Key Points
In murine AML models, Herc1 modulates nucleoside analog resistance and levels of deoxycitidine kinase, their rate-limiting metabolic enzyme.
HERC1 is highly expressed in human AML and its elevated expression is associated with worse prognosis.
Visual Abstract
For several decades, induction therapy with nucleoside analogs, in particular cytarabine (Ara-C) and, to a lesser extent, fludarabine, has been the standard of care for patients diagnosed with acute myeloid leukemia (AML). However, the antitumor efficacy of nucleoside analogs is often limited by intrinsic and acquired drug resistance, thereby leading to poor therapeutic response and suboptimal clinical outcomes. In this study, we used genome-wide CRISPR-based pharmacogenomic screening to map the genetic factors that modulate the response to nucleoside analogs in AML and identified the E3 ubiquitin ligase, Herc1, as a key modulator of Ara-C response in mouse AML models driven by the KMT2A/MLLT3 fusion or by the constitutive coexpression of Hoxa9 and Meis1, both in vitro and in vivo. Loss of HERC1 enhanced nucleoside analog–induced cell death in both murine and human AML cell lines by compromising cell cycle progression. In-depth proteomic analysis and subsequent validation identified deoxycytidine kinase as a novel target of Herc1 in both mouse AML models. We observed that HERC1 is overexpressed in AML when compared with other cancer types and that higher HERC1 expression was associated with shorter overall survival in patients with AML in the The Cancer Gene Atlas program (TCGA) and BEAT-AML cohorts. Collectively, this study highlights the importance of HERC1 in the response of AML cells to nucleoside analogs, thereby establishing this E3 ubiquitin ligase as a novel predictive biomarker and potential therapeutic target for the treatment of AML.
Introduction
For patients affected by acute myeloid leukemia (AML), particularly those younger than 65 years of age, backbone induction chemotherapy consists of a combination of a nucleoside analog, such as the deoxycytidine analog cytarabine (Ara-C) or the deoxyadenosine analog fludarabine (Flu), and an anthracycline (eg, idarubicin or daunorubicin).1 Unfortunately, half of adult patients relapse after standard-of-care chemotherapy within 3 years,1-4 and the overall 5-year survival rate in adults is around 30%,5 thereby highlighting the critical need to better understand the determinants of chemoresponsiveness in AML cells.
Nucleoside analogs exert their cytotoxic effects (1) by disrupting normal DNA synthesis through direct incorporation into extending DNA strands and (2) by destabilizing the optimal intracellular pool of deoxyribonucleotides (dNTPs), thereby leading to the stalling of replication forks and the formation of double-stranded DNA breaks.6,7 Intracellularly, nucleoside analogs undergo sequential phosphorylation steps initiated by deoxycytidine kinase (DCK) for conversion into their active triphosphate forms.7,8 Conversely, the sterile alpha motif and histidine–aspartate (HD) domain–containing protein 1 (SAMHD1) hydrolyzes dNTPs into their constituent triphosphate and 2ʹ-deoxynucleoside, thereby acting as a sensor for the maintenance of dNTP pools. Previous studies have shown that DCK and SAMHD1 activities are important determinants of the sensitivity to nucleoside analogs in hematological malignancies.9-11
To comprehensively map the genetic factors that influence the response of AML cells to nucleoside analogs, we performed 2 complementary in vitro genome-wide CRISPR-Cas9 screens using syngeneic mouse models of AML that are driven by the oncogenic fusion protein KMT2A/MLLT3 or overexpression of HoxA9 and Meis1. Aside from the expected targets Dck, which emerged among the top sensitizing genes, and Samhd1, which scored among the top genes that provide resistance to Ara-C, we identified the E3 ubiquitin (Ub) ligase, homologous to the E6AP carboxyl terminus (HECT), and regulator of chromosome condensation 1 (RCC1)-like domain–containing protein 1 (Herc1) as a modulator of Ara-C response in vitro and in vivo. HERC1 is aberrantly expressed in myeloid-related disorders, such as AML and chronic myeloid leukemia,12 and a role for HERC1 in the response to UV-induced apoptosis has been described previously.13 However, the role of HERC1 in the response of AML to Ara-C so far has not been described. Altogether, this study demonstrated that HERC1 is a modulator of the cytotoxic response to Ara-C in AML cells and highlights the modulation of DCK protein stability as a mechanism to control the cytotoxic effect of nucleoside analogs.
Methods
In vitro CRISPR screening
The KMT2A/MLLT3 (also known as MLL/AF9, herein abbreviated as MA) and HoxA9/Meis1 (HM) mouse leukemia models, generated by retroviral transduction of hematopoietic progenitor cells from a Rosa26-CAG-Cas9/GFP knock-in mouse (Stock 024858, Jackson laboratories), have been described previously.14 Cell lines were propagated in RPMI-1640 media supplemented with recombinant murine stem cell factor (mSCF) produced by a Chinese hamster ovary cell line, 5 ng/mL murine interleukin 3 (mIL3) (BioLegend), 10% fetal bovine serum (Wisent), and penicillin/streptomycin (Lonza). For the genome-wide CRISPR/Cas9 screens, 4.8 × 107 cells from each MA-Cas9 and HM-Cas9 line were spinfected with the GeCKO v2 library (Addgene, catalog no. 1000000052) in the presence of 8 μg/mL Polybrene (Sigma-Aldrich, catalog no. TR-1003) using freshly prepared virus at a multiplicity of infection of ∼0.25, as previously described,14 followed by 72 hours of selection with 10 μg/mL of puromycin (BioShop). Cells were passaged daily, and cell number was maintained at 5 × 105 cells per mL. Ara-C or vehicle treatment was started at day 3 after puromycin selection with sublethal doses of Ara-C (MA-Cas9 IC50 [40 nM] and HM-Cas9 [80 nM]) for 5 days. Before and after Ara-C exposure, cells were collected for genomic DNA extraction using phenol/chloroform (Thermo Fisher), then single guide RNA (sgRNA) libraries were amplified using a 1-step polymerase chain reaction (PCR) protocol.58 Sequencing was performed using an Illumina HiSeq 2000. For every biologic and technical replicate, the read count files were generated from the Fastq files using the count function of the MAGeCK algorithm.15 To determine the technical performance of the screen, technical replicates were correlated using Pearson and Spearman correlations. The gene scores that represented depletion or enrichment (NormZ scores) were calculated using the drugZ algorithm.16 Pathway enrichment analysis was performed using STRING database.17
All animal experiments were approved by the institutional review board of McGill University.
