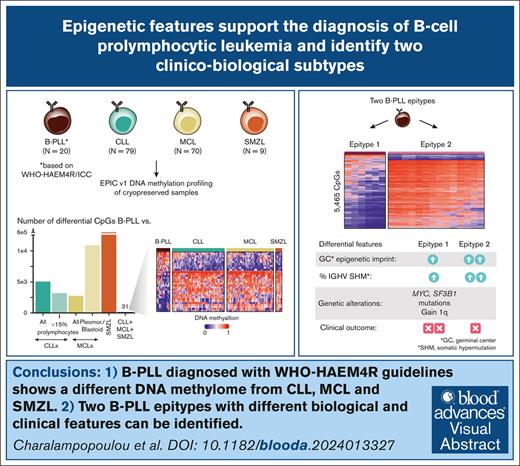
Key Points
B-PLL has a differential DNA methylation signature compared with CLL, MCL, and SMZL.
Identification of 2 potential B-PLL subtypes with different biological and clinical features.
Visual Abstract
The recognition of B-cell prolymphocytic leukemia (B-PLL) as a separate entity is controversial based on the current classification systems. Here, we analyzed the DNA methylome of a cohort of 20 B-PLL cases diagnosed according to the guidelines of the International Consensus Classification/Fourth revised edition of the World Health Organization Classification, and compared them with chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), splenic marginal zone lymphoma (SMZL), and normal B-cell subpopulations. Unsupervised principal component analyses suggest that B-PLL is epigenetically distinct from CLL, MCL, and SMZL, which is further supported by robust differential methylation signatures in B-PLL. We also observe that B-PLL can be segregated into 2 epitypes with differential clinicobiological characteristics. B-PLL epitype 1 carries lower immunoglobulin heavy variable somatic hypermutation and a less profound germinal center–related DNA methylation imprint than epitype 2. Furthermore, epitype 1 is significantly enriched in mutations affecting MYC and SF3B1, and displays DNA hypomethylation and gene upregulation signatures enriched in MYC targets. Despite the low sample size, patients from epitype 1 have an inferior overall survival than those of epitype 2. This study provides relevant insights into the biology and differential diagnosis of B-PLL, and potentially identifies 2 subgroups with distinct biological and clinical features.
Introduction
B-cell prolymphocytic leukemia (B-PLL) is a very rare and clinically aggressive mature B-cell disorder characterized by a percentage of prolymphocytes exceeding 55% of the lymphoid cells in the peripheral blood and presenting with lymphocytosis, cytopenia(s), and splenomegaly.1 Although it was first described in the 1970s,2,3 it was not recognized as a separate entity by the World Health Organization (WHO) classification of tumors until 2001.4 From the genetic perspective, B-PLL frequently shows MYC rearrangements and gains, TP53 deletions and mutations, as well as complex karyotypes.1,5 However, whether the clinicobiological features of B-PLL are sufficiently different from other B-cell malignancies such as chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), and splenic marginal zone lymphoma (SMZL) to justify its designation as an independent entity is a matter of international debate.6,7 Indeed, the fifth WHO classification, whose online version was recently released, no longer recognizes B-PLL as a separate entity and proposes to reclassify cases within the umbrella term “other lymphoid tumors.”8 According to this classification, B-PLLs expressing CD5 are classified either as MCL variants or as a prolymphocytic progression of CLL/small lymphocytic lymphoma when there is >15% prolymphocytes in the peripheral blood/bone marrow. CD5− cases should be classified in the new category of “splenic B-cell lymphoma/leukemia with prominent nucleoli,” which encompasses the previously designated hairy cell leukemia variant and CD5− B-PLL. In contrast, B-PLL is still recognized in the International Consensus Classification (ICC),9 which is based on the previous criteria of the revised fourth edition of the World Health Organization Classification of Hematologic Neoplasms (WHO HAEM4R)10 and emphasizes the need to perform a careful differential diagnosis assessment to exclude other lymphoid neoplasms, especially CLL, MCL, and SMZL.
Complementary to genetic changes, DNA methylation features have greatly contributed to our understanding of the cellular origin, pathogenetic mechanism, and clinical behavior of B-cell neoplasms.11 For instance, 3 epigenetic subtypes (ie, epitypes) with different clinicobiological features have been identified in CLL, which show imprints of germinal center (GC)-inexperienced (low-programmed naive-like CLL [n-CLL]) and various degrees of GC-experienced mature B cells (moderate GC imprint in intermediate CLL, and strong GC imprint in high-programmed/memory-like CLL).11-14 Likewise, 2 epitypes have been identified in MCL based on their resemblance to GC-inexperienced (C1-MCL) and GC-experienced B cells (C2-MCL), which show a major overlap with conventional MCL (cMCL) and leukemic nonnodal MCL (nnMCL),15 respectively. Moreover, 2 epigenetic subtypes have been identified in Waldenström macroglobulinemia (WM) based on similarities to memory B cells (MBCs) and plasma cells (PCs).16 Finally, a study on promoter DNA methylation in SMZL reported the presence of 2 epitypes with distinct clinicobiological characteristics.17 In addition to epigenetics-based disease subtyping, the DNA methylome of B-cell tumors also reflects their proliferative history and shows de novo changes affecting key pathways.18
Here, we have analyzed the DNA methylome of a B-PLL cohort diagnosed according to the ICC/WHO-HAEM4R guidelines, and compared it with CLL, MCL, SMZL, and normal B-cell subpopulations, aiming to shed light on its biological nature as well as the differential diagnosis with other mature B-cell tumors.
