iPSC-derived primitive and definitive RBCs display different characteristics, red cell antigen expression, and disease-modeling capabilities.
Isogenic iPSC-derived definitive erythroblasts closely mimic primary fetal liver–derived erythroblasts.
Visual Abstract
During development, erythroid cells are produced through at least 2 distinct hematopoietic waves (primitive and definitive), generating erythroblasts with different functional characteristics. Human induced pluripotent stem cells (iPSCs) can be used as a model platform to study the development of red blood cells (RBCs) with many of the differentiation protocols after the primitive wave of hematopoiesis. Recent advances have established that definitive hematopoietic progenitors can be generated from iPSCs, creating a unique situation for comparing primitive and definitive erythrocytes derived from cell sources of identical genetic background. We generated iPSCs from healthy fetal liver (FL) cells and produced isogenic primitive or definitive RBCs which were compared directly to the FL-derived RBCs. Functional assays confirmed differences between the 2 programs, with primitive RBCs showing a reduced proliferation potential, larger cell size, lack of Duffy RBC antigen expression, and higher expression of embryonic globins. Transcriptome profiling by scRNA-seq demonstrated high similarity between FL- and iPSC-derived definitive RBCs along with very different gene expression and regulatory network patterns for primitive RBCs. In addition, iPSC lines harboring a known pathogenic mutation in the erythroid master regulator KLF1 demonstrated phenotypic changes specific to definitive RBCs. Our studies provide new insights into differences between primitive and definitive erythropoiesis and highlight the importance of ontology when using iPSCs to model genetic hematologic diseases. Beyond disease modeling, the similarity between FL- and iPSC-derived definitive RBCs expands potential applications of definitive RBCs for diagnostic and transfusion products.
Introduction
Red blood cells (RBCs) are currently used as an important biomedical product, both for transfusion medicine and for diagnostic purposes. COVID-19–related blood shortages,1 increasing demand,2 safety challenges, and requirements for specific RBC phenotypes3 have created a global need for alternatives to volunteer blood donation, including ex vivo and in vitro manufacturing of RBCs.4-6 Human induced pluripotent stem cells (iPSCs) are an attractive source for the generation of blood products because they can effectively differentiate into erythroid cells, potentially providing an unlimited source of RBCs.7,8 Moreover, as they are amenable to gene editing, iPSCs can be engineered to express specific blood types9 (including “universal” donor type O-10) or receptors for drug delivery,11,12 in addition to providing a platform for disease-modeling and basic research.
During development, erythroid cells are produced through at least 2 hematopoietic waves, primitive and definitive, which generate RBCs with different functional characteristics. The first wave generates primitive erythroblasts that are detectable in the yolk-sac blood islands of the human conceptus as early as day 18 of gestation.13 Primitive RBCs are relatively large, nucleated, transient, and express embryonic globins (ε- and ζ-globin).14 Definitive RBCs arise from different subsets of hematopoietic progenitor cells (HPCs) that are formed later in the arterial vasculature at various sites in the embryo15-17 and subsequently colonize the fetal liver at 4 postconception weeks (PCWs). Definitive RBCs are smaller, can enucleate, and predominantly express fetal globin subunits, such as γ-globin to fulfill the oxygen requirements of the growing fetus.18
Many current methodologies that generate iPSC-derived RBCs (iRBCs) mimic the first wave of embryonic hematopoiesis with the production of yolk sac-like primitive HPCs or generate a mixed population of primitive and definitive progenitors19-22 that can be further differentiated into erythroblasts. These models have been extremely valuable in characterizing pathological phenotypes, initiating scale-up manufacturing processes and developing diagnostic applications.9,23-26 However, primitive iRBCs may not recapitulate some fetal-specific phenotypes27 and their limited proliferation potential, antigen expression, embryonic globin content, and larger size can be a hinderance to clinical translation. Although the switch between embryonic and fetal erythropoiesis has been extensively studied in mice,16,28,29 many aspects of this process during human development remain elusive. Understanding how RBCs are generated through development in humans is a critical step in the production of pure functional iRBCs for therapeutic, diagnostic, and research use.
Recent advances in the directed differentiation of pluripotent stem cells have identified methodology that better recapitulates definitive, fetal-like hematopoiesis with protocols available to generate predominantly primitive or definitive HPCs.22,30-33 A direct comparison of the erythroid potential of these iPSC-derived HPCs with primary fetal erythropoiesis has not been done, limiting our knowledge on how closely this cellular model recapitulates human development.
To gain insight into this process, we reprogrammed healthy fetal liver (FL) cells into iPSC lines and generated isogenic primitive and definitive HPCs. Using the same primary FL sample, we differentiated the FL- and iPSC-derived HPCs into red cells and compared them with functional assays, immunophenotyping, and transcriptomics, including scRNA-seq. This experimental design eliminates phenotypic variability caused by differences in genetic background and allowed the first direct comparison of iPSC-derived erythroid cells with genetically identical erythroid cells generated from primary FL. To highlight the importance of ontology in disease modeling, we also characterized a loss-of-function mutation in the erythroid master regulator KLF1 and found that certain phenotypes are only seen in the definitive RBC program. By using an isogenic FL sample, this study validates iPSC-derived models of definitive erythropoiesis and deciphers differences between embryonic (primitive) and fetal (definitive) erythropoiesis in the human system.
Methods
Cell sources
Fetal liver
A healthy 18 PCW FL sample was obtained after informed consent under a protocol approved by the University of Pennsylvania Institutional Review Board, protocol #824810.