Validation of CRISPR screen using single sgRNAs
sgRNA sequences were designed using the Broad Institute Genetic Perturbation Platform (supplemental Table 8) and subcloned into pKLV2-U6gRNA5(BbsI)-PGK-puro2AmCherry-W or puro2ABFP-W vectors (gift from Kosuke Yusa, Addgene; catalog no. 67974 and 67977) as previously described.18 Virus particles produced from transfected HEK293T cells were used to spinfect AML cells. Two days after spinfection, cells were selected using 10 μg /mL puromycin for 5 days. mCherry+ single cells were sorted into 96-well plates, and genotyping PCR was performed using primers targeting the target sites (supplemental Table 9).
Multicolor competition assays
Multicolor competition assays were performed to verify the impact of gene knockouts on fitness defects in the presence of Ara-C treatment as previously described.19 In these assays, the target cells expressed mCherry, whereas control cell lines expressed EGFP and/or BFP. Cells were treated with Ara-C (50 nM) for a minimum of 14 days and were assessed using flow cytometry every 4 days on the BD LSRFortessa instrument.
Cell viability assays
A total of 10 000 cells were seeded in 96-well plates and subjected to serial dilutions of Ara-C, gemcitabine (Gem), Flu, or doxorubicin (all from Selleckchem) for 72 hours before performing a CellTiter-Glo (CTG) assay according to manufacturer’s instructions. Luminescence was measured using the FLUOstar OPTIMA bioluminescent reader (BMG Labtech).
Apoptosis and cell cycle profiling
A total of 150 000 cells were seeded in 6-well plates and treated with Ara-C at concentrations of 200 nM (MA), 400 nM (HM), and 1 μg (U937) for 24 to 48 hours. Cells were then washed and stained with APC annexin V (BioLegend, catalog no. 64094) and DAPI (4′,6-diamidino-2-phenylindole) for 15 minutes. For intracellular caspase-3 staining, cells were treated with BD Cytofix/Cytoperm buffer, according to the manufacturer’s instructions, and then stained with caspase-3 AF647 antibody (BD Bioscience) for 30 minutes and washed with BD Cytoperm buffer. For cell cycle analysis, 1 × 106 MA cells were treated with 200 nM Ara-C or an equivalent volume of water diluted in culture media. Cells were then permeabilized with 70% ethanol, washed twice with phosphate-buffered saline and stained in 2× propidium iodide buffer (40 μg propidium iodide, 0.4 mg RNase, and 0.4% Triton X-100) for 30 minutes at 37°C. The samples were analyzed using the BDFortessa instrument.
RNA sequencing (RNA-seq)
The isolation of total RNA from Herc1 knockout (MA clone number 2) and control cells (MA sgCtrl) was carried out using a combination of TRIzol (Invitrogen) and the RNeasy Mini Kit (Qiagen, catalog no. 74106). Complementary DNA (cDNA) libraries were prepared using the KAPA RNA HyperPrep Ribo Erase Kit and were sequenced using the Illumina Nextseq500 platform. Sequences were trimmed using Trimmomatic version 0.35.20 Alignment to the reference mouse genome (GRCm38) was performed using STAR v2.7.1a,21 and gene annotation from GENCODE vM25, based on Ensembl 100, was used to provide read counts and transcripts per million values. In addition, RSEM22 was employed to compute normalized gene and transcript-level expression and provided the fragments per kilobase million and transcripts per million values for the stranded RNA libraries. DESeq2 version 1.30.1 was then employed to normalize read counts.23
TMT-labeling and mass spectrometry (MS)
Herc1 knockout (MA clone number 2) and control cells (MA sgCtrl) were lysed using RIPA buffer (50 mM tris-hydrochloride pH 7.6, 150 mM sodium chloride, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate), supplemented with the Mini Protease Inhibitor Cocktail (Roche, catalog no. 11836153001), for 30 minutes on ice, and samples were then sonicated for 5 seconds at 30% amplitude for 3 cycles. Samples were treated with TMT-16plex reagents (ThermoFisher Scientific) according to the manufacturer’s instructions. Labeled peptides were fractionated using Pierce High pH Reversed-Phase Peptide Fractionation Kit into 8 fractions. Each fraction was re-solubilized in 0.1% aqueous formic acid and 2 μg of each was loaded onto an Orbitrap Fusion instrument (ThermoFisher Scientific) that was operated in DDA-MS3 mode for 3 hours.24 To translate .raw files into protein identifications and TMT reporter ion intensities, Proteome Discoverer 2.2 (ThermoFisher Scientific) was used with the built-in TMT Reporter ion quantification workflows. The default settings were applied with trypsin as the enzyme specificity. Spectra were matched against the mouse protein FASTA database obtained from UniProt (2022). Dynamic modifications were set as oxidation (M) and acetylation on protein N-termini. Cysteine carbamidomethyl was set as a static modification, together with the TMT tag on both peptide N-termini and K residues. All results were filtered to a 1% false discovery rate.
Western blotting
Cells were washed using cold phosphate-buffered saline and lysed using RIPA buffer (25 mM tris-hydrochloride pH 7.6 [BioShop], 150 mM sodium chloride [BioShop], 1% NP-40 Surfact-Amps Detergent Solution [ThermoFisher Scientific], 1% sodium deoxycholate [ThermoFisher Scientific] and 0.1% sodium dodecyl sulfate BioShop]) containing a protease inhibitor cocktail (Thermo Scientific). After 30 minutes on ice, samples were sonicated at 30% for 5 seconds for 3 rounds. The protein concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Scientific). Samples were reduced using 4× Laemmli sample buffer and β-mercaptoethanol, followed by heating at 70°C for 10 minutes. A total of 20 mg of whole cell extracts was fractionated using NuPAGE 4% to 12% Bis-Tris mini protein gels (Invitrogen) and then transferred to a polyvinylidene difluoride (Bio-Rad) membrane. The following antibodies were used in this study: dCK (ThermoFisher, catalog no. PA5-27787), α-tubulin (Cell Signaling, 11H10, catalog no. 21255,), vinculin (Cell Signaling, catalog no. 13901), and horseradish peroxidase-conjugated anti-rabbit (Cell Signaling, catalog no. 7074S).