Methods
Selection of B-PLL cases
We gathered a total of 20 B-PLL cases, which were diagnosed according to the criteria established in the WHO-HAEM4R classification. All patients provided written informed consent, the study was performed in accordance with the Declaration of Helsinki and was approved by the local ethics review boards. Genetic and immunophenotypic data are available from 17 cases previously published.5,19 Detailed immunophenotypic and clinical features of all the B-PLL cases are shown in supplemental Table 1. None of the cases had a previous history of any other B-cell neoplasm nor showed any molecular hallmark of hairy cell leukemia variant and SMZL such as MAP2K1 and KLF2 mutations, respectively, or MCL such as CCND1, CCND2, and CCND3 chromosomal rearrangements. Careful cytological examination showed lymphoid cells with prominent nucleoli and lack of cytoplasmic projections in all cases.1 To further support the differential diagnosis between B-PLL and MCL or CLL, we assessed in B-PLL the expression of previously reported genes specific to MCL and CLL20 using RNA-sequencing (RNA-seq) data from 11 B-PLL cases,5 5 MCL21 (2 cMCL and 3 nnMCL), and 294 CLL22 cases (supplemental Figure 1A). In addition, we compared the mutational landscape of B-PLL with that of previously described MCL23 and CLL24 cohorts (supplemental Figure 1B). A summary of the biological and clinical characteristics of B-PLL as compared with CLL, MCL, and SMZL is provided in supplemental Table 2.
DNA methylation profiling
We used Illumina EPIC Version 1 (EPIC V1) arrays to generate DNA methylation profiles of the 20 B-PLLs, as well as from splenic tumor material of 9 SMZLs. These SMZLs were diagnosed according to current guidelines25 of the University Hospital Dorset (immunoglobulin heavy variable [IGHV] sequencing analysis [IGHV1-02∗04, n = 6/9], deletion 7q status [n = 4/9 positive], and KLF2 [n = 3/9 mutated] and NOTCH2 [n = 3/9 mutated]). Moreover, we used previous EPIC array data from 78 CLL26 (27 n-CLL, 10 i-CLL, 39 m-CLL, and 2 unclassified), 70 MCL23 (49 C1-MCLs (39 with classical morphology, 10 with pleomorphic/blastoid), 19 C2 MCLs, and 2 unclassified; supplemental Table 3). Although the Illumina arrays are robust and show little slide-number batch effects, we compared the methylation profiles of cases from the same disease epitype located in different slides. We found a minimal number of differentially methylated CpGs indicating that the batch effect in our series is negligible as compared with the biological differences (data not shown). We additionally used HumanMethylation450 BeadChip array (450k) data of 476 CLL cases and 5 CLL cases containing between 16% and 44% prolymphocytic cells (CLL/PL22; supplemental Table 4), as well as EPIC V1 data of 2 sequential samples of a CLL case that underwent a clonally related prolymphocytic progression (CLL-pPL).27 Finally, we also included EPIC V1 data of normal B-cell subpopulations including 2 naive B cells (NBCs), 1 GC cells, 3 MBCs, and 1 PC27 (Figure 1A) as well as complementary 450k data of 6 hematopoietic precursor cells, 12 pre–B cells, 4 intermediate B cells, 15 NBCs, 9 GC cells, 8 tonsil PCs, 10 MBCs, and 3 bone marrow PCs28 to extend our normal B-cell data set for phylogenetic analyses.
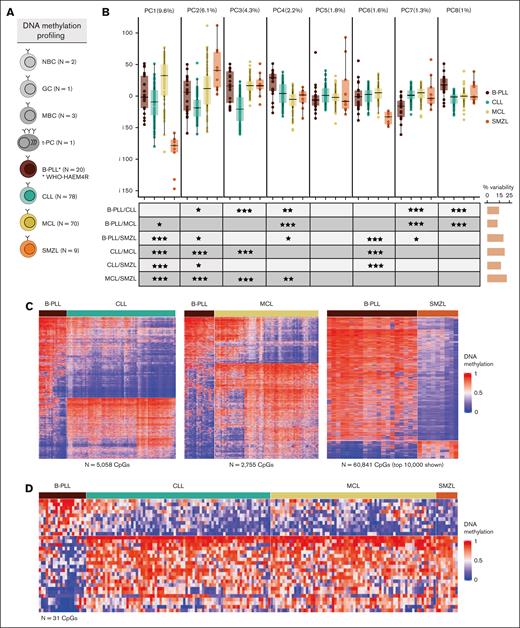
Differential DNA methylation patterns of B-PLL compared with CLL, MCL, and SMZL. (A) Number of samples with DNA methylation data available. (B) Boxplots of the first 8 PCAs including B-PLL, CLL, MCL, and SMZL cases. At bottom, the table shows the false discovery rate (FDR)–corrected P values of Wilcoxon test of pair-wise comparisons across entities. On the right, the bars represent the total percentage of the variability showing significant differences for each PCA comparison. (C) Heat maps of the differentially methylated CpGs between B-PLL and CLL (n = 5058), B-PLL and MCL (n = 2755), and B-PLL and SMZL (n = 60 841). In the case of SMZL, the top 10 000 CpGs with the highest standard deviation are represented. (D) Heat map of the 31 simultaneous differentially methylated CpGs comparing B-PLL to CLL, MCL, and SMZL. FDR levels of significance: FDR < 0.001 = ∗∗∗, FDR < 0.01 = ∗∗, and FDR < 0.05 = ∗.