The FL iPSC line was generated by infection of isolated FL mononuclear cells with integration-free Sendai virus containing 4 reprogramming genes (OCT4/SOX2/KLF4/MYC).34,35 Two independent clones were isolated, expanded, and checked for normal karyotype. Pluripotency was confirmed by flow cytometry and trilineage differentiation. Sendai clearance was checked at different passages using qPCR,36 see supplemental Figure 1.
To predict antigen expression, blood group antigen genotyping was performed on the PreciseType Human Erythrocyte Antigen BeadChip (Immucor).
Other iPSC lines
Hematopoietic and erythroid differentiation of iPSCs
For primitive hematopoietic differentiation, we used a previously established protocol.32,33 Definitive hematopoietic progenitors were generated as described,30,31,39 with minor modifications. Erythroid differentiation of hematopoietic progenitors was performed as previously described.9 Detailed information about culture conditions, flow cytometry analysis, cytospin preparation, colony assays, globin analysis, and quantitative RT-PCR are provided in supplemental Methods.
Single-cell RNA sequencing
Experimental details and bioinformatic analyses are described in supplemental Methods.
Results
Generation of primitive and definitive HPCs from a FL–derived iPSC line
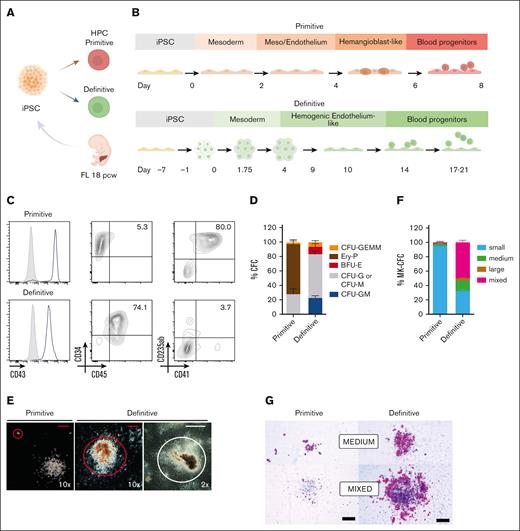
A major roadblock to the use of iRBCs for clinical and research applications is our limited understanding of the fidelity of in vitro cell models to recapitulate in vivo hematopoiesis. Specifically, a thorough validation between primary FL- and iPSC-derived erythroblasts has been hindered by intrinsic genetic variability among iPSC lines40 and technical challenges in obtaining a pure population of definitive HPCs to generate erythrocytes.21 To compare primary FL- and iPSC-derived RBCs, iPSC lines were generated from an 18 PCW karyotypically normal FL sample (Figure 1A; supplemental Figure 1A-D) and compared to RBCs cultured from the same FL.
Generation of primitive and definitive HPCs from a FL–derived iPSC line. (A) Study design overview; iPSCs were produced from an 18 PCW healthy FL sample, and primitive and definitive HPCs were generated. (B) Schematic representation of the differentiation protocols and timeline used to generate primitive or definitive HPCs. (C) Representative flow cytometry analysis of primitive (top) and definitive (bottom) iPSC-derived HPCs. Percentage of positive cells is indicated; negative control is shown in gray. (D) Colony-forming cell frequency in primitive and definitive HPC (n = 6-9). (E) Representative images (bright field) of different progenitor-derived colonies at different magnification. Red colonies are circled. Red scale bar is 250 μm and white scale bar is 2 mm. (F) Megakaryocytic colony-forming cell frequency in primitive and definitive HPC (n = 2-4). G) Representative images (bright field) of megakaryocite colonies. Black scale bar is 200 μm.
Generation of primitive and definitive HPCs from a FL–derived iPSC line. (A) Study design overview; iPSCs were produced from an 18 PCW healthy FL sample, and primitive and definitive HPCs were generated. (B) Schematic representation of the differentiation protocols and timeline used to generate primitive or definitive HPCs. (C) Representative flow cytometry analysis of primitive (top) and definitive (bottom) iPSC-derived HPCs. Percentage of positive cells is indicated; negative control is shown in gray. (D) Colony-forming cell frequency in primitive and definitive HPC (n = 6-9). (E) Representative images (bright field) of different progenitor-derived colonies at different magnification. Red colonies are circled. Red scale bar is 250 μm and white scale bar is 2 mm. (F) Megakaryocytic colony-forming cell frequency in primitive and definitive HPC (n = 2-4). G) Representative images (bright field) of megakaryocite colonies. Black scale bar is 200 μm.
To generate primitive and definitive HPCs, iPSCs were differentiated according to previously described protocols (Figure 1B; supplemental Methods) that specifically pattern primitive-posterior or definitive-lateral plate mesoderm.30-32 As expected, both primitive and definitive iPSC-derived HPCs expressed the pan-hematopoietic marker CD43 and the progenitor marker CD34 (Figure 1C). Primitive HPCs also expressed CD41 and CD235ab markers typical of either megakaryocytes or erythrocytes that have been previously described to mark primitive HPCs.41 In contrast, definitive HPCs mainly coexpressed CD34 and CD45 with no additional lineage-commitment markers, without transitioning through a CD41+CD235ab+ stage as they differentiate (data not shown).