RT-qPCR
RNA was extracted using the RNeasy kit (Qiagen) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). Reverse transcriptase quantitative PCR (RT-qPCR) was performed using the Luna Universal qPCR Master Mix with 2 μL of cDNA product diluted 1:10. The SYBR mode setting on the Applied Biosystems 7500 Fast instrument was selected and RT-qPCR was performed according to the Luna Universal qPCR Master Mix Protocol (NEB). The following primer sequences were used: Dck (5ʹ-CCTCTGAGGATTGGGAAGTGG-3ʹ/5ʹ- CACCGCTCTTCTGAGACGTT-3ʹ), Gapdh (5ʹ-GCACAGTCAAGGCCGAGAAT-3ʹ/5ʹ-GCCTTCTCCATGGTGGTGAA-3ʹ), B2m (5ʹ-ACGTAACACAGTTCCACCCG-3ʹ/5ʹ- CAGTCTCAGTGGGGGTGAAT-3ʹ).
In vivo experiments
All in vivo experiments were approved by the McGill University Animal Care Committee. Herc1 knockout MA and HM cells were mixed 1:1 with sgCtrl cells and 1 000 000 cells were transplanted into sublethally irradiated (4.5 Gy) C57BL/6 mice at 3 to 12 months of age. Peripheral blood was collected from the saphenous vein 14 to 21 days after transplantation and analyzed using flow cytometry (BD LSRFortessa) to confirm the presence of leukemic cells, after which mice were treated with 100 mg/kg of Ara-C for 5 constitutive days. Mice were euthanized using isoflurane/CO2 inhalation, followed by bone marrow extraction for analysis.
Statistical analysis
The data for experiments in which the half maximal inhibitory effect (IC50), apoptosis, cell cycle, and EdU incorporation were measured are presented as mean ± standard error of the mean with data from at least 3 independent experiments, as described in the legends of figures 2,3. All data sets were tested for normal distribution using the Shapiro-Wilk test. Statistical significance was determined using the tests indicated in the legends. All statistical analyses were performed using Prism v9 (GraphPad software).
Analysis of public data sets
For analysis of human AML survival, we used the online resources cBioPortal (https://www.cbioportal.org) for BEAT-AML RNA-seq analysis (556 patients); KMPlot (http://www.kmplot.com) for GSE1159, GSE121417, GSE37642, GSE6891, and GSE8970 microarray data; and BloodSpot (https://www.bloodspot.eu) for TCGA data (GSE13159; 172 patients) using the medians of gene expression as cutoffs for the analysis of overall survival. For the correlation analysis, we used the TCGA-LAML cohort (https://portal.gdc.cancer.gov/) and the Cancer Cell Line Encyclopedia (CCLE) data set (https://depmap.org/portal/, Public 23Q2). For the analysis of patients with AML (n = 177) and the cell line proteomics (n = 26 × 4 biologic replicates) data sets, we accessed the pride data sets PXD023201 and PXD022894. For this data set, access to RNA-seq (n = 136) data were also available in EGAD00001008484.
Results
CRISPR screening identified Herc1 as a modulator of Ara-C in AML cells
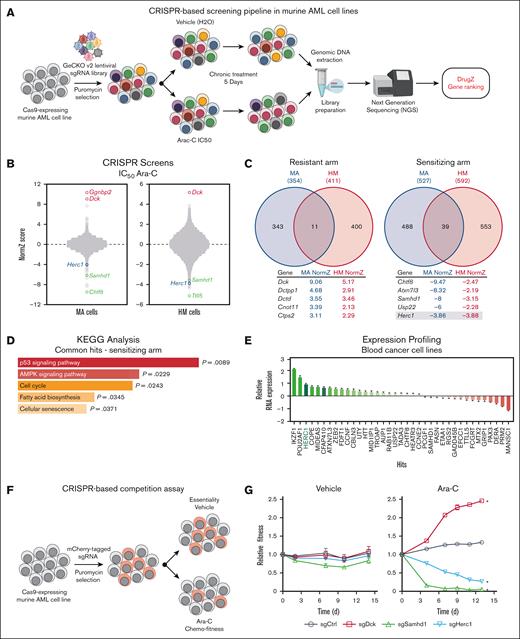
To better understand the factors that modulate Ara-C response in AML cells, we employed 2 different Cas9-transgenic murine AML mouse lines that recapitulate the main clinical features observed in AML patients in vivo,25,26 namely (1) the MA cell line in which overexpression of the fusion protein KMT2A/MLLT3 (MLL/AF9) in sorted murine multipotent progenitors drives AML development26 and (2) the HM cell line in which the transcription factors Hoxa9 and Meis1 are overexpressed in murine lineage-negative progenitors.27 We performed CRISPR-based genome-wide screening in both murine AML cell lines by transducing them with the GeCKO v2 sgRNA library28 (Figure 1A). Both the MA and HM transduced cell lines were subsequently treated with either the Ara-C IC50 or with vehicle (H2O) for 5 days before being processed for next-generation sequencing analysis. The DrugZ algorithm was used to monitor the relative abundance of a given sgRNA and to identify genes whose knockout confer either resistance (red) or sensitivity (green) to Ara-C (−2 < NormZ score < 2).16 In both cell lines, the deoxycytidine kinase Dck emerged as one of the top resistant genes, whereas the dNTPase enzyme Samhd1 scored as one of the top sensitizers (Figure 1B; supplemental Table 1), validating our CRISPR-based screening approach.
CRISPR screening identifies Herc1 as a modulator of Ara-C response in murine AML cells. (A) Schematic of our CRISPR/Cas9–based screening pipeline developed in murine AML cells. (B) Representation of CRISPR/Cas9–based dropout screen performed in MA and HM cells in the presence of Ara-C (IC50). (C) Representation of overlapping genes that are providing sensitivity (NormZ score, <2) and resistance to Ara-C (>2). (D) Pathway enrichment analysis of sensitizing genes using the KEGG database. (E) Expression analysis of the 39 overlapping sensitizers in the hematopoietic and lymphoid tissues from the CCLE database (n = 188). (F) Schematic of CRISPR-based competition assay. (G) Competitive growth assay ± Ara-C (50 nM) or H2O (vehicle) in MA cells. Data are represented as the ratio of mCherry+ normalized to day 0 (t3 independent transductions). Significance was determined using 2-way analysis of variance (ANOVA), followed by a Dunnett test. ∗P ≤ .05. AMPK, adenosine monophosphate-activated protein kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes.
