TO THE EDITOR:
Mutations in the HBB gene cause β-thalassemia by inhibiting the production of the β-globin subunit of adult hemoglobin (HbA, α2β2).1 Consequently, excess free α-globin forms cytotoxic precipitates that cause maturation delay and apoptosis of erythroid precursors (ineffective erythropoiesis [IE]) and shortened lifespan of circulating red blood cells (RBC) (hemolysis). The severity of β-thalassemia varies according to the causal HBB mutation and modifier alleles that influence the ratio of α-globin to β-like globins (mainly β- and γ-), which form HbA or fetal hemoglobin (α2γ2), respectively. Patients with transfusion-dependent (TD) β-thalassemia, also called β-thalassemia major, express little or no β-like globin and require regular RBC transfusions. Patients with non–transfusion-dependent (NTD) β-thalassemia, also called β-thalassemia intermedia, express residual β-globin and/or γ-globin and require transfusions sporadically. Both TD β-thalassemia and NTD β-thalassemia are common disorders that cause substantial morbidity and premature mortality.
Mouse models for β-thalassemia are helpful for elucidating disease pathophysiology and developing new therapies.2 These models were created by generating targeted mutations in the mouse β-globin (Hbb) genes. While humans have a single HBB gene on each chromosome, mice express β-globin from 2 tandem, ancestrally duplicated Hbb genes.3-5 Common inbred strains of mice used for laboratory research harbor one of 2 different Hbb haplotypes, termed single and diffuse (supplemental Figure 1A). The Hbb single haplotype, normally found in C57BL/6 mice, contains Hbb-bs and Hbb-bt, which encode identical β-globin proteins. The diffuse haplotype, found in 129 and BALB/c strains, contains Hbb-b1 and Hbb-b2, which encode proteins that differ from each other and from β-globin single (supplemental Figure 1B-C). The Hbb-b1 and Hbb-b2 genes are also referred to as Hbb-major (βmaj) and Hbb-minor (βmin) because they generate ∼80% and 20% of RBC β-globin protein, respectively.6
The commonly studied Hbbth3/+ mouse harbors a heterozygous 20 kb deletion that eliminates both Hbb genes in cis (supplemental Figure 1A), resulting in β-globin haploinsufficiency with a phenotype that resembles human NTD β-thalassemia.7 Commercially available Hbbth3/+ mice are hybrids of C57BL/6 and 129 strains, which harbor Hbb single and diffuse haplotypes, respectively (Jackson Laboratories, catalog no. 003253; B6;129-Hbb-b1tm1Unc Hbb-b2tm1Unc/J). Thus, this β-thalassemic strain is hemizygous for a wild-type Hbb haplotype, which can be single or diffuse. Considering that the severity of α-thalassemia and normal RBC traits in mice are both influenced by these Hbb haplotypes,8,9 we tested their effects on β-thalassemic phenotypes by generating C57BL/6 Hbbth3/+ strains that were congenic for hemizygous Hbb single or diffuse haplotypes (supplemental Methods; supplemental Figure 1A).