Differential DNA methylation patterns of B-PLL compared with CLL, MCL, and SMZL. (A) Number of samples with DNA methylation data available. (B) Boxplots of the first 8 PCAs including B-PLL, CLL, MCL, and SMZL cases. At bottom, the table shows the false discovery rate (FDR)–corrected P values of Wilcoxon test of pair-wise comparisons across entities. On the right, the bars represent the total percentage of the variability showing significant differences for each PCA comparison. (C) Heat maps of the differentially methylated CpGs between B-PLL and CLL (n = 5058), B-PLL and MCL (n = 2755), and B-PLL and SMZL (n = 60 841). In the case of SMZL, the top 10 000 CpGs with the highest standard deviation are represented. (D) Heat map of the 31 simultaneous differentially methylated CpGs comparing B-PLL to CLL, MCL, and SMZL. FDR levels of significance: FDR < 0.001 = ∗∗∗, FDR < 0.01 = ∗∗, and FDR < 0.05 = ∗.
DNA methylation data analyses
DNA methylation data were analyzed as previously decribed.18 Briefly, we removed CpGs with a P value ≤.01 in >10% of the samples as well as single-nucleotide polymorphism–associated CpGs, sexual chromosome CpGs, and individual-specific CpGs. After all the filtering steps, the data were normalized using the subset-quantile within array normalization algorithm. CpGs were annotated using the annotation package IlluminaHumanMethylationEPICanno.ilm10b4.hg19. CpGs whose methylation are modulated during B-cell differentiation were selected from a previous publication using 450k Illumina data28 or calculated for EPIC array-specific CpGs using an absolute DNA methylation difference of at least 0.25 between NBCs and MBCs. Previously generated chromatin states of the GM12878 B-cell cell line29 were used to annotate CpGs to regulatory regions. CLL and MCL cases were further subdivided into epitypes using a previous DNA methylation classifier of B-cell tumors.18 Unsupervised analyses of all the DNA methylation data were performed using principal component analyses (PCA). Differential methylation (DM) analyses were performed using limma with false discovery rate of <0.05 and methylation difference of at least 0.25 as thresholds. Gometh was used to identify the functions of genes showing DM.
We used a permutation analysis to test the robustness of the DM signatures between B-PLL and CLL or MCL. The permutation analysis was carried out by performing 1000 DM analyses with distinct sets of randomly selected CLL or MCL cases in each permutation. The number of randomly selected CLL or MCL cases in each permutation was the same as the number of B-PLL cases. In that way, we obtained the distribution of the expected number of DM CpGs assuming that our B-PLL cases were, in reality, misdiagnosed CLL or MCL cases, and compared it with the observed number of DM CpGs in B-PLL vs CLL or MCL. The P value was calculated as the number of permutations resulting in lower number of DM CpGs than the observed divided by the total number of permutations.
Phylogenetic analyses with DNA methylation
Phylogenetic analyses were performed as in Oakes et al.13 Briefly, phylogenetic trees were inferred by the minimal evolution method using the fastme.bal function in the R package ape. Phylogenies were generated by applying the minimal evolution algorithm on Euclidean distance matrices based on continuous methylation values of 1112 CpGs present in the normalized data matrix.
Gene expression analyses
We used previous RNA-seq data of 11 B-PLL cases5 to perform differential expression analyses with DESeq2 package considering as differentially expressed the genes with false discovery rate of <0.05. Gene set enrichment analysis was done with the fgsea package using the Hallmark gene sets of the Broad Institute. For the gene expression comparisons performed with B-PLL, MCL, and CLL data together, we transformed the normalized expression values to percentiles to minimize the possible batch effects from RNA-seq data of distinct studies and data modalities.
TF binding analysis
Transcription factor (TF) binding analysis was performed with PWMEnrich package in windows of ±100 base pairs around the target CpGs, as described previously.18 TFs were considered significant if the binding site was present in at least 10% of the sequences and the P value was <.05.
Additional statistical analyses
Genetic and clinical associations were tested using 2-tailed Fisher test and the log-rank test, respectively. All analyses were performed in R software.