To test the multilineage potential of the primitive and definitive HPCs, colony-forming unit (CFU) assays were performed (Figure 1D). Primitive HPCs generated small erythroid colonies (Ery-P; Figure 1E) and myeloid colonies (granulocytes, CFU-G or macrophages, CFU-M). Definitive progenitors generated larger colonies, including erythroid BFU-E, mixed granulomyelocytic colonies (CFU-GM), and very large multilineage colonies (CFU-GEMM, granulocyte, erythroid, macrophage, and megakaryocyte). In addition, megakaryocytic colony potential was assayed, as megakaryocytes from primitive and definitive origins exhibit substantial diversity in size and proliferation.42,43 Significant differences in colony composition and morphology (Figure 1E-G) were observed, with primitive HPCs generating smaller, uni-lineage colonies compared with those generated by definitive HPCs. Taken together, these data show that iPSC-derived primitive and definitive HPCs have different characteristics, display distinct cellular markers, and give rise to different progeny.
Characterization of primitive and definitive RBC
To assess the functional differences between primitive and definitive iRBCs, HPCs were expanded in erythroid liquid cultures. Definitive iRBCs were more proliferative, expanding more than 60-fold by day 15 of differentiation (Figure 2A), whereas primitive iRBCs stopped dividing at approximately day 10 of differentiation and progressively underwent cell death. The kinetics of erythroid marker expression were markedly different between primitive and definitive erythroblasts. The primitive iRBCs highly expressed the erythroid markers CD36 (fatty acid translocase), CD71 (transferrin receptor), and CD235a (Glycophorin A) by day 3 of differentiation. The definitive iRBCs expressed these markers, but their expression peaked after day 9 of differentiation (Figure 2B). Terminally differentiated FL iRBCs, both definitive and primitive, were examined for maturation markers Band3 and CD49d to assure that samples were at similar stages of RBC maturation (supplemental Figure 2A). Cytospin and flow assays of erythroid cultures demonstrated low rates of enucleation (5%-8%) in the FL RBCs and definitive iRBCs, but no enucleation was detected in the primitive iRBCs (supplemental Figure 2B-C). Lastly, cell morphology and size were different between the developmental programs, with primitive iRBCs being larger as would be expected for this population (Figure 2C; supplemental Figure 3A).
Functional characterization of primitive and definitive iRBCs. (A) Erythroid expansion of primitive and definitive HPCs in erythroid liquid cultures. (B) Representative histograms of CD36, CD71, and CD235a expression of primitive (top) or definitive erythroblasts during erythroid differentiation. Differentiation days are indicated on the right and negative control is shown in gray. (C) Cytospins showing May-Grünwald Giemsa staining of erythroid cells from primitive and definitive cultures at day 6 and day 12 of differentiation. Scale bars represent 20 μm. (D) Relative abundance of β-like transcripts (β+ε+γg+γa) at day 12 of erythroid liquid culture (mean ± SD, n = 3-4). (E) HPLC analysis of globin monomers in differentiated erythroblasts, relative quantification of α-like and β-like subunits are shown (primitive and definitive, mean ± SD, n = 3; FL n = 1). (F) HPLC analysis of globin tetramers in differentiated erythroblasts, relative quantification of adult hemoglobin (adult, α2β2), HbF (fetal, α2 γ2), and embryonic (Gower 1 ζ2ε2 and Gower 2, α2ε2) globins (mean ± SD). HPLC, high-performance liquid chromatography.
Functional characterization of primitive and definitive iRBCs. (A) Erythroid expansion of primitive and definitive HPCs in erythroid liquid cultures. (B) Representative histograms of CD36, CD71, and CD235a expression of primitive (top) or definitive erythroblasts during erythroid differentiation. Differentiation days are indicated on the right and negative control is shown in gray. (C) Cytospins showing May-Grünwald Giemsa staining of erythroid cells from primitive and definitive cultures at day 6 and day 12 of differentiation. Scale bars represent 20 μm. (D) Relative abundance of β-like transcripts (β+ε+γg+γa) at day 12 of erythroid liquid culture (mean ± SD, n = 3-4). (E) HPLC analysis of globin monomers in differentiated erythroblasts, relative quantification of α-like and β-like subunits are shown (primitive and definitive, mean ± SD, n = 3; FL n = 1). (F) HPLC analysis of globin tetramers in differentiated erythroblasts, relative quantification of adult hemoglobin (adult, α2β2), HbF (fetal, α2 γ2), and embryonic (Gower 1 ζ2ε2 and Gower 2, α2ε2) globins (mean ± SD). HPLC, high-performance liquid chromatography.
A major feature that distinguishes primitive and definitive erythroblasts is globin gene expression. In humans, primitive erythroid cells express predominantly ε-globin and ζ-globin, whereas definitive erythroid cells switch to γ-globin and α-globin production.18 To assess globin content at the transcriptional and protein levels, FL cells and iPSCs were terminally differentiated to primitive and/or definitive RBCs. In the primitive iRBCs, the ε-globin transcripts represented 42.2% ± 2.3 of total β-like transcripts compared with 7.9% ± 3.8 in the definitive iRBCs and 2.8% ± 1.1 in FL-derived RBCs, respectively (Figure 2D). At the protein level, a strong reduction of ε-globin and ζ-globin was accompanied by an increase of γ-globin and α-globin in both FL- and iPSC–derived definitive RBCs, indicating that these cells underwent the embryonic-to-fetal globin switch (Figure 2E; supplemental Figure 2D). This was also confirmed by hemoglobin tetramer analysis (Figure 2F; supplemental Figure 2E), in which definitive RBCs predominantly produced fetal hemoglobin (HbF, α2 γ2) compared with primitive RBCs that primarily produced embryonic forms (Gower 1, ζ2ε2 and Gower 2, α2ε2). Similar results were also obtained in other PSC lines (supplemental Figure 2F), validating the robustness of the definitive differentiation protocol in generating iRBCs that express fetal globins comparable with FL-derived RBCs.