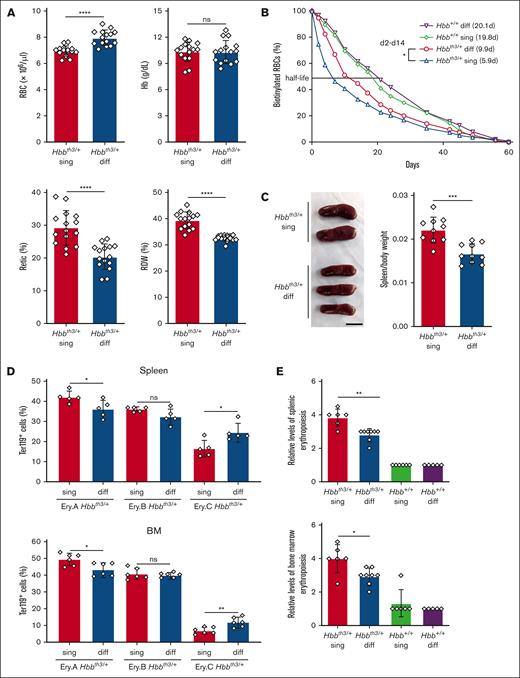
Compared with Hbbth3/+ single mice, Hbbth3/+ diffuse mice exhibited improved RBC indices, including a 15% increase in RBC count (P < .0001), a 14% increase in hematocrit, a 30% reduction in reticulocyte count (P < .0001) and a 17% reduction in the red cell distribution width (P < .0001) (Figure 1A; supplemental Table 1). In contrast, these indices were not significantly different between wild-type (Hbb+/+) congenic C57BL/6 mice with Hbb single or diffuse haplotypes (supplemental Table 1). Biotin-labeling studies revealed circulating RBC half-lives of 5.9 days for Hbbth3/+ single mice and 9.9 days for Hbbth3/+ diffuse mice, compared with ∼20 days for wild-type Hbb+/+ mice that were homozygous for either haplotype (Figure 1B). Compared with Hbbth3/+ single mice, Hbbth3/+ diffuse mice exhibited reduced spleen-to-body weight ratios (Figure 1C), improved maturation of erythroid precursors (Figure 1D; supplemental Figure 2A) and reduced erythroid hyperplasia in spleen and bone marrow (Figure 1E; supplemental Figure 2B-C; supplemental Table 2). Thus, the Hbb diffuse haplotype is associated with reductions in hemolysis and IE in Hbbth3/+ mice compared with Hbbth3/+ mice with Hbb single haplotype.
Reduced hemolysis and IE in Hbbth3/+ mice with the Hbb diffuse haplotype. (A) Erythroid indices (y-axis) according to genotype (x-axis) in 8-week-old mice. Hbbth3/+ single (sing), n = 15; Hbbth3/+ diffuse (diff), n = 15. (B) Circulating RBC survival determined after biotin labeling. Graph shows mean values ± SD for 7 mice each with the indicated genotypes. Calculated RBC half-life in days are shown in parentheses. Differences between Hbbth3/+ sing and Hbbth3/+ diff mice were significant at all time points between days 2 and 14, with a false discovery rate of 0.05 by the Benjamini and Hochberg method. (C) Representative spleens from mice of the indicated genotypes. The scale bar represents 1 cm. Graph on right shows spleen-to-body weight ratios. n = 10 mice for each genotype. (D) Developmental stage distribution of erythroblasts in spleen (left panel) and bone marrow (right panel) determined by flow cytometry for surface antigens and forward scatter (FSC). Ery.A (Ter119highCD71highFSChigh), Ery.B (Ter119highCD71highFSClow) and Ery.C (Ter119highCD71lowFSClow) represent increasingly mature erythroblast populations.10 Representative flow cytometry plots are shown in supplemental Figure 2A. Hbbth3/+ sing, n = 5 or 6; Hbbth3/+ diff, n = 5 or 6. (E) Semiquantitative evaluation of splenic and bone marrow erythropoiesis in H&E-stained sections (supplemental Figure 2B and C) assigned by a blinded, board–certified veterinary pathologist (H.S.). Graphs show the levels of erythroid precursors on a 5-point scale (arbitrary units), with 0 being the lowest. n = 6-8 mice for each genotype. All graphs show data as mean value ± SD. Bar graphs were analyzed by 2-sample t test. ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001. Hb, hemoglobin; H&E, hematoxylin and eosin; ns, not significant; Retic, reticulocyte; RDW, red cell distribution width; SD, standard deviation.
