that use NAD+ as a substrate.28,29 Cancer cells do not efficiently use the de novo or Preiss-Handler pathways for NAD+ production due to low levels of nicotinic acid and tryptophan and lack of expression of key enzymes in these pathways. Consequently, cancer cells generate NAD+ mainly through the NAD+ salvage pathway.30 NAMPT is the rate-limiting enzyme in the NAD+-salvage pathway and has been extensively researched as a potential drug target for targeting cancer cell metabolism.31 By inhibiting NAMPT, the salvage pathway is halted, resulting in decreased NAD+ levels, followed by depletion of adenosine triphosphate, and, eventually, death of cancer cells.30 Our results show higher dependence of AML cells on NAMPT than that of healthy cells, as demonstrated by their significantly higher sensitivity to NAMPT inhibition.
Several NAMPT inhibitors have been developed and have shown encouraging results in terms of anticancer efficacy in different in vitro and in vivo models of solid32-35 and hematological malignancies.36 These inhibitors include daporinad (APO866, FK866),37 GMX1778 (CHS 828),38 LSN3154567,39 KPT-9274 (padnarsertib),40 and, most recently, OT82.41 However, when evaluated in phase 1/2 clinical trials, NAMPT inhibitors have been reported to have limited drug efficacy profiles and severe toxic side effects, including thrombocytopenia, anemia, and gastrointestinal symptoms.42-46 Research in rodent models have also shown that NAMPT inhibition causes retinopathy.47 Because dose-limiting toxicities have so far prevented NAMPT inhibitors from achieving desirable therapeutic effects and clinical approval, our identification of AML with -7/-7q as an exceptionally responsive patient population could improve the clinical profile of NAMPT inhibitors, notably by using an actionable therapeutic window that limits side effects while maintaining a high level of efficacy.
Hypothesizing that the observed sensitivity of AML with -7/-7q to NAMPT inhibitors is due to the loss of 1 copy of the NAMPT gene, termed NAMPT haploinsufficiency, we investigated the role of the location of the del(7q) breakpoint on drug sensitivity in AML. By analyzing copy number profiles defined by exome sequencing in del(7q) AML samples, we found that in 2 patients, the del(7q) breakpoint was on the telomeric side of the NAMPT locus, maintaining diploid NAMPT. These samples did not show the increased sensitivity to daporinad observed in other samples with NAMPT haploinsufficiency, and their response was comparable with AML samples without -7/-7q. These results demonstrate that del(7q) AML are sensitive to inhibition of NAMPT when the deletion breakpoint is located on the centromeric side of the NAMPT locus at 7q22.3, causing NAMPT haploinsufficiency. Therefore, a precise characterization of del(7q) is necessary to identify patients who can benefit from NAMPT-targeted therapy. Conventional cytogenetic analysis and fluorescence in situ hybridization are used routinely in the diagnosis of AML.48 These methods lack the resolution to accurately detect the 7q breakpoint location. However, the breakpoint location can be determined using high-resolution methods such as whole-genome sequencing.49
When we investigated the effect of NAMPT inhibition on different cell populations in BM samples from patients with -7/-7q AML by flow cytometry–based drug sensitivity testing, we observed that differentiated CD34+CD38+ myeloblasts were significantly more sensitive to NAMPT inhibition in -7/-7q AML samples than in AML without -7/-7q. In contrast, less differentiated CD34+CD38– myeloblasts from -7/-7q AML did not exhibit increased sensitivity to NAMPT inhibition. At the gene expression level, our single-cell analysis of primary AML samples showed that CD34+CD38+ myeloblasts express higher levels of NAMPT than CD34+CD38– myeloblasts. As expected, an AML sample with monosomy 7 had notably low NAMPT gene expression. Taken together, our results suggest CD34+CD38+ myeloblasts depend on NAMPT for survival. In the context of -7/-7q AML, reduced NAMPT expression due to haploinsufficiency renders these cells highly vulnerable to NAMPT inhibition.