Red cell antigen expression and typing
In addition to globins, expression of blood group antigens is developmentally regulated and differs in embryonic, fetal, and adult RBCs.44,45 In particular, many blood antigens that are determined by immunodominant sugars are not fully glycosylated at birth. Blood group antigen expression was assessed on FL-derived RBCs and iRBCs by flow cytometry and correlated to RBC antigen phenotypes determined by genotyping the iPSC line (supplemental Table 3). As expected, the FL-derived RBCs and both primitive and definitive iRBCs expressed Kell (early embryonic) but not BCAM/Lu (after birth) (Figure 3A). Duffy was expressed exclusively on the FL- and iPSC–derived definitive RBCs and not the primitive iRBCs.
Red cell antigen expression in different developmental programs. (A) Expression of different red cell antigens by flow cytometry in primitive, definitive, and FL-derived RBC. Negative control is shown in gray. (B) Representative pictures of gel card assay using primitive and definitive isogenic iRBCs. RBCs were incubated with IgG against specific red cell antigens or control IgG. A positive reaction (agglutination) is indicated by the retention of RBCs at the top of the gel, whereas a negative reaction results in RBCs pelleted at the bottom of the tube. Expected outcome based on iPSC genotype is indicated.
Red cell antigen expression in different developmental programs. (A) Expression of different red cell antigens by flow cytometry in primitive, definitive, and FL-derived RBC. Negative control is shown in gray. (B) Representative pictures of gel card assay using primitive and definitive isogenic iRBCs. RBCs were incubated with IgG against specific red cell antigens or control IgG. A positive reaction (agglutination) is indicated by the retention of RBCs at the top of the gel, whereas a negative reaction results in RBCs pelleted at the bottom of the tube. Expected outcome based on iPSC genotype is indicated.
To assess functional expression of red cell antigens by primitive and definitive iRBCs, a standard blood bank assay of gel column agglutination was used. Previous work from our group demonstrated that primitive iRBCs could be utilized in this assay to identify uncommon antibodies against specific Rh antigens in alloimmunized patients.9 To test if Duffy expression could be functionally used in an agglutination assay, primitive and definitive iRBCs were incubated with monoclonal typing antibodies and tested on gel columns (Figure 3B). Both iRBCs showed the predicted agglutination pattern for the Kell system (K-k+), but only the definitive iRBCs showed agglutination to Duffy (Fya+ Fyb+). As S and s antigens are highly glycosylated, agglutination was much higher on the definitive than primitive iRBCs. These results identify blood group antigens as specific markers of definitive erythropoiesis and expand the possibilities of using iRBCs for diagnostic applications.
scRNA-seq of primitive, definitive, and FL-derived RBCs
To determine if our iRBC model of in vitro erythroid development truly recapitulates fetal definitive erythropoiesis, FL- and iPSC-derived RBCs were analyzed using single-cell RNA sequencing (scRNA-seq). The FL- and iPSC-derived HPCs were used to generate RBCs using the same differentiation media and analyzed on day 6 to prevent excess hemoglobinization (Figure 4A). Erythroid surface marker expression was comparable (Figure 4B), due to stage matching the erythroblasts from the different sources. FL- and iPSC-derived definitive RBCs were smaller, displayed slightly higher CD36 levels during differentiation, and showed a similar morphology compared with primitive iRBCs (Figure 4B; supplemental Figure 3A-B). Following data processing and quality control (supplemental Figure 4A), a total of 11 598, 9547, and 8808 cells were captured for FL-derived RBCs, primitive iRBC and definitive iRBCs, respectively.
Transcriptional analysis by scRNA-seq of primitive, definitive, and FL-derived RBCs. (A) Schematic outline of scRNA-seq experiment. (B) Flow cytometry histograms of erythroid markers at the time of library preparation in primitive (pink), definitive (green), and FL-derived (blue) RBCs. Negative control is shown in gray. (C) Uniform manifold approximation and projection of primitive (n = 9547), definitive (n = 8808), and FL-derived (n = 11 598) RBCs. Subsets of individual samples are shown on the left (primitive iRBCs, pink; definitive iRBCs, green; and FL-derived RBCs, blue). (D) Distinct clusters of primitive, definitive, and FL-derived RBCs as defined by Seurat. (E) Dot plot showing expression of selected marker genes in each cluster. The dot size represents the percentage of cells within a cell cluster in which the gene was detected, and the dot color intensity represents the relative normalized average expression level of that marker. (F) Differential expression of a selection of erythroid genes in primitive, definitive, and FL-derived RBCs. (G) Correlation heat map of FL and definitive and primitive-derived RBCs in erythroid clusters. (H) Histogram showing the percentage of cells in each erythroid clusters in different samples. (I) Heat map of the top 25 differentially expressed genes between primitive and FL-definitive clusters.