Results
B-PLL shows a DNA methylation signature different from CLL, MCL, and SMZL
We initially performed an unsupervised PCA including cases from all 4 diseases and analyzed separately each of the first 8 principal components to systematically evaluate similarities and differences among them (Figure 1B; supplemental Figure 2). We performed pair-wise entity comparisons of the values of each component, and we detected significant differences between B-PLL vs CLL, MCL, and SMZL in 5, 4, and 5 components, respectively. Next, we performed DM analyses and we detected that B-PLL shows 5058 DM CpGs vs CLL, 2755 vs MCL, and 60841 vs SMZL (Figure 1C; supplemental Tables 5-7). In the case of B-PLL vs CLL and B-PLL vs MCL, we further validated the robustness of the signatures by permutation analyses, which aimed at calculating the number of DM CpGs that would be obtained by chance if the B-PLL were misdiagnosed CLL or MCL cases (see “Methods”). These analyses indicated that the number of observed DM CpGs between B-PLL and CLL or MCL are very unlikely to be obtained by chance (P < .001; supplemental Figure 3). Despite the fact that most DNA methylation changes in lymphoid tumors do not have a direct impact on the expression of underlying genes,11 we performed a Gene Ontology analysis of the DM CpGs between B-PLL and each of the other diseases and normal B cells. Although we did not obtain highly significant enrichments, hypomethylated CpGs in B-PLL as compared with CLL, MCL, and SMZL were related to genes involved in functions associated with different pathways and the immune system (supplemental Table 8). We also identified a signature of 31 CpGs that can differentiate B-PLL vs CLL, MCL, and SMZL (Figure 1D; supplemental Table 9). Collectively, these analyses suggest that B-PLL, diagnosed on the basis of the ICC/WHO-HAEM4R criteria, has a different epigenetic make-up from CLL, MCL, and SMZL, and the identified methylation patterns may set the basis for an epigenetic-based differential diagnostic approach.
B-PLLs are epigenetically distinct from CLLs with prolymphocytoid cells, a prolymphocytic progression of CLL, and blastoid/pleomorphic MCLs
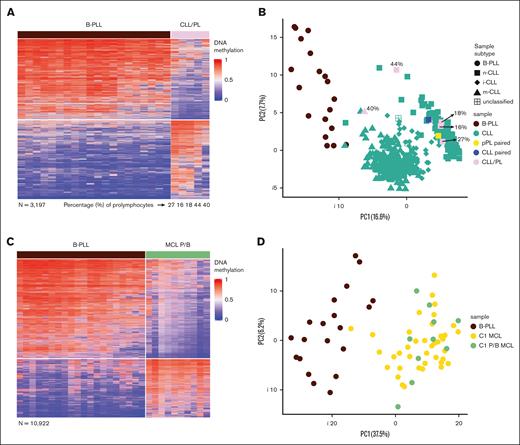
As previously mentioned, the WHO-HAEM5 classification states that some B-PLL cases should be considered prolymphocytic progressions of CLL or blastoid MCL.8 Therefore, we performed DM analysis between B-PLL and 5 CLL cases with percentage of prolymphocytes between 16% and 44% (CLL/PL) and we identified 3197 DM CpGs (supplemental Table 10; Figure 2A). Moreover, we identified 2906 CpGs comparing 476 CLLs with a lower percentage of prolymphocytes (<10%) vs B-PLL (supplemental Table 11). We then performed a PCA with those CpGs including the 5 CLL/PL cases to evaluate whether they cluster with CLL or B-PLL. We observed that the CLL/PL cases are separated from B-PLL cases and cluster with their respective CLL epitype regardless of the percentage of prolymphocytes (Figure 2B). Finally, within the same PCA we additionally represented 2 sequential samples from a highly informative CLL case, which represents a bona fide clonally related prolymphocytic progression of a previous CLL with classic morphology (CLL-pPL).27 In this case, neither the progressed nor the initial sample cluster with the B-PLLs but show a CLL-related DNA methylome (Figure 2B). Overall, these analyses indicate that de novo B-PLLs (ie, lacking evidence of a previous B-cell tumor) are epigenetically distinct from CLLs with prolymphocytoid cells or even a prolymphocytic progression of a previous CLL. These observations seem to question the assumption that B-PLLs are prolymphocytic progressions of CLL, as proposed by the WHO-HAEM5 classification.
Differential DNA methylation patterns of B-PLL compared with CLL/PL and MCL P/B. (A) Heat map of the differentially methylated CpGs between B-PLL and CLL/PL (n = 3197). (B) PCA of the differentially methylated CpGs between B-PLL and CLL with <10% prolymphocytoid cells (n = 2906), including also the CLL/PL (showing the percentage of cells with prolymphocytoid morphology) and the paired CLL/pPL case. The percentage of prolymphocytes of each of the CLL/PL samples is indicated. (C) Heat map of the differentially methylated CpGs between B-PLL and MCL P/B (n = 10 922). (D) PCA of the differentially methylated CpGs between B-PLL and C1 MCL with classic morphology (n = 8809) including in the plot the C1 MCL P/B.