The results of our study indicate that targeting both NAMPT and BCL2 may represent a promising approach for treating patients with AML with -7/-7q chromosomal abnormalities and an undifferentiated disease phenotype. Our flow cytometry–based drug sensitivity assay revealed that undifferentiated CD34+CD38– myeloblasts in AML samples with -7/-7q are less responsive to NAMPT inhibition alone compared with more differentiated CD34+CD38+ myeloblasts. However, CD34+CD38– myeloblasts have been shown to be dependent on BCL2 for survival.50 Our experiments combining a NAMPT inhibitor (KPT-9274) with the BCL2-specific inhibitor venetoclax resulted in near-total cell death of the leukemic blasts in these samples due to an additive effect between the 2 agents. These results support the idea that cotargeting the NAMPT and BCL2 pathways may be an effective strategy for eliminating AML cells with -7/-7q chromosomal abnormalities. Additional studies are needed to validate these findings in larger patient populations and investigate the safety and efficacy of this dual-therapy approach. Previously, metabolic assays with KPT-9274 and venetoclax showed synergistic effects, but these experiments were done in bulk readout; hence, cell population-specific effects might not have been detected.40 A previous study also demonstrated the efficacy of NAMPT inhibition in the context of relapsed AML and venetoclax resistance but did not evaluate a combination of the 2 agents.51 NAMPT inhibitors have also been investigated in combination with SREBP2 or stearoyl-CoA desaturase inhibitors, which resulted in increased NAMPT inhibitor efficacy in the leukemic stem cell population.52
Our results identify NAMPT inhibition as a novel therapeutic vulnerability in AML cells with -7/-7q chromosomal abnormalities. Instances of chromosomal abnormality conferring increased susceptibility to a particular treatment have been previously described, notably in MDS. Specifically, patients with MDS carrying -5q have shown exceptional sensitivity to the immunomodulatory drug lenalidomide. This observation was supported by several studies conducted in vitro and in vivo, as well as in clinical trials, ultimately resulting in the approval of the drug for clinical use in these patients.53
In conclusion, our findings demonstrate that AML cells with -7/-7q are highly sensitive to NAMPT inhibition, suggesting that NAMPT inhibitors have the potential to be an effective targeted therapy for AML with -7/-7q. Additionally, our results propose NAMPT inhibitors as a suitable combination partner for venetoclax for -7/-7q AML. Further research is needed to investigate the clinical potential of NAMPT inhibitors in the treatment of patients with -7/-7q AML. In addition, because monosomy 7 and deletion 7q are observed in other blood disorders, including MDS, the efficacy of NAMPT inhibitors is worth evaluating in these patients.
Acknowledgments
The authors thank the Helsinki Biobank for the control samples and the Finnish Hematology Registry and Clinical Biobank for providing patient samples and data. Drug plate preparation was carried out at the FIMM High Throughput Biomedicine Unit and single-cell sequencing at the FIMM Single-Cell Analytics Unit, which are hosted by the University of Helsinki and supported by HiLIFE and Biocenter Finland. J.S. and N.I. are PhD candidates at the University of Helsinki. This work is submitted in partial fulfillment of the requirement for the PhD. The authors express their gratitude to Minna Suvela and Alun Parsons for their role in processing the samples for the study. The authors acknowledge the invaluable contributions of Daniela Mendoza Ortiz in data analysis. The results published here are in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
This work was financially supported by the University of Helsinki, Cancer Foundation Finland, the Academy of Finland grants 334781 and 1320185 (C.A.H.), grant 334273, the Sigrid Jusélius Foundation, the Magnus Ehrnrooth Foundation (S.E.), and the Finnish Cultural Foundation (D.M.).
Authorship
Contribution: S.E. and C.A.H. conceived the project and provided leadership; S.E., J.S., and N.I. analyzed and interpreted the data; D.M. provided the cell line data; N.I. and M.V.-K. performed single-cell transcriptome analysis; M.K., B.T.G., and K.P. collected the samples and interpreted clinical data; S.E., J.S., N.I., D.M., and C.A.H. wrote the manuscript; and all authors critically read the manuscript, provided constructive comments, and agreed to the content.
Conflict-of-interest disclosure: B.T.G. has served as a consultant for BerGenBio and Pfizer Inc; holds stock options in privately-held companies, Alden Cancer Therapy and KinN Therapeutics; and has provided consultancy services to Novartis. M.K. is an adviser to Novartis, Faron Pharmaceuticals, Bristol Myers Squibb, Pfizer, and AbbVie; has received consultancy fees, research funding, and speaker’s bureau involvement from AbbVie; and received consultancy fees from Astellas Pharma. K.P. has been awarded honoraria from Pfizer, Novartis, Incyte, Bristol Myers Squibb, Astellas, and AbbVie, and has received research funding from Celgene/Bristol Myers Squibb, Incyte, Pfizer, and Novartis. C.A.H. has received research funding from Oncopeptides, IMI2 projects HARMONY and HARMONY PLUS, WNTResearch, Orion, Kronos Bio, Novartis, Celgene, and Zentalis Pharmaceuticals; has been awarded honoraria from Amgen; and received consultancy fees from Autolus. The remaining authors declare no competing financial interests.
Correspondence: Caroline Heckman, Institute for Molecular Medicine Finland, University of Helsinki, Tukholmankatu 8, PO Box 20, FI-00290 Helsinki, Finland; email: caroline.heckman@helsinki.fi; and Samuli Eldfors, Massachusetts General Hospital Cancer Center, Building 149, 13th St, Charlestown, MA 02129; email: seldfors@mgh.harvard.edu.
References
Author notes
S.E. and J.S. contributed equally to this study.
Presented in oral abstract form at the 65th annual meeting of the American Society of Hematology, New Orleans, LA, 15 November 2022.
Previously generated CTG–based drug sensitivity results from samples from patients with AML and healthy controls are available at https://doi.org/10.1158/2159-8290.CD-21-0410 (supplemental Table 7). Gene expression data from the FIMM AML cohort used in this study are available at https://doi.org/10.5281/zenodo.7274740.
All other original data are available upon reasonable request from the corresponding authors, Caroline A. Heckman (caroline.heckman@helsinki.fi) and Samuli Eldfors (seldfors@mgh.harvard.edu).
The full-text version of this article contains a data supplement.