CRISPR screening identifies Herc1 as a modulator of Ara-C response in murine AML cells. (A) Schematic of our CRISPR/Cas9–based screening pipeline developed in murine AML cells. (B) Representation of CRISPR/Cas9–based dropout screen performed in MA and HM cells in the presence of Ara-C (IC50). (C) Representation of overlapping genes that are providing sensitivity (NormZ score, <2) and resistance to Ara-C (>2). (D) Pathway enrichment analysis of sensitizing genes using the KEGG database. (E) Expression analysis of the 39 overlapping sensitizers in the hematopoietic and lymphoid tissues from the CCLE database (n = 188). (F) Schematic of CRISPR-based competition assay. (G) Competitive growth assay ± Ara-C (50 nM) or H2O (vehicle) in MA cells. Data are represented as the ratio of mCherry+ normalized to day 0 (t3 independent transductions). Significance was determined using 2-way analysis of variance (ANOVA), followed by a Dunnett test. ∗P ≤ .05. AMPK, adenosine monophosphate-activated protein kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes.
To take into consideration the heterogeneity of the disease, we focused our attention on hits that were commonly identified in both the MA and HM cell lines (Figure 1C). Importantly, only 11 genes were providing resistance to Ara-C in both cell lines when knocked out (Figure 1C, left panel), whereas 39 genes were common sensitizers in the murine AML cells (Figure 1C, right panel). KEGG analysis of these common sensitizers identified p53 signaling (P = .0089), adenosine monophosphate-activated protein kinase signaling (P = .0229), and the cell cycle pathway (P = .0243) as the top enriched pathways (Figure 1D; supplemental Table 2). To gain further insight into these hits, we examined their expression profile in a panel of 188 cancer cell lines according to tissue of origin. The transcriptional regulators IKZF1, POU2AF1, and ZEB2, which have been proposed to play a key role in the pathogenesis of and therapeutic response in AML,29-31 were among the most overexpressed hits in human blood cancer cell lines (Figure 1E; supplemental Table 3). Of note, the E3 Ub ligase HERC1, which scored in the top 25 sensitizer genes in both CRISPR screens (MA: position 21, NormZ = −3.86; HM: position 6, NormZ = −3.88; Figure 1C; supplemental Table 1), emerged as the third most expressed hit in this panel of human blood cancer cell lines (Figure 1E; supplemental Table 3).
Using a well-established CRISPR-based competition assay32 in which Cas9-expressing MA cells were transduced with a vector that co-expressed both sgRNA and mCherry (Figure 1F), we monitored the relative abundance of mCherry+ cells in control (vehicle) and Ara-C–treated conditions (IC50) using flow cytometry. We incorporated sgRNAs in this experimental design that targeted well-established regulators of the Ara-C response (eg, Samhd1, Dck), alongside top hits from our CRISPR screens (eg, Chtf8, Herc1) and a nontargeting sgRNA as control. Although targeting Dck in MA cells provided resistance to Ara-C, knocking out Samhd1 hypersensitized MA cells to this chemotherapeutic agent (Figure 1G), thereby validating our competition assay. Loss of Chtf8 impaired the proliferation of MA cells in both the vehicle and Ara-C conditions (supplemental Figure 1A), suggesting that Chtf8 is essential for murine AML cells. In contrast, deletion of Herc1 impaired the growth of MA cells in the presence of Ara-C with no significant impact in the presence of the vehicle (Figure 1G), suggesting a potential specific role for this E3 Ub ligase in the modulation of the Ara-C response in vitro. Because Herc1 possesses a druggable enzymatic activity, we focused our attention on this hit in the rest of the investigations.
Loss of Herc1 increases the sensitivity of murine AML cell to nucleoside analogs in vitro
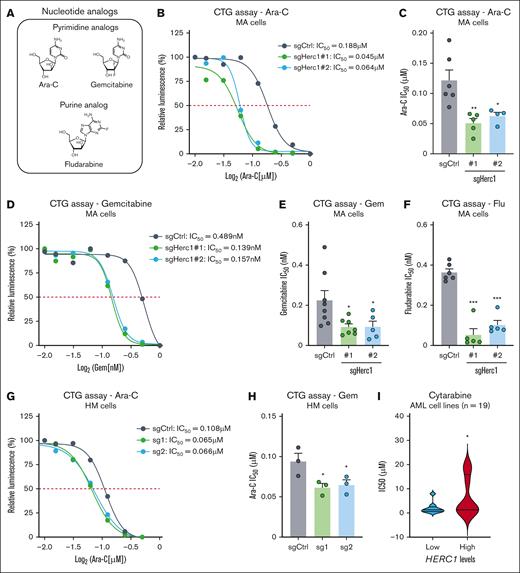
To further characterize the role of Herc1 in the modulation of nucleoside analogs (Figure 2A), we generated Herc1 knockout clones (1 and 2) from murine MA cells (supplemental Figure 1B-C), and we monitored their response to Ara-C using the CTG assay. We observed that loss of Herc1 led to an approximately twofold decrease in the IC50 of MA cells to Ara-C (supplemental Figure 2B-C). We extended our analysis to another pyrimidine analog, Gem (Figure 2A), that has been tested as salvage therapy for the treatment of patients with relapsed AML and that has shown minimal activity,33,34 and we noticed a similar trend in Herc1-depleted MA cells exposed to Gem when compared with control cells of an ∼2.4-fold decrease in their IC50 to Gem (Figure 2D-E). Similarly, Herc1 knockout MA cells were hypersensitized to the purine analog Flu (Figure 2A) with an approximately sevenfold and ∼3.5-fold decrease in the IC50 for Flu for clone number 1 and clone number 2, respectively (Figure 2F). Importantly, we did not observe any significant difference in the response of Herc1-depleted MA cells to doxorubicin when compared with control cells (supplemental Figure 1D), suggesting that Herc1 specifically modulates the response of MA cells to nucleoside analogs. Next, we extended our approach to the HM cell line in which we targeted Herc1 with 2 distinct sgRNAs (sg1 and sg2) (supplemental Figure 1B,E). In agreement with the results obtained in MA cells, targeting Herc1 hypersensitized HM cells to Ara-C when compared with the control cells, leading to an ∼1.5-fold decrease in the IC50 for Ara-C for both sgRNAs against Herc1 (Figure 2G-H). We extended our analysis to the CCLE, which contains gene expression data from 1296 different cancer cell lines and their pharmacologic profiles for 24 distinct anticancer drugs, thereby enabling predictive modeling of anticancer drug sensitivity.35 In line with our observations, we observed that high RNA expression of HERC1 correlated with significantly higher cytarabine IC50 in human AML cell lines (n = 19) (low HERC1 expressors: mean IC50 = 1.965 μM; high HERC1 expressors: mean IC50 = 6.665 μM; Figure 2I). Altogether, our data indicate that targeting Herc1 hypersensitizes murine AML cells to nucleoside analogs in vitro.