Differential DNA methylation patterns of B-PLL compared with CLL/PL and MCL P/B. (A) Heat map of the differentially methylated CpGs between B-PLL and CLL/PL (n = 3197). (B) PCA of the differentially methylated CpGs between B-PLL and CLL with <10% prolymphocytoid cells (n = 2906), including also the CLL/PL (showing the percentage of cells with prolymphocytoid morphology) and the paired CLL/pPL case. The percentage of prolymphocytes of each of the CLL/PL samples is indicated. (C) Heat map of the differentially methylated CpGs between B-PLL and MCL P/B (n = 10 922). (D) PCA of the differentially methylated CpGs between B-PLL and C1 MCL with classic morphology (n = 8809) including in the plot the C1 MCL P/B.
Similarly, the comparison of B-PLL vs 10 pleomorphic/blastoid MCL cases with EPIC data led to a clear signature of 10 922 DM CpGs (supplemental Table 12; Figure 2C). Considering that all pleomorphic/blastoid cases belong to the C1 epitype, we then compared 39 classical morphology C1 MCLs with B-PLL and identified 8809 DM CpGs (supplemental Table 13). The PCA of those CpGs clearly indicated that the 10 blastoid C1-MCL cases do cluster with C1-MCLs with classical morphology rather than with B-PLLs (Figure 2D). These data indicate that the B-PLLs included in this study show a different DNA methylome than blastoid MCLs, which, in turn, are epigenetically similar to MCLs with classical morphology.
Identification of 2 B-PLL epitypes with different levels of GC-related epigenetic imprinting
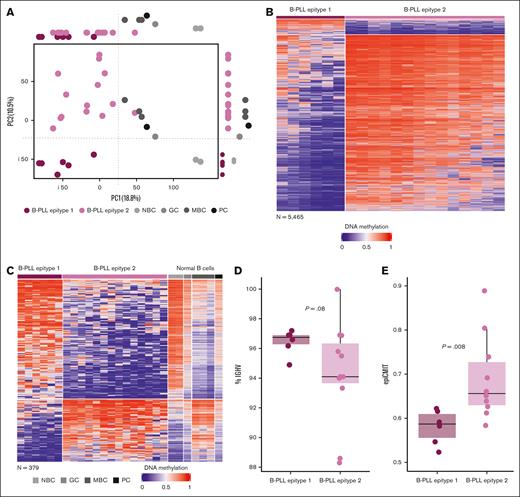
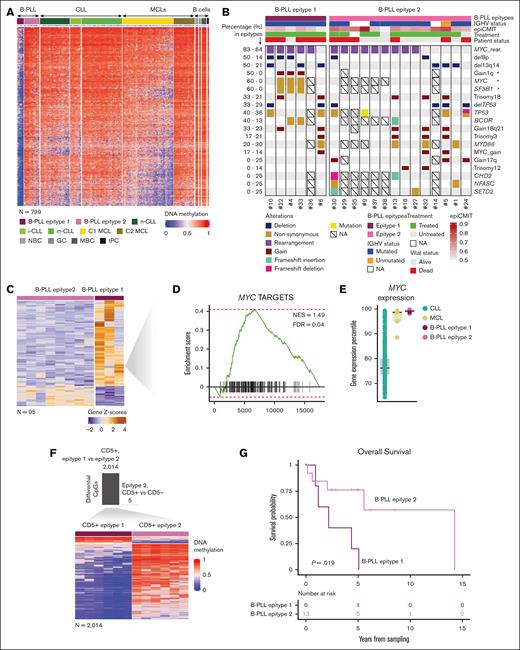
Because epigenetic variability in CLL and MCL is, in part, related to different epitypes with methylation imprints of pre- and post-GC B cells,11-15 we further explored whether this was also the case for B-PLL. We then performed a PCA with B-PLL and normal B-cell subsets (Figure 3A; supplemental Figure 4) and showed that PC1 separated B-PLL and normal B cells, whereas PC2 suggested the presence of 2 epitypes of B-PLL aligned with variability in normal B-cell subsets. One cluster (n = 6, epitype 1) was epigenetically closer to NBCs whereas the other (n = 14, epitype 2) was closer to GC-experienced B cells (Figure 3A). This finding suggests that B-PLL could be segregated into 2 subgroups related to B cells at different maturation stages. Next, we compared their DNA methylome and identified 5465 DM CpGs (Figure 3B; supplemental Table 14). Of those, 5036 and 429 CpGs were hypomethylated and hypermethylated in epitype 1 as compared with epitype 2, respectively. Additionally, when we examined those CpGs in the context of normal B-cell differentiation, we noticed that 9.5% of CpGs hypomethylated and 81% of CpG hypermethylated in epitype 1 were B cell–related (supplemental Table 14), suggesting that the hypermethylation signature mostly contains a B-cell differentiation epigenetic imprint. Evaluating those B-cell–related CpGs in the context of NBC and MBC, we identified that 249 of 429 (58%) CpGs hypermethylated in epitype 1 and 130 of 5036 (2.6%) CpGs hypomethylated in epitype 1 show a clear signature linking epitype 1 to NBC (ie, GC-inexperienced cells) and epitype 2 to MBC (ie, GC-experienced; Figure 3C; supplemental Table 15). Because B-PLL almost always shows IGHV somatic hypermutation (supplemental Table 1),5 it is unlikely that some cases arise from GC-inexperienced cells. Therefore, to evaluate the overall cellular origin of B-PLL, we used CpGs distinguishing GC-experienced vs GC-inexperienced B cells that were previously applied to trace the origin of CLL.13 A phylogenetic analysis (see “Methods”) with those CpGs revealed that B-PLL as a whole shows an epigenetic imprint associated with the GC-experienced cells (supplemental Figure 5), consistent with our initial PCA analysis (PC1). Thus, the most plausible explanation is that although all B-PLLs derive from GC experienced B-cells, the epitypes may be subjected to different levels of GC exposure. Epitype 1 shows a partial GC epigenetic programming, maintaining thereby some similarities with GC-inexperienced cells, whereas epitype 2 shows a more extensive GC programming. This interpretation is concordant with the fact that epitype 1 has a tendency toward lower IGHV somatic hypermutation (mean germ line identity of 96.45% vs 94.32%, respectively, P = .08; Figure 3D) and a significantly lower proliferative history measured by the epiCMIT mitotic clock (P = .00818; Figure 3E), similar to n-CLL and C1/cMCL epitypes, which show earlier cells of origin.11,18