Reduced hemolysis and IE in Hbbth3/+ mice with the Hbb diffuse haplotype. (A) Erythroid indices (y-axis) according to genotype (x-axis) in 8-week-old mice. Hbbth3/+ single (sing), n = 15; Hbbth3/+ diffuse (diff), n = 15. (B) Circulating RBC survival determined after biotin labeling. Graph shows mean values ± SD for 7 mice each with the indicated genotypes. Calculated RBC half-life in days are shown in parentheses. Differences between Hbbth3/+ sing and Hbbth3/+ diff mice were significant at all time points between days 2 and 14, with a false discovery rate of 0.05 by the Benjamini and Hochberg method. (C) Representative spleens from mice of the indicated genotypes. The scale bar represents 1 cm. Graph on right shows spleen-to-body weight ratios. n = 10 mice for each genotype. (D) Developmental stage distribution of erythroblasts in spleen (left panel) and bone marrow (right panel) determined by flow cytometry for surface antigens and forward scatter (FSC). Ery.A (Ter119highCD71highFSChigh), Ery.B (Ter119highCD71highFSClow) and Ery.C (Ter119highCD71lowFSClow) represent increasingly mature erythroblast populations.10 Representative flow cytometry plots are shown in supplemental Figure 2A. Hbbth3/+ sing, n = 5 or 6; Hbbth3/+ diff, n = 5 or 6. (E) Semiquantitative evaluation of splenic and bone marrow erythropoiesis in H&E-stained sections (supplemental Figure 2B and C) assigned by a blinded, board–certified veterinary pathologist (H.S.). Graphs show the levels of erythroid precursors on a 5-point scale (arbitrary units), with 0 being the lowest. n = 6-8 mice for each genotype. All graphs show data as mean value ± SD. Bar graphs were analyzed by 2-sample t test. ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001. Hb, hemoglobin; H&E, hematoxylin and eosin; ns, not significant; Retic, reticulocyte; RDW, red cell distribution width; SD, standard deviation.
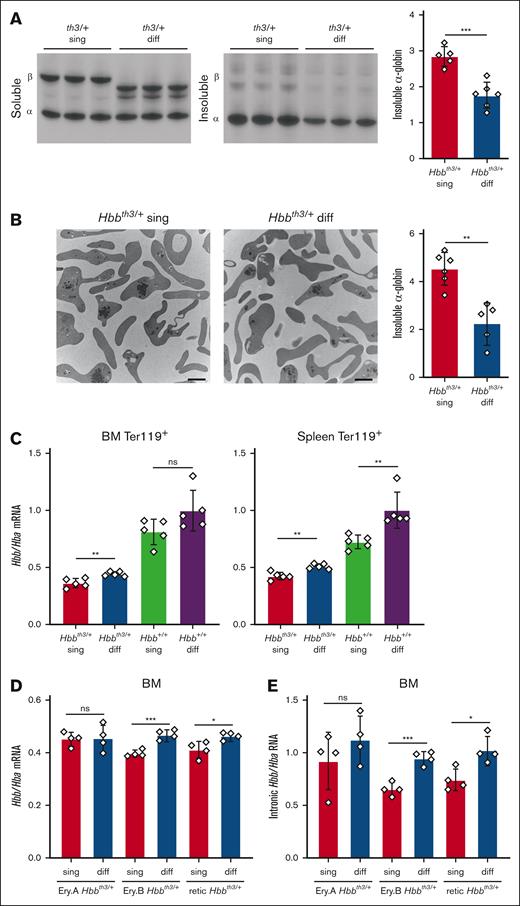
Biochemical fractionation followed by triton acid urea gel electrophoresis revealed a 60% increase of insoluble α-globin in RBC lysates from Hbbth3/+ single mice compared with Hbbth3/+ diffuse mice (P < .001) (Figure 2A). Similarly, transmission electron microscopy with quantitative image analysis revealed a 100% increase of α-globin precipitates in reticulocytes from Hbbth3/+ single mice compared with Hbbth3/+ diffuse mice (P < .01) (Figure 2B). These findings indicate that the β-globin to α-globin imbalance in Hbbth3/+ mice is increased with the Hbb single haplotype compared with the Hbb diffuse haplotype. This imbalance could result from reduced stability of the β-globin single protein or a reduction in its expression compared with β-globin diffuse.