Transcriptional analysis by scRNA-seq of primitive, definitive, and FL-derived RBCs. (A) Schematic outline of scRNA-seq experiment. (B) Flow cytometry histograms of erythroid markers at the time of library preparation in primitive (pink), definitive (green), and FL-derived (blue) RBCs. Negative control is shown in gray. (C) Uniform manifold approximation and projection of primitive (n = 9547), definitive (n = 8808), and FL-derived (n = 11 598) RBCs. Subsets of individual samples are shown on the left (primitive iRBCs, pink; definitive iRBCs, green; and FL-derived RBCs, blue). (D) Distinct clusters of primitive, definitive, and FL-derived RBCs as defined by Seurat. (E) Dot plot showing expression of selected marker genes in each cluster. The dot size represents the percentage of cells within a cell cluster in which the gene was detected, and the dot color intensity represents the relative normalized average expression level of that marker. (F) Differential expression of a selection of erythroid genes in primitive, definitive, and FL-derived RBCs. (G) Correlation heat map of FL and definitive and primitive-derived RBCs in erythroid clusters. (H) Histogram showing the percentage of cells in each erythroid clusters in different samples. (I) Heat map of the top 25 differentially expressed genes between primitive and FL-definitive clusters.
The FL- and iPSC-derived definitive samples overlapped considerably on the uniform manifold approximation and projection, whereas the primitive samples remained distinct (Figure 4C). After quality control (supplemental Figure 4A) and clustering, 14 cell clusters were identified (Figure 4D) and differentially expressed genes were calculated (supplemental Figure 4B). All samples were comprised predominantly of erythroid cells, with small populations of megakaryocytes, myeloid, and mast cells also identified based on marker gene expression (Figure 4D-E; supplemental Figure 4B-C) and morphological examination (data not shown). Notably, only FL- and iPSC–derived definitive samples contained a distinct mast-cell population, in line with previous observations that identified an erythroid-megakaryocyte-mast progenitor in the FL.46-48 Cell-cycle analysis revealed that primitive cells were less proliferative, with most cells in G1, whereas FL- and iPSC–derived definitive RBCs were predominantly cycling (G2M and S; supplemental Figure 4D). Erythroid maturation staging was confirmed using previously published gene lists49,50 and by CytoTRACE analysis51 (supplemental Figure 4E-F). Ten erythroid clusters were identified according to differentiation markers and cell-cycle stage, 3 of which were comprised mainly of primitive iRBCs (P_Ery_Mid_G2M, P_Ery_Late and P_Ery_Ter) and 7 that contained both FL- and iPSC–derived definitive RBCs (Ery_Early, Ery_Mid_S, Ery_Mid_G2M, Ery_Late1, Ery_Late2, Ery_Ter1, Ery_Ter2) (Figure 4D-E; supplemental Figure 4E).
An examination of individual erythroid genes identified core erythroid markers and transcription factors, such as Glycophorin B (GYPB), GATA-binding protein 1 (GATA1), and nuclear factor-erythroid 2 (NFE2) that were expressed at similar levels in all samples (Figure 4F, top panel; supplemental Figure 4G, KLF1), confirming our stage-matching strategy. As expected, the expression of globin genes remained significantly different, with embryonic globins such as ζ (HBZ) highly expressed in primitive iRBCs, and fetal or adult globins (γ, HBG1 and β, HBB, respectively) expressed more abundantly in FL- and iPSC-derived RBCs (Figure 4F, bottom panel). To quantify similarity between the different samples, a Pearson correlation analysis was performed. Importantly, a higher correlation was observed between the FL- and iPSC-derived definitive RBCs (0.96) compared with the primitive iRBCs (0.63) (Figure 4G). Notably, no FL cells and only a small portion of definitive iRBCs (1.5%) were observed in the primitive erythroid clusters (Figure 4H), indicating the purity of samples.
Differentially expressed genes s within primitive and FL-definitive erythroid clusters were identified (Figure 4I and supplemental Data). In primitive iRBCs, the top 25 upregulated genes included HBZ, as expected, and genes involved in purine metabolism (ADA), response to stress (HSF1, ATF5) and redox regulation of the cell (PRXL2A). In the FL and definitive erythroid clusters, there was higher expression of cell-cycle genes (megakaryocite I67, TK1, CCNA2, MYBL, and RRM2), confirming their proliferative state, in addition to β-globin (HBB), HES6, and CD36 as previously observed by flow cytometry analysis. Importantly, the expression of the known erythroid fetal gene SOX6, was only detected in definitive and FL RBCs (supplemental Figure 4H), as was increased expression of genes involved in globin switching such as ZBTB7A52 and NFI-factors.53,54
We used a recently published human yolk-sac data set55 to further explore similarities between our iPSC-derived RBCs and human primitive erythropoiesis. To compare the 2 datasets, we projected our scRNA-seq data onto the yolk-sac atlas and assigned each cell of our dataset to the most similar cluster in the reference. We then calculated prediction scores within our erythroid clusters and observed a higher value for the primitive sample compared to the definitive and FL erythroblasts (supplemental Figure 4I-J), indicating a higher level of similarity between primitive and human yolk-sac red cells.
Our results suggest that human FL- and iPSC–derived definitive RBCs share similar transcriptomic and cellular composition, whereas primitive iRBCs remain distinct.
Regulon and gene network analysis
To better understand the transcriptional differences across erythroid developmental programs, we used the single-cell regulatory network inference and clustering algorithm56,57 to infer transcription factor activity and gene-regulatory networks (GRNs) in our samples (Figure 5A). A total of 361 active transcriptional regulons were identified. This analysis further validated previous cluster identities, with nonerythroid groups enriched for known transcriptional regulons specific to megakaryocytes58 (ETV6, FLI1, and MEIS1), granulocytes59 (CEBPE and IRF8), and mast cells60 (GATA2 and BATF) (supplemental Figure 5A).