Identification of 2 B-PLL epitypes with signatures related to normal B-cell differentiation. (A) PCA analysis showing PC1 and PC2 with B-PLL and normal B-cell samples. On its side, the boxplots display differences between B-PLL epitypes and normal B cells. (B) Heat map of the differentially methylated CpGs (n = 5465) between B-PLL epitypes. (C) Heat map of the differentially methylated CpGs between B-PLL epitypes related to methylation patterns of NBCs and MBCs (n = 379, of which 249 are hypermethylated and 130 hypomethylated in epitype 1). (D) Percentage of IGHV gene identity B-PLL epitypes. (E) Magnitude of the epiCMIT scores between B-PLL epitypes.
Identification of 2 B-PLL epitypes with signatures related to normal B-cell differentiation. (A) PCA analysis showing PC1 and PC2 with B-PLL and normal B-cell samples. On its side, the boxplots display differences between B-PLL epitypes and normal B cells. (B) Heat map of the differentially methylated CpGs (n = 5465) between B-PLL epitypes. (C) Heat map of the differentially methylated CpGs between B-PLL epitypes related to methylation patterns of NBCs and MBCs (n = 379, of which 249 are hypermethylated and 130 hypomethylated in epitype 1). (D) Percentage of IGHV gene identity B-PLL epitypes. (E) Magnitude of the epiCMIT scores between B-PLL epitypes.
(Epi)genetic characterization, CD5 expression, and clinical relevance of the B-PLL epitypes
We further characterized the DM CpGs between B-PLL epitypes. Epitype 2 did not show any specific de novo methylation signature. However, epitype 1 hypomethylation was mostly occurring de novo, as both normal B cells and epitype 2 were methylated at those CpGs. Moreover, 799 of 5036 of the hypomethylated CpGs in B-PLL epitype 1 (Figure 4A; supplemental Table 16) were also de novo compared with CLL and MCL epitypes. These CpGs were mainly located in low CpG-content regions and enriched in regulatory regions and transcription-related chromatin states (supplemental Figure 6A-B). A TF analysis using the genomic regions associated to these CpGs (“Methods”) showed a significant enrichment in binding motifs of several TF families (supplemental Figure 6C; supplemental Table 17). Of all the TFs identified, BHLHA15/MIST1 was also found to be significantly overexpressed in epitype 1 B-PLL (P = .016, supplemental Figure 6D), suggesting that it could be involved in the pathogenesis of this epitype. At the genetic level, epitype 1 was enriched in MYC (P = .034) and SF3B1 (P = .027) mutations and 1q gain (P = .024) (Figure 4B). Further supporting the existence of these 2 B-PLL epitypes, we identified 95 differentially expressed genes (Figure 4C; supplemental Table 18). Overexpressed genes in epitype 1 were found to be enriched in MYC targets (Figure 4D), although MYC expression was consistently high in all B-PLL cases (Figure 4E). In addition, because the WHO-HAEM5 classification highlights the importance of CD5 expression to reclassify B-PLL cases, we also explored CD5 expression in the 2 B-PLL epitypes. CD5 was expressed in all 6 epitype 1 and 6 of 14 epitype 2 B-PLLs (P = .04), which could suggest a relationship between CD5 and methylation patterns. Nonetheless, when comparing the presence or absence of CD5 within epitype 2 cases, we only detect 5 DM CpGs. In contrast, 2014 CpGs were differential comparing CD5+ cases between epitypes (Figure 4F; supplemental Table 19). These data strongly suggest that CD5, although enriched in epitype 1, is unrelated to the DM pattern observed between B-PLL epitypes. Furthermore, they suggest that the decision of the WHO classification of segregating B-PLLs into other entities based on the expression of CD5 is not supported by a differential DNA methylation signature. Finally, despite the small sample size inherent to this rare leukemia, epitype 1 showed a shorter overall survival than epitype 2 (Figure 4G), although the proportion of cases with TP53 abnormalities was similar in both epitypes (Figure 4B).