Targeting of Herc1 increases the sensitivity of murine AML cell to nucleoside analogs in vitro. (A) Representation of nucleoside analogs used for cell viability assays. (B) MA cells were seeded in quadruplicates into 96-well plates and treated with varying concentrations of Ara-C. The number of viable cells was measured after 72 hours using the CTG Luminescent Cell Viability Assay. One of the independent experiments is shown. (C) IC50 concentrations of Ara-C of 6 independent repeated experiments in MA with either sgHerc1 number 1 or sgHerc1 number 2 analyzed together or separate with sgCtrl. Significance was determined using 1-way ANOVA followed by Dunnett’s multiple comparisons test. ∗P ≤ .05. (D) MA cells were seeded in quadruplicates into 96-well plates and treated with varying concentrations of Gem. The number of viable cells was measured after 72 hours using the CTG Luminescent Cell Viability Assay. One of independent experiments is shown. (E) IC50 concentrations of Gem of 8 independent repeated experiments in MA with either sgHerc1 number 1 and sgHerc1 number 2 analyzed together or separate with sgCtrl. Significance was determined using one-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤ .05. (F) IC50 concentrations of Flu of 8 independent repeated experiments in MA with either sgHerc1 number 1 or sgHerc1number 2, analyzed together or separate with sgCtrl. Significance was determined using a 1-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤ .05. (G) HM cells were seeded in quadruplicates into 96-well plates and treated with varying concentrations of Ara-C. The number of viable cells was measured after 72 hours using the CTG Luminescent Cell Viability Assay. One of independent experiments is shown. (H) IC50 concentrations of Ara-C of 3 independent repeated experiments in HM. Significance was determined using a 1-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤ .05. (I) HERC1 expression was analyzed in a panel of human AML cell lines (n = 19) and linked to their respective cytarabine IC50. Cell lines were arbitrarily split into low (n = 9) and high (n = 10) expressers of HERC1. Significance was determined using an unpaired 1-tailed t test. ∗P ≤ .05.
Targeting of Herc1 increases the sensitivity of murine AML cell to nucleoside analogs in vitro. (A) Representation of nucleoside analogs used for cell viability assays. (B) MA cells were seeded in quadruplicates into 96-well plates and treated with varying concentrations of Ara-C. The number of viable cells was measured after 72 hours using the CTG Luminescent Cell Viability Assay. One of the independent experiments is shown. (C) IC50 concentrations of Ara-C of 6 independent repeated experiments in MA with either sgHerc1 number 1 or sgHerc1 number 2 analyzed together or separate with sgCtrl. Significance was determined using 1-way ANOVA followed by Dunnett’s multiple comparisons test. ∗P ≤ .05. (D) MA cells were seeded in quadruplicates into 96-well plates and treated with varying concentrations of Gem. The number of viable cells was measured after 72 hours using the CTG Luminescent Cell Viability Assay. One of independent experiments is shown. (E) IC50 concentrations of Gem of 8 independent repeated experiments in MA with either sgHerc1 number 1 and sgHerc1 number 2 analyzed together or separate with sgCtrl. Significance was determined using one-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤ .05. (F) IC50 concentrations of Flu of 8 independent repeated experiments in MA with either sgHerc1 number 1 or sgHerc1number 2, analyzed together or separate with sgCtrl. Significance was determined using a 1-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤ .05. (G) HM cells were seeded in quadruplicates into 96-well plates and treated with varying concentrations of Ara-C. The number of viable cells was measured after 72 hours using the CTG Luminescent Cell Viability Assay. One of independent experiments is shown. (H) IC50 concentrations of Ara-C of 3 independent repeated experiments in HM. Significance was determined using a 1-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤ .05. (I) HERC1 expression was analyzed in a panel of human AML cell lines (n = 19) and linked to their respective cytarabine IC50. Cell lines were arbitrarily split into low (n = 9) and high (n = 10) expressers of HERC1. Significance was determined using an unpaired 1-tailed t test. ∗P ≤ .05.
Targeting the E3 Ub ligase Herc1 exacerbates Ara-C-induced apoptosis in AML cells
We further investigated the contribution of Herc1 to the modulation of the Ara-C response by monitoring cell death using the presence of cells with fractional DNA content, designated the sub-G1 population, as proxy.36 This analysis revealed that targeting Herc1 led to a significant increase in the sub-G1 population following treatment of MA cells with Ara-C (200 nM; Figure 3A). It should be noted that we did not observe a significant impact on Herc1-depleted MA cells at steady state (Figure 3A), highlighting that Herc1 specifically contributed to the response to nucleoside analogs. Flow cytometry analysis of murine MA cells by annexin V/DAPI staining confirmed that there was a significant increase in the proportion of Herc1-depleted cells that were undergoing apoptosis when compared with control cells (Figure 3B-C). We made similar observations for the HM cells in that we noticed an approximately threefold increase in the proportion of apoptotic cells that were depleted of Herc1 and exposed to Ara-C treatment when compared with controls (Figure 3D). Furthermore, annexin V/DAPI profiling of the U937 promonocytic human myeloid leukemia cell line revealed that loss of HERC1 led to an ∼1.5-fold increase in the proportion of apoptotic cells following exposure to Ara-C (Figure 3E; supplemental Figure 1F), suggesting that HERC1 plays a role in the response of both human and murine AML cells to Ara-C.
Targeting the E3 Ub ligase Herc1 exacerbates Ara-C-induced apoptosis in AML cells. (A) MA cells were plated for 24 hours in triplicates into 6-well plates and treated with ±Ara-C (200 nM) or H2O (vehicle). The values are presented as means ± standard error of the mean (SEM) (n = 3). Significance was determined using a 2-way ANOVA analysis, followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05. (B) Representative flow cytometry analysis of MA cells treated with ±Ara-C (200 nM) or H2O (vehicle) for 24 hours and stained with annexin V/DAPI. (C) Representation of the annexin V/DAPI analysis displayed in panel B for MA cells. The values are means ± SEM for n = 4 independent replicates. Significance was determined using a 1-way ANOVA, followed by Holm-Šídák multiple comparisons test. ∗∗P ≤ .005; ∗∗∗∗P ≤ .0001. (D) Representation of the annexin V/DAPI analysis displayed in panel B for HM cells. Values are means ± SEM for n = 3 independent replicates. Significance was determined using a 1-way ANOVA, followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05. (E) Representation of the annexin V/DAPI analysis displayed in panel B for U937 cells. Values are means ± SEM for n = 4 independent replicates. Significance was determined using a 1-way ANOVA, followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05. (F) Representative flow cytometry analysis of MA cells treated with Ara-C (200 nM) or H2O (vehicle) for 0 hours, 6 hours, and 12 hours and stained with propidium iodide or DAPI for cell cycle analysis. (G) Representation of the cell cycle analysis displayed in panel B for MA cells with ±Ara-C (200 nM) or water (Vehicle) for 0 hours and 6 hours (n = 3 independent experiments with 3 technical replicates) and for 12 hours (1 independent experiment, 3 technical replicates). Significance was determined using mixed-effects analysis followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05; ∗∗P < .01; ∗∗∗∗P < .0001.