Reduced free α-globin in Hbbth3/+ mice with the Hbb diffuse haplotype. (A) Soluble and insoluble globins in RBCs determined by biochemical fractionation and triton acid urea (TAU) gel electrophoresis. Representative TAU gel images are shown in the left panels and the results of multiple experiments are summarized in the graph on the right. The y-axis represents relative staining intensities of insoluble α-globin on TAU gels, measured by automated image analysis and expressed in arbitrary units. Hbbth3/+ sing, n = 5; Hbbth3/+ diff, n = 6. (B) Transmission electron microscopy of flow cytometry–purified reticulocytes showing electron-dense, α-globin inclusions. Representative micrographs are shown in the 2 left panels. The scale bars represent 2 μm. The graph on the right shows the results of quantitative automated image analysis of multiple mice with relative levels of α-globin inclusions indicated on the y-axis using an arbitrary scale. Hbbth3/+ sing, n = 6; Hbbth3/+ diff, n = 5. (C) Hbb/Hba mRNA ratios in Ter119+ erythroblasts from bone marrow (BM, left) or spleen (right) of mice with the indicated genotypes, determined by RNA-sequencing analysis. Five mice were assessed per genotype. (D) Hbb/Hba mRNA ratios in EryA and EryB erythroblasts isolated from BM and reticulocytes (retic) isolated from peripheral blood, determined by RNA-sequencing analysis. Four mice were assessed per genotype. (E) Intronic Hbb/Hba RNA ratios in the samples described for panel D. All graphs show data as mean value ± SD. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ns, not significant; SD, standard deviation.
Reduced free α-globin in Hbbth3/+ mice with the Hbb diffuse haplotype. (A) Soluble and insoluble globins in RBCs determined by biochemical fractionation and triton acid urea (TAU) gel electrophoresis. Representative TAU gel images are shown in the left panels and the results of multiple experiments are summarized in the graph on the right. The y-axis represents relative staining intensities of insoluble α-globin on TAU gels, measured by automated image analysis and expressed in arbitrary units. Hbbth3/+ sing, n = 5; Hbbth3/+ diff, n = 6. (B) Transmission electron microscopy of flow cytometry–purified reticulocytes showing electron-dense, α-globin inclusions. Representative micrographs are shown in the 2 left panels. The scale bars represent 2 μm. The graph on the right shows the results of quantitative automated image analysis of multiple mice with relative levels of α-globin inclusions indicated on the y-axis using an arbitrary scale. Hbbth3/+ sing, n = 6; Hbbth3/+ diff, n = 5. (C) Hbb/Hba mRNA ratios in Ter119+ erythroblasts from bone marrow (BM, left) or spleen (right) of mice with the indicated genotypes, determined by RNA-sequencing analysis. Five mice were assessed per genotype. (D) Hbb/Hba mRNA ratios in EryA and EryB erythroblasts isolated from BM and reticulocytes (retic) isolated from peripheral blood, determined by RNA-sequencing analysis. Four mice were assessed per genotype. (E) Intronic Hbb/Hba RNA ratios in the samples described for panel D. All graphs show data as mean value ± SD. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ns, not significant; SD, standard deviation.
All mouse β-globins contain a conserved cysteine at position 94 (supplemental Figure 1B), similar to human β-globin protein. Diffuse β-globins also contain a reactive cysteine at 14, which may protect against hemoglobin oxidation and denaturation under oxidizing conditions, such as β-thalassemia.11 In addition, the Hbb genes are expressed at slightly higher levels in mouse strains harboring the diffuse haplotype compared to those with Hbb single.9 In agreement, RNA-sequencing analysis revealed greater Hbb/Hba messenger RNA (mRNA) ratios in Ter119+ erythroblasts from the spleen or bone marrow of wild-type (Hbb+/+) and β-thalassemic (Hbbth3/+) mice with the diffuse Hbb haplotype, as compared with those with Hbb single (Figure 2C). Similarly, Hbb/Hba mRNA ratios were higher in flow cytometry–purified, developmental stage–matched EryB (Ter119+/CD71high/FSClow) erythroblasts and circulating reticulocytes from Hbbth3/+ diffuse mice compared with those from Hbbth3/+ single mice (Figure 2D). We detected similar differences in the ratios of intron-containing RNAs (Figure 2E), suggesting that Hbb single genes are transcribed at lower rates than Hbb diffuse genes.