Identification of active regulons and gene signatures in primitive and definitive iRBCs. (A) Heat map of top 10 differentially expressed regulons across Seurat erythroid clusters. (B) Heat map of a selection of differentially active regulons in erythroid clusters. The color intensity represents the average regulon activity, z-score normalized by row. (C) Violin plot illustrating regulon activity of BCL11A and SOX6 in erythroid clusters. (D) Split violin plot of BCL11A, SOX6, and KLF1 regulon activity in FL-definitive erythroid clusters. Definitive iRBCs are shown in green and FL sample is shown in blue. Adjusted P-values are calculated using Wilcoxon rank sum test with Bonferroni correction (§Padj < 1e-4, #Padj < 1e-8). (E) GSEA analysis of FL and definitive and primitive-derived RBC. Normalized enrichment scores of significant pathways (Padj < .01) are shown.
Identification of active regulons and gene signatures in primitive and definitive iRBCs. (A) Heat map of top 10 differentially expressed regulons across Seurat erythroid clusters. (B) Heat map of a selection of differentially active regulons in erythroid clusters. The color intensity represents the average regulon activity, z-score normalized by row. (C) Violin plot illustrating regulon activity of BCL11A and SOX6 in erythroid clusters. (D) Split violin plot of BCL11A, SOX6, and KLF1 regulon activity in FL-definitive erythroid clusters. Definitive iRBCs are shown in green and FL sample is shown in blue. Adjusted P-values are calculated using Wilcoxon rank sum test with Bonferroni correction (§Padj < 1e-4, #Padj < 1e-8). (E) GSEA analysis of FL and definitive and primitive-derived RBC. Normalized enrichment scores of significant pathways (Padj < .01) are shown.
Within the erythroid clusters, we observed high activity of many core erythroid transcription factors (TFs) (GATA1, LMO2, NFE2, KLF1, and TAL1; supplemental Figure 5B). In the 3 primitive erythroid clusters, 148 GRNs with a regulon specificity score above 0.1 were observed, mostly active in the more differentiated cluster (P_Ery_Ter), of which 14 were specifically active in primitive iRBCs (Figure 5B and extended data). Among these regulons we found genes implicated in endoplasmic reticulum (ER) stress response (CREB3L1, ATF4, and HSF1) and stress–induced globin regulation (ATF4,61 HSF1,62 and HNF4A63).
The FL- and iPSC–derived definitive RBC clusters showed more complexity, in line with a heterogeneous and multistep maturation process of these cells. We inferred TF regulon activity in specific phases of erythropoiesis (early, mid, late, and terminal) corresponding to previously identified erythroid clusters (Figures 4D and 5B). In early phases of differentiation, we observed specific activity of known GRNs including LIN28B,64 MAZ (a direct binder of α-globin promoter involved in human erythroid traits),65 and BCLAF1 (implicated in fetal hematopoiesis66,67). Mid and late erythroid clusters showed increased activity of regulons associated with cell cycle and genomic stability (BRCA1), chromatin 3D structure (CTCF), and oxidative stress regulation (GABPB1), also in addition to erythroid regulators BCL11A and SOX6 (Figure 5C), known to be important in fetal and adult erythropoiesis.68 However, the definitive iRBCs showed reduced activity of these regulons compared to FL RBCs from the same cluster, whereas other erythroid core factors like KLF1 were similar (Figure 5D), possibly explaining the noncomplete silencing of ζ- and ε-globin genes observed in the definitive iRBCs. During terminal erythroid differentiation, we observed higher activity for many GRNs including NFIX,69 EPO-responsive chromatin remodeler YY1 and known regulators of terminal erythropoiesis and enucleation such as FOXO3.70
To identify changes in biological processes across different samples, we also performed gene set enrichment analysis (GSEA) in erythroid subsets (Figure 5E). Primitive RBCs showed an enrichment in TGF-β signaling and platelet genes (hallmark coagulation, extended data), suggesting a more promiscuous transcriptional program indicative of the erythro-megakaryocytic nature of embryonic HPCs. In contrast, definitive iPSC-derived and FL RBCs were enriched in cell-cycle pathways (E2F targets, G2M checkpoints, and MYC targets), mTorc1 signaling and the unfolded protein response. Moreover, definitive and FL RBCs upregulated fatty acid metabolism and glycolysis, a feature commonly found in proliferating cells.71 Taken together, our data suggest core erythroid similarities between primitive and definitive iRBCs but also highlight a differential activation of transcriptional units and biological processes between the 2 developmental programs.
Disease modeling with different developmental programs
To highlight the importance of cell ontology in disease modeling, studies were undertaken to characterize a loss-of-function mutation in the erythroid master regulator Krüppel-like factor 1 (KLF1). This transcription factor regulates erythroid cell specification 72 and may have differential involvement in primitive and definitive erythropoiesis as KLF1-null mice die in utero by E15.5 due to severe anemia.73,74 In addition, heterozygous carriers of KLF1 mutations produce well studied erythroid-specific phenotypes, such as low CD44 expression and the In(Lu) blood type.75,76
To investigate the role of KLF1 in these developmental programs, iPSC lines expressing a known pathogenic missense mutation (L300P77) in the DNA-binding domain of KLF1 was generated. Both heterozygous and homozygous mutations were created using base editing technology78 (supplemental Figure 6A). Primitive and definitive HPCs followed by expansion to iRBCs were generated from the isogenic control and mutant lines (Figure 6A). No differences were observed in HPC yield between any of the lines (supplemental Figure 6B; data not shown) and only primitive KLF1L300P/L300P HPCs had a reduction in CD235ab expression with a slight increase in megakaryocytic markers (supplemental Figure 6C).