Targeting the E3 Ub ligase Herc1 exacerbates Ara-C-induced apoptosis in AML cells. (A) MA cells were plated for 24 hours in triplicates into 6-well plates and treated with ±Ara-C (200 nM) or H2O (vehicle). The values are presented as means ± standard error of the mean (SEM) (n = 3). Significance was determined using a 2-way ANOVA analysis, followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05. (B) Representative flow cytometry analysis of MA cells treated with ±Ara-C (200 nM) or H2O (vehicle) for 24 hours and stained with annexin V/DAPI. (C) Representation of the annexin V/DAPI analysis displayed in panel B for MA cells. The values are means ± SEM for n = 4 independent replicates. Significance was determined using a 1-way ANOVA, followed by Holm-Šídák multiple comparisons test. ∗∗P ≤ .005; ∗∗∗∗P ≤ .0001. (D) Representation of the annexin V/DAPI analysis displayed in panel B for HM cells. Values are means ± SEM for n = 3 independent replicates. Significance was determined using a 1-way ANOVA, followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05. (E) Representation of the annexin V/DAPI analysis displayed in panel B for U937 cells. Values are means ± SEM for n = 4 independent replicates. Significance was determined using a 1-way ANOVA, followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05. (F) Representative flow cytometry analysis of MA cells treated with Ara-C (200 nM) or H2O (vehicle) for 0 hours, 6 hours, and 12 hours and stained with propidium iodide or DAPI for cell cycle analysis. (G) Representation of the cell cycle analysis displayed in panel B for MA cells with ±Ara-C (200 nM) or water (Vehicle) for 0 hours and 6 hours (n = 3 independent experiments with 3 technical replicates) and for 12 hours (1 independent experiment, 3 technical replicates). Significance was determined using mixed-effects analysis followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05; ∗∗P < .01; ∗∗∗∗P < .0001.
Mechanistically, upon phosphorylation of Ara-C by the DCK, it competes directly with dCTP for incorporation into newly synthesized DNA, thereby causing a delay in replication fork progression and interfering with cellular proliferation. To better understand the role of Herc1 in this process, we analyzed the progression of MA cells through the cell cycle in the presence or absence of Ara-C. As previously observed,37 Ara-C treatment induced a marked decrease in the proportion of cells in the G1 phase and a concomitant increase in the proportion of cells in the S phase at 6 hours and 12 hours after exposure (Figure 3F-G). Strikingly, loss of Herc1 increased the proportion of cells in the G1 phase at the early time point (6 hours) but had a limited impact on the cell cycle distribution of MA cells after 12 hours of treatment with Ara-C (Figure 3F-G), suggesting that Herc1 may be important in the early stage of the Ara-C response.
Herc1 controls Dck protein levels in murine AML cells
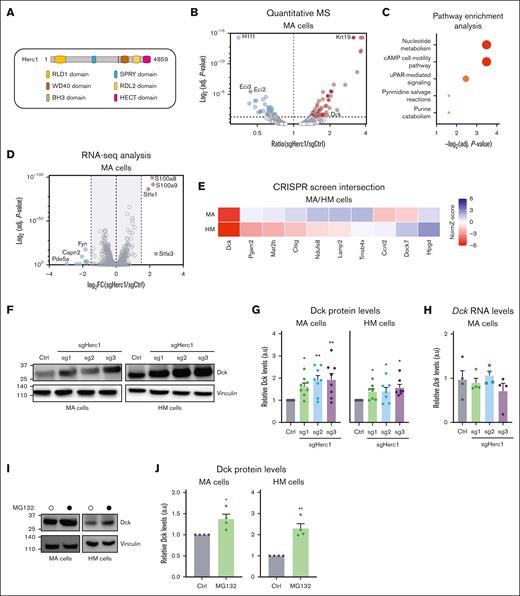
HERC1 is the largest member of the HERC family (4861 amino acids in Homo sapiens and 4859 amino acids in Mus musculus) and is structurally characterized by a HECT domain at its C-terminus and by a single SPRY (spl A and ryanodine receptor), WD40 (G protein β subunit like repeats), 2 RCC1-like domains, and a putative BH3 (Bcl-2 homology domain 3) (Figure 4A). HERC1 acts like an E3 Ub ligase through its HECT domain38, thereby promoting the ubiquitination and degradation of many cellular proteins related to DNA damage repair, cell proliferation, and migration.12,13,39-42 To gain better insight into the mechanisms through which Herc1 impacts the Ara-C response in murine AML cells, we used a MS-based proteomic approach and quantified proteins that were stabilized upon Herc1 loss in MA cells. In total, we detected 3511 proteins within sgHerc1 and sgCtrl MA cells (supplemental Table 4), including 106 that were significantly up- or downregulated in Herc1-depleted cells (P >.05; Figure 4B; supplemental Table 5). Pathway enrichment analysis identified nucleotide metabolism, including pyrimidine salvage reactions (eg, Dck, Uck2) and purine catabolism (eg, Gda, Gpx1), as significantly enriched in this subset of proteins (Figure 4C). To ensure that these changes at the protein level were not caused by transcriptional dysregulation, we performed a systematic RNA-seq analysis of both Herc1-depleted and control MA cells (supplemental Table 6). A limited portion of targets (10/106) were significantly up- or downregulated at the RNA level (log2 fold change ± 1.5; P < .05; Figure 4D), including Staf1, Staf3, S100a8, and S100a9. We subsequently focused our attention on the 40 MS targets that were significantly enriched in Herc1-depleted MA cells but that were not caused by major transcriptional changes in our RNA-seq analysis (supplemental Table 7).