Epigenetic and genetic characterization of the differential signature of the B-PLL epitypes. (A) Heat map of the de novo epitype 1 specific methylation signature (n = 799 CpGs). (B) Oncoplot showing genetic alterations of B-PLL cases. (C) Heateukemic nonnodal map of 97 differentially expressed genes between B-PLL epitypes. (D) Gene set enrichment analysis (GSEA) showing enrichment of MYC targets in epitype 1 B-PLL (E) MYC expression percentile of B-PLL, CLL, and MCL cases (F) Bar plot of the numbers of differentially methylated CpGs between CD5+ epitype 2 and CD5− epitype 2 (top) and heat map of DM CpGs between CD5+ epitype 1 and CD5+ epitype 2 (n = 2014; bottom) (G) Kaplan-Meier curves for overall survival between B-PLL epitypes.
Epigenetic and genetic characterization of the differential signature of the B-PLL epitypes. (A) Heat map of the de novo epitype 1 specific methylation signature (n = 799 CpGs). (B) Oncoplot showing genetic alterations of B-PLL cases. (C) Heateukemic nonnodal map of 97 differentially expressed genes between B-PLL epitypes. (D) Gene set enrichment analysis (GSEA) showing enrichment of MYC targets in epitype 1 B-PLL (E) MYC expression percentile of B-PLL, CLL, and MCL cases (F) Bar plot of the numbers of differentially methylated CpGs between CD5+ epitype 2 and CD5− epitype 2 (top) and heat map of DM CpGs between CD5+ epitype 1 and CD5+ epitype 2 (n = 2014; bottom) (G) Kaplan-Meier curves for overall survival between B-PLL epitypes.
Discussion
This project on the DNA methylome of B-PLL was initiated when the disease was still recognized as a separate entity in the WHO-HAEM4R classification. However, when the new WHO-HAEM5 and ICC classifications were published in 2022,8,9 an international debate started on whether B-PLL truly exists or not. Whereas the ICC maintained B-PLL as a separate entity, the new WHO classification eliminated this leukemia and cases were fragmented to become variants or progressive forms of MCL, CLL, or SMZL.8 Therefore, we started our analysis by evaluating, in the first place, whether B-PLL, as diagnosed following the ICC/WHO-HAEM4R criteria, was epigenetically distinct from MCL, CLL, and SMZL. After exhaustive unsupervised and supervised statistical analyses, our results suggest that the 20 B-PLL cases studied herein show an epigenetic configuration that is overall different from the other 3 entities. The differential features of B-PLL vs each of the 3 entities is discussed below.
In the case of MCL, initial reports on B-PLL included some cases that, in reality, were misdiagnosed MCLs, because they had the CCND1::IGH translocation.30 In our series, all the B-PLL lack any of the CCND1, 2, or 3 rearrangements, which is a key criterion to differentiate B-PLL from MCL in the ICC/WHO-HAEM4R classifications. Also, they do not show overexpression of any of the CCN family genes excluding the presence of cryptic CCND translocations that have been reported previously in MCL.31,32 Moreover, B-PLLs and MCL show differential mutational patterns and gene expression profiles, and present 2755 DM CpGs. We could also show that MCL cases with blastoid/pleomorphic morphology present a clear DM signature from B-PLL, further ruling out that, as the WHO-HAEM5 classification states, some B-PLL cases are actually MCLs with blastoid morphology.8 Therefore, it seems that none of the B-PLLs included in our study are misdiagnosed MCLs.
Regarding CLL, the WHO-HAEM5 classification asserts that CD5+ B-PLL cases represent prolymphocytic progressions of CLL. We included in our analyses CLL cases with >15% and <55% of cells with prolymphocytoid morphology and observed that these cases were epigenetically distinct from B-PLL and similar to CLLs with typical morphology. Therefore, prolymphocytic progressions of CLL maintain a CLL-related methylome and do not resemble B-PLLs. Moreover, we further analyzed a highly informative case in which a classic CLL progressed into a clonally related CLL with prolymphocytic morphology.27 Both CLL and CLL-pPL cells showed a DNA methylation pattern consistent with CLL and not with B-PLL. This scenario is similar to the Richter transformation of CLL into diffuse large B-cell lymphoma (DLBCL). Such DLBCLs arising from a previous CLL seem to maintain a CLL epigenetic imprint and can be differentiated from de novo DLBCL.33 Additionally, the B-PLLs studied herein and CLLs show differential mutational landscapes and expression of key CLL genes. Overall, we could not find any molecular evidence that B-PLLs merely represent progressed forms of CLL.
As compared with SMZL, B-PLLs were epigenetically very different. However, we recognize that the SMZL sample size was low, and therefore, a more detailed comparison of B-PLL vs a larger SMZL series is needed to properly characterize the potential overlap between these 2 entities.