In summary, we generated and compared C57BL/6 β-thalassemic Hbbth3/+ mice that were congenic for hemizygous Hbb single or diffuse haplotypes and showed that these are genetic modifiers of β-thalassemia. The Hbb single haplotype, normally present in the C57BL/6 strain, is associated with more severe β-thalassemia, at least in part due to its reduced mRNA expression, resulting in increased levels of free α-globin. In addition, cysteine at position 14 of diffuse β-globins, which is not present in single β-globin (supplemental Figure 1B), may confer protection against the denaturing effects of β-thalassemia–associated oxidant stress.11 Although humans do not harbor homologs for mouse Hbb single and diffuse haplotypes, our findings are consistent with the general concept that genetic variants, both linked and unlinked to the HBB locus, modify the severity of β-thalassemia.12,13
From a practical perspective, commercially available Hbbth3/+ β-thalassemic mice are of mixed C57BL/6 and 129 backgrounds and therefore, hemizygous for either the Hbb single or diffuse haplotype, with reduced hemolysis and IE occurring with the latter. Because this variability is likely to complicate the interpretation of experimental perturbations, most studies will be improved by utilizing Hbbth3/+ mice with the same Hbb haplotype. Either haplotype may have distinct advantages or disadvantages, depending on experimental design and objectives. For example, Hbb single is associated with more severe β-thalassemia, which may enhance the sensitivity for testing new drugs or other therapeutic interventions in adult animals. On the other hand, the single haplotype is more likely to reduce the perinatal survival of Hbbth3/+ mice that harbor deleterious modifier alleles introduced during genetic screens (eg, Lechauve et al14). Regardless of the Hbb haplotype present, it is most important to control for this variable in studies utilizing β-thalassemic mice.
Acknowledgments: The authors are grateful to Joseph Emmons and staff (St. Jude Veterinary Pathology Core), Camenzind Robinson and staff (St. Jude Cellular Imaging Shared Resources), Geoffrey Neale and Sanchit Trivedi (St. Jude Hartwell Center), and Richard Ashmun and staff (St. Jude Flow Cytometry Core). The authors thank Keith A. Laycock (St. Jude Department of Scientific Editing) for scientific editing of the manuscript. This work was funded by National Heart, Lung, and Blood Institute grant R01HL165798 (M.J.W.) and American Lebanese Syrian Associated Charities.
Contribution: C.L., J.K., E.K., and M.J.W. conceived the project and designed and interpreted experiments; C.L., J.K., A.G.F., T.M., X.Q., and H.-M.H. performed experiments; L.P. analyzed RNA-sequencing data; M.W.D., K.M., and R.T. assisted with animal breeding; H.S. performed histopathological analysis; and C.L., J.K., H.-M.H., and M.J.W. wrote the manuscript.
Conflict-of-interest disclosure: M.J.W. is a consultant for Novartis, Vertex Therapeutics, and bluebird bio. The remaining authors declare no competing financial interests.
The current affiliation for C.L. is Sanofi, Waltham, MA.
Correspondence: Christophe Lechauve, Genomic Medicine Unit, Rare Blood Disorders, Sanofi, 225 2nd Ave, Waltham, MA 02451; email: christophe.lechauve@sanofi.com; and Mitchell J. Weiss, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, MS 355, Memphis, TN 38105; email: mitch.weiss@stjude.org.
References
Author notes
RNA-sequencing data are available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) through accession numbers GSE260900 and GSE260901. Other original data that are not publicly accessible, are available upon reasonable request from the corresponding author, Mitchell Weiss (mitch.weiss@stjude.org).
The full-text version of this article contains a data supplement.