Multiomic analysis identifies the Dck as a downstream target of Herc1. (A) Representation of the functional domains of HERC1 protein. (B) Volcano plot showing protein abundance in sgHerc1-MA cells vs sgCtrl MA cells. P values were calculated using t tests and were corrected for multiple hypothesis testing with Benjamin-Hochberg method. The dashed line represents P = .05. (C) Pathway enrichment analysis of 106 proteins that were significantly abundant in sgHerc1-MA cells. (D) Volcano plot showing differential expressed genes in sgHerc1-MA cells vs sgCtrl MA cells. The dashed line represents log2 fold change (FC) of ±1.5. (E) Integration of transcriptomic and proteomic data with the CRISPR/Cas9 screen to identify potential targets of Herc1. Potential candidates that passed the following criteria of (1) greater protein abundance (>1.5-fold) (shown in panel B) and (2) no corresponding change in mRNA transcript abundance (<1.5-fold) (shown in panel D). (F) Assessment of DCK protein levels in Herc1–gene edited cells. Representative western blot of whole cell lysates shows the DCK protein level in MA and HM-sgCtrl and sgHerc1 knockout cells using 3 different sgRNAs. (G) Quantification of DCK western blot as shown in panel F. Proteins are represented in arbitrary units (a.u.), normalized against sgCtrl. Values are means ± SEM of n = 8 (MA) and n = 7 (HM) independent replicates. Significance was determined using Wilcoxon signed-rank test. ∗P ≤ .05; ∗∗P < .01. (H) Quantitative PCR shows the mRNA levels of Dck in MA cells. Values are means ± SEM, (n = 4). Significance was determined using a 1-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤.05; ∗∗P < .01. (I) Assessment of DCK protein levels in MA and HM cells treated with the proteasome inhibitor MG132 (10 μM, 8 hours). Representative western blot of whole cell lysates shows DCK protein level in MA and HM cells. (J) Quantification of DCK western blot as shown in panel I. Proteins are presented in a.u., normalized to the vinculin loading control. Values are means ± SEM (n = 4). Significance was determined using an unpaired 2-tailed t test. ∗P ≤ .05; ∗∗P < .01.
Multiomic analysis identifies the Dck as a downstream target of Herc1. (A) Representation of the functional domains of HERC1 protein. (B) Volcano plot showing protein abundance in sgHerc1-MA cells vs sgCtrl MA cells. P values were calculated using t tests and were corrected for multiple hypothesis testing with Benjamin-Hochberg method. The dashed line represents P = .05. (C) Pathway enrichment analysis of 106 proteins that were significantly abundant in sgHerc1-MA cells. (D) Volcano plot showing differential expressed genes in sgHerc1-MA cells vs sgCtrl MA cells. The dashed line represents log2 fold change (FC) of ±1.5. (E) Integration of transcriptomic and proteomic data with the CRISPR/Cas9 screen to identify potential targets of Herc1. Potential candidates that passed the following criteria of (1) greater protein abundance (>1.5-fold) (shown in panel B) and (2) no corresponding change in mRNA transcript abundance (<1.5-fold) (shown in panel D). (F) Assessment of DCK protein levels in Herc1–gene edited cells. Representative western blot of whole cell lysates shows the DCK protein level in MA and HM-sgCtrl and sgHerc1 knockout cells using 3 different sgRNAs. (G) Quantification of DCK western blot as shown in panel F. Proteins are represented in arbitrary units (a.u.), normalized against sgCtrl. Values are means ± SEM of n = 8 (MA) and n = 7 (HM) independent replicates. Significance was determined using Wilcoxon signed-rank test. ∗P ≤ .05; ∗∗P < .01. (H) Quantitative PCR shows the mRNA levels of Dck in MA cells. Values are means ± SEM, (n = 4). Significance was determined using a 1-way ANOVA, followed by Dunnett multiple comparisons test. ∗P ≤.05; ∗∗P < .01. (I) Assessment of DCK protein levels in MA and HM cells treated with the proteasome inhibitor MG132 (10 μM, 8 hours). Representative western blot of whole cell lysates shows DCK protein level in MA and HM cells. (J) Quantification of DCK western blot as shown in panel I. Proteins are presented in a.u., normalized to the vinculin loading control. Values are means ± SEM (n = 4). Significance was determined using an unpaired 2-tailed t test. ∗P ≤ .05; ∗∗P < .01.
If Herc1 activity mediates the response to Ara-C by decreasing the abundance of a given protein that promotes the cytotoxic effect of Ara-C, we hypothesized that targeting this given substrate by CRISPR should also impact the Ara-C response. Therefore, to identify which of Herc1’s protein targets were relevant to the Ara-C response, we intersected them with our CRISPR-based genome-wide screens data and identified 10 targets that had a significant score in at least 1 murine AML cell line (NormZ score ± 1.5; Figure 4E). Interestingly, Dck emerged as the only substrate identified by MS that provided resistance to Ara-C in both MA and HM cells (Figure 4E). Furthermore, in agreement with the MS data, Herc1 loss led to a significant increase in the steady state Dck protein levels in MA and HM cells (Figure 4F-G) without affecting Dck messenger RNA (mRNA) levels (supplemental Figure 2A). Because Herc1 targets proteins for degradation through the Ub-proteasome system, we therefore explored whether inhibiting this pathway will impact Dck protein levels. Indeed, treating both MA and HM cells with MG132 led to a significant increase in the Dck protein levels in both murine AML cell lines (supplemental Figure 2B). Altogether, these data suggest that Herc1 modulates Ara-C response by controlling Dck protein levels at steady state in murine AML cells.
Targeting Herc1 modulates Ara-C response in vivo
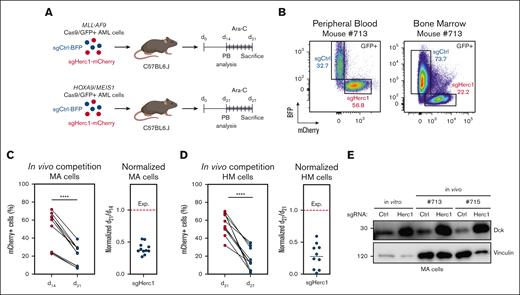
To evaluate the preclinical relevance of our findings, we targeted Cas9/GFP-expressing MA or HM cells with either sgHerc1 coupled to mCherry or sgCtrl coupled to BFP and transplanted them in a 1:1 ratio into syngeneic, sublethally irradiated C57BL6.J mice (Figure 5A). MA and HM cells were allowed to engraft and were analyzed in extracted peripheral blood (at 14 days and 21 days, respectively) before the mice were treated with Ara-C for 6 days (Figure 5A). The mice were subsequently sacrificed, and bone marrow was harvested for flow cytometry analysis (Figure 5B). In these settings, we observed a significant decrease in the percentage of mCherry+ cells that expressed sgHerc1 in both models (Figure 5C-D). Although the expected normalized ratio of mCherry+ cells between day 21 and day 14 should be ∼1, we observed median ratios of 0.48 and 0.28 (Figure 5C), suggesting that Herc1-depleted MA and HM cells are hypersensitive to Ara-C treatment in vivo. Importantly, cells sorted for BFP or mCherry expression before injection or after in vivo Ara-C treatment preserved their differential Dck protein levels (Figure 5E).