Functional characterization of primitive and definitive RBCs from KLF1 edited iPSC lines. (A) Schematic representation of the experimental layout. (B) Erythroid expansion of primitive or definitive erythroid liquid cultures (mean ± SD, n = 2-5). (C) Percentage of erythroid colonies derived from primitive and definitive HPCs from wild type (WT) and KLF1 edited lines (mean ± SD, n = 2-9, 1-way analysis of variance, Tukey test; ∗∗P < .01 and ∗∗∗P < .001). (D) Expression of KLF1-dependent or independent red cell antigens by flow cytometry in primitive or definitive day 12 RBCs from WT or KLF1WT/L300P lines. (E) Relative abundance of β-like transcripts (β+ε+γg+γa) at day 12 of erythroid liquid culture (mean ± SD, n = 3-5, ∗P < .05, paired 2-tailed t test WT vs KLF1WT/L300P). (F-G) HPLC quantification of globin subunits (F) and tetramers (G) in primitive and definitive RBCs from WT or KLF1WT/L300P iPSC lines (mean ±SD, n = 2-4, ∗ = P < .05, paired 2-tailed t test WT vs KLF1WT/L300P). HPLC, high-performance liquid chromatography.
Functional characterization of primitive and definitive RBCs from KLF1 edited iPSC lines. (A) Schematic representation of the experimental layout. (B) Erythroid expansion of primitive or definitive erythroid liquid cultures (mean ± SD, n = 2-5). (C) Percentage of erythroid colonies derived from primitive and definitive HPCs from wild type (WT) and KLF1 edited lines (mean ± SD, n = 2-9, 1-way analysis of variance, Tukey test; ∗∗P < .01 and ∗∗∗P < .001). (D) Expression of KLF1-dependent or independent red cell antigens by flow cytometry in primitive or definitive day 12 RBCs from WT or KLF1WT/L300P lines. (E) Relative abundance of β-like transcripts (β+ε+γg+γa) at day 12 of erythroid liquid culture (mean ± SD, n = 3-5, ∗P < .05, paired 2-tailed t test WT vs KLF1WT/L300P). (F-G) HPLC quantification of globin subunits (F) and tetramers (G) in primitive and definitive RBCs from WT or KLF1WT/L300P iPSC lines (mean ±SD, n = 2-4, ∗ = P < .05, paired 2-tailed t test WT vs KLF1WT/L300P). HPLC, high-performance liquid chromatography.
As expected, the KLF1L300P/L300P homozygous mutation led to limited expansion, lack of hemoglobinization, and reduced expression of red cell markers in both primitive and definitive iRBCs (Figure 6B; supplemental Figure 6D). A strong reduction in KLF1 expression was also observed during erythroid differentiation of mutant lines, suggesting abnormal differentiation (supplemental Figure 6E). This impairment in erythropoiesis was confirmed in colony-forming cell -assays, where an almost complete loss of erythroid colonies was observed (Figure 6C). In comparison, the KLF1wild type(WT)/L300P heterozygous mutation led to an intermediate phenotype, with significantly reduced numbers of erythroid colonies (Figure 6B-C). With the limited generation of iRBCs from the KLF1L300P/L300P line, further studies were performed comparing WT and KLF1WT/L300P iRBCs only.
During development, the expression of many erythroid-specific antigens is regulated by KLF1, including CD44 and Duffy.76 To assess expression of KLF1-independent (CD47) and KLF1-dependent (CD44 and Duffy) red cell markers, primitive, and definitive WT and mutant iRBCs were analyzed by flow cytometry. As expected, no difference in CD47 expression was observed (Figure 6D), whereas CD44 expression was reduced in both primitive and definitive mutant iRBCs, Duffy expression was reduced in definitive mutant iRBCs only.
In addition to expression of red cell antigens, KLF1 is also involved in globin expression and switching during development. In primitive mutant iRBCs, the embryonic (ε-globin) and fetal (γ-globin) globin composition was unaffected (Figure 6E). In definitive mutant compared to WT iRBCs, a relative increase in the expression of embryonic globins was observed at the expense of fetal globins, indicating a different ontogenetic involvement of KLF1 at the globin loci. This observation was confirmed by high-performance liquid chromatography, with a significant increase of ε-globin in single-chain analysis and concomitant reduction of HbF (Figure 6F-G). Our findings show that iPSCs can model erythroid defects caused by pathogenic KLF1 mutations, such as reduced proliferation, hemoglobinization, and CD44 expression; however, only definitive iRBCs could capture differences in globin switching and Duffy expression in KLF1 mutants.
These data highlight the importance of cell ontology in disease modeling and demonstrate the strength of an appropriate platform to identify specific developmental features of a disease phenotype.
Discussion
Due to their unlimited proliferation and differentiation capabilities, iPSCs are a promising source to generate blood products ex vivo.79 The first methods to generate erythroblasts from iPSCs mainly recapitulated primitive erythropoiesis, producing iRBCs with embryonic characteristics.19,20,32,33 Utilizing manipulation of signaling inputs during mesoderm patterning, more recent methods have emerged to generate pure primitive or definitive HPCs22,30,31 but the iRBCs generated from these protocols have not been studied in depth.