Overall, this initial part of our analyses seems to indicate that B-PLL, diagnosed on the basis of the ICC/WHO-HAEM4R classification, is epigenetically different from MCL, CLL, and SMZL. Additionally, we identified a DNA methylation signature that segregates B-PLL from the other 3 entities, which can potentially be used to develop molecular biomarkers that may contribute to the differential diagnosis of B-PLL.
In addition to identifying an epigenetic signature of B-PLL as a whole, we detected that the disease can potentially be divided into 2 epigenetic subtypes. This finding is aligned with findings in MCL, CLL, and WM, in which different epitypes linked to distinct B-cell maturation stages are described.11-16 As compared with epitype 2, B-PLL epitype 1 showed a less profound GC-related epigenetic imprint, a tendency toward lower IGHV somatic hypermutation, and a lower proliferative history. These findings may suggest that B-PLL epitype 1 derive from post-GC B cells that underwent a moderate exposure to the (epi)mutagenic environment of the GC whereas epitype 2 shows a stronger GC imprint (ie, various rounds of recirculation between light and dark zone). Additionally, epitype 1 was the only of the 2 that presented a de novo specific hypomethylation signature enriched for BHLHA15/MIST1, a TF also overexpressed in this epitype and previously reported to be related to lower overall survival in cervical cancer.34 At the genetic level, epitype 1 was enriched in 1q gains as well as MYC and SF3B1 mutations, and showed an upregulation of genes related to MYC pathways. MYC is a bona fide poor prognostic factor in cancer,35 whereas SF3B1 is a known unfavorable marker in CLL36 associated with the intermediate epitype12,22 and stereotyped subset no. 2.22,37 All these molecular features of epitype 1 may account for the poorer clinical outcome of this epitype as compared with epitype 2.
In conclusion, our study provides fresh insights into the molecular features of B-PLL carefully diagnosed based on the ICC/WHO-HAEM4R criteria. We did not find epigenetic evidence that B-PLLs, or a fraction of cases, resemble MCL, CLL, or SMZL. Instead, it seems that B-PLL is a de novo disease independent from the other 3 entities. We are aware that the relatively small sample size imposes a careful interpretation of the data, and in particular the presence of 2 epitypes, needs to be validated in the future. Moreover, a comprehensive analysis of the B-PLL methylome in the context of a broader range of B-cell lymphoproliferative disorders is needed. We expect that the data provided by this study will significantly contribute to the current discussion on B-PLL, with the hope that an international consensus will be reached.
Acknowledgments
This work was partially developed at the Centro Esther Koplowitz (Barcelona, Spain).
This research was funded by the Accelerator award Cancer Research UK/Italian Association for Cancer Research/Spanish Association Against Cancer joint funder partnership (J.I.M.-S.), Generalitat de Catalunya Suport Grups de Recerca AGAUR (2021-SGR-1343 [J.I.M.-S.], 2021-SGR-01172 [E.C.], and 2021-SGR-01293 [S.B.]), la Caixa Foundation (CLLEvolution, LCF/PR/HR17/52150017 [HR17-00221LCF] and CLLSYSTEMS - LCF/PR/HR22/52420015 [HR22-00172] Health Research 2017 and 2022 Programs, [E.C.]), Force Hemato (grant reference: 03-2022), French Innovative Leukemia Organization group, Association des Cytogénéticiens de Langue Française (grant 2022) and SIRIC-CURAMUS (Cancer United research Associating Medecine, University and Society; grant reference: INCa-DGOS-INSERM_12560, INCa-DGOS-INSERM-ITMO Cancer_18010, and INCa-DGOS-INSERM-ITMO Cancer_18002). M.D.-F. is supported by a postdoctoral grant from the Spanish Association Against Cancer. J.C.S. was supported by research grants from the Kay Kendall Leukemia Fund, and Cancer Research UK (Early Cancer Research Initiative Network on MBL-M3 Accelerator award C42023/A29370).
Authorship
Contribution: S.C. analyzed and interpreted the data, and wrote the manuscript; E. Chapiro provided samples and data, and assisted in data interpretation; F.N., A.M.-F., M.A., M.R., D.R.-W., O.B., S.A.S., H.P., R.W., C.C.O., J.C.S., and T.Z. provided essential data and tools; S.B., E. Campo, and E.M. provided samples and assisted in data interpretation; and M.D.-F., F.N.-K., and J.I.M.-S. jointly designed the research, supervised the study, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Florence Nguyen-Khac, Hôpital Pitié-Salpêtrière, Service d’Hématologie Biologique, 83 Boulevard de l’Hôpital, 75013 Paris, France; email: florence.nguyen-khac@aphp.fr; and José I. Martín-Subero, Institut d’Investigacions Biomèdiques August Pi i Sunyer, Rosselló 149-153, Barcelona 08036, Spain; email: imartins@recerca.clinic.cat.
References
Author notes
M.D.-F., F.N.-K., and J.I.M.-S. contributed equally to this work as joint senior authors.
DNA methylation data from B-PLL samples have been deposited at the Gene Expression Omnibus under the accession number GSE279602.
The full-text version of this article contains a data supplement.