In this study, we sought to define the purity and ability of primitive and definitive iRBC populations to recapitulate human developmental erythropoiesis and compare them to isogenic FL-derived RBCs. Our data show significant differences between the 2 iRBC programs, with definitive iRBCs being smaller and more proliferative then primitive iRBCs, expressing definitive red cell antigens, and undergoing embryonic-to-fetal globin switching. The definitive iRBCs were very similar to genetically-matched FL-derived RBCs in regard to gene expression as well as globin composition. Although very similar, the definitive iRBCs are not identical to FL RBCs, as they show lower expression of some key genes and regulatory networks involved in globin switching such as BCL11A and SOX6, which may inhibit these cells from complete silencing of embryonic globin expression. Future optimization efforts focused on increasing expression of these factors during erythroid differentiation will further improve the quality of iPSC-derived RBCs.
Transcriptional regulatory analysis of erythroid populations uncovered a number of factors which are differentially active throughout red cell maturation and across developmental origins. Although core erythroid factor GRNs (supplemental Figure 5B) are similarly active in all samples, definitive and FL RBCs show a more complex, dynamic, and stage-specific utilization of transcriptional units. Many regulatory networks involved in this maturation process include histone-modifying enzymes (HDAC2, KDM5A), chromatin insulators (CTCF), and chromatin remodelers (EP300, HMGA1). Investigation of chromatin accessibility and architecture in primitive and definitive iRBCs at specific maturation stages can be the topic of further studies and unravel additional differences between these developmental programs.
A major outcome of assessing blood antigen expression during our erythroid differentiations is the identification of the RBC antigen Duffy (ACKR1) that can distinguish definitive human erythroblasts. Interestingly, ACKR1 was also one of the top 100 upregulated genes in definitive vs primitive mouse erythroblasts identified by a previous study.80 We previously generated iRBCs with specialized Rh phenotypes including high and low prevalence Rh antigens (hrS, Goa, V/VS), which are difficult but important to identify because they may lead to delayed hemolytic transfusion reactions.3,81 Although the primitive iRBCs generated express Rh and Kell antigens at sufficient levels for diagnostic testing, the ability to express additional blood group antigens including Duffy and MNS, enhances the diagnostic scope of these in vitro-derived iRBCs to identify clinically significant antibodies and improve transfusion safety.82
The generation of definitive fetal liver–like erythroblasts from iPSCs further enhances the use of these cells for disease modeling. We demonstrated that iRBCs harboring pathologic mutations in KLF1 reveal specific phenotypes only when a definitive differentiation protocol is used. This finding has important consequences both for basic research and drug discovery approaches, because pathological phenotypes or drug targets might be evident only when a specific developmental stage is modeled. This could be the case for many hematologic disorders that start in fetal life including transient myeloproliferative disorder,83 thrombocytopenia-absent radius84 (OMIM #274000), Diamond-Blackfan anemia,85 or Hb Bart's hydrops fetalis.86
Overall, our work demonstrates that definitive in vitro differentiation protocols faithfully mimic human FL erythropoiesis and uncover a larger set of phenotypes when using iPSCs for hematopoietic disease modeling. These findings pave the way for a better understanding of erythropoiesis in human development, and they will help to generate a more uniform population of the most mature RBCs to be used as a clinical diagnostic reagent and potentially, as future transfusion products.
Acknowledgments
The authors extend special thanks to the members of the U01 NHLBI Progenitor Cell Translational Consortium, especially Gordon Keller for sharing the definitive hematopoietic progenitor cell protocol. Sequencing was conducted at the Center for Applied Genomics at the Children’s Hospital of Philadelphia.
This work was supported by National Heart, Lung, and Blood Institute U01 HL134696 (S.T.C., P.G., and D.L.F.) and DK 100854 (S.T.C.). The fetal liver sample was reprogrammed by the CHOP Human Pluripotent Stem Cell Core through the support of a Distinguished Chair of Pediatrics (S.T.C.).
Authorship
Contribution: G.P. conceived the study, designed and performed experiments, analyzed data, and wrote the manuscript; J.G.K. performed experiments and analyzed data; C.C.N. performed megakaryocyte experiments; J.H.S. and C.S.T. performed analysis on scRNA-seq data; H.H.A. performed gel card experiments; O.A. performed high-performance liquid chromatography analyses; P.A.G. collected the fetal liver sample; J.A.M. reprogrammed fetal liver cells; C.W. performed experiments; D.L.F. and K.T. provided scientific advice and edited the manuscript; and S.T.C. and P.G. conceived the study, designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: S.T.C. has filed a US patent (application number 16/757,815), entitled “Engineered Red Blood Cells Having Rare Antigen Phenotypes” in the name of The Children’s Hospital of Philadelphia and New York Blood Center, Inc (national stage entry of international application number PCT/US2018/057932); the manuscript describes using the erythroid differentiation protocol of the engineered red blood cells covered in patient application. The remaining authors declare no competing financial interests.
Correspondence: Paul Gadue, Department of Pathology and Laboratory Medicine, Children's Hospital of Philadelphia, Philadelphia, PA 19104; email: gaduep@chop.edu; and Giulia Pavani, Department of Pathology and Laboratory Medicine, Children's Hospital of Philadelphia, Philadelphia, PA 19104; email: pavanig@chop.edu.
References
Author notes
Raw sequencing data and processed reads associated with this work have been deposited in the Gene Expression Omnibus under accession number GSE242101.
Any other data relevant to the study are available upon reasonable request from the corresponding authors, Paul Gadue (gaduep@chop.edu) and Giulia Pavani (pavanig@chop.edu).
The full-text version of this article contains a data supplement.