Targeting Herc1 modulates Ara-C response in vivo. (A) Schematic of the in vivo competition experiment. sgHerc1-mCherry cells and sgCtrl-BFP cells were combined in a 1:1 ratio before transplantation. A mix of 1 × 106 cells was transplanted into sublethally irradiated C57BL6.J mice. MA and HM cells were analyzed at the indicated timepoints. (B) Representation of flow cytometry analysis of the peripheral blood (PB) and bone marrow (BM) before and after Ara-C treatment, as described in panel A, in a MA mouse who underwent transplantation. The in vivo competition experiment for MA (C) and HM cells (D). Every value represents the relative abundance of sgHerc1-mCherry AML cells in a mouse (MA, n = 11; HM, n = 10) at given timepoints. Significance was determined using paired t test. ∗∗∗∗P < .0001. MA and HM cells were normalized to pretreatment abundance in the PB. The results are from 2 independent experiments. (E) Assessment of Dck in Herc1–gene edited cells in vitro and 2 mice. BFP+ and mCherry+ cells were sorted before western blot analysis.
Targeting Herc1 modulates Ara-C response in vivo. (A) Schematic of the in vivo competition experiment. sgHerc1-mCherry cells and sgCtrl-BFP cells were combined in a 1:1 ratio before transplantation. A mix of 1 × 106 cells was transplanted into sublethally irradiated C57BL6.J mice. MA and HM cells were analyzed at the indicated timepoints. (B) Representation of flow cytometry analysis of the peripheral blood (PB) and bone marrow (BM) before and after Ara-C treatment, as described in panel A, in a MA mouse who underwent transplantation. The in vivo competition experiment for MA (C) and HM cells (D). Every value represents the relative abundance of sgHerc1-mCherry AML cells in a mouse (MA, n = 11; HM, n = 10) at given timepoints. Significance was determined using paired t test. ∗∗∗∗P < .0001. MA and HM cells were normalized to pretreatment abundance in the PB. The results are from 2 independent experiments. (E) Assessment of Dck in Herc1–gene edited cells in vitro and 2 mice. BFP+ and mCherry+ cells were sorted before western blot analysis.
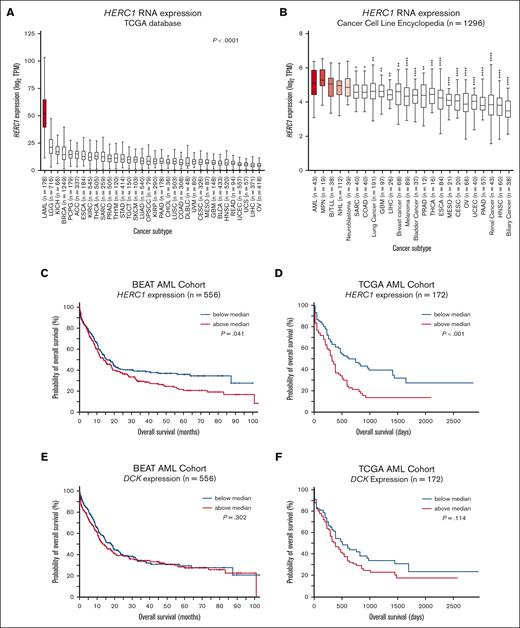
HERC1 expression is linked to DCK expression and has prognostic impact in human AML
To investigate the clinical relevance of our findings, we analyzed the expression pattern of HERC1 in a series of tumor tissues using the TCGA database. Strikingly, we noted that HERC1 is significantly upregulated in AML (TCGA-LAML, n = 178) when compared with 33 other cancer subtypes, including several hematological malignancies (eg, diffuse large B-cell lymphoma) and solid tumors (eg, lower grade glioma) (Figure 6A), suggesting a potential role for HERC1 in human AML. We made similar observations when referencing the CCLE (n = 1296) in which it was observed that HERC1 expression was significantly higher in a panel of AML cell lines (n = 43) than in 19 other panels of cancer cell lines, except those originating from myeloproliferative neoplasm, B- and T-lymphoblastic leukemia/lymphoma, non-Hodgkin lymphoma, and neuroblastoma (Figure 6B). These data argue for a strong dependency of AML cells on HERC1; however, our CRISPR screens in both the murine AML models14 argue against HERC1 being an essential rather than a context-specific gene in AML. We therefore investigated the prognostic potential of HERC1 expression on patient outcome in a series of AML cohorts (eg, BEAT-AML, TCGA-LAML, TARGET, GSE1159, GSE12417, GSE37642, GSE6891). Among these, high expression levels of HERC1 correlated with a significantly poorer overall survival among patients with AML in both the BEAT-AML (n = 556; P = .041) and the TCGA-LAML (n = 172; P < .001) data sets (Figure 6C-D). In contrast, a similar analysis in both cohorts of patients showed that DCK mRNA expression did not correlate with patient overall survival (Figure 6E-F). Confirming the relationship between HERC1 and DCK protein expression in human AML, a proteome data set of 26 human AML cell lines (n = 4 biological replicates per cell line)43 showed an inverse correlation between HERC1 and DCK protein abundance in AML that was driven by KMT2A-translocation or NRAS mutations (supplemental Figure 2A). This relationship operates at the protein level because, at the mRNA level, the relationship in human AML is reversed with the expression of HERC1 and DCK mRNA being strongly and positively correlated (supplemental Figure 2B). Overall, these findings in human AML suggest that HERC1 and DCK are co-regulated and that HERC1, rather than DCK, mRNA expression has prognostic potential in AML.
HERC1 expression has prognostic potential in human AML. (A) HERC1 expression was analyzed in a total of 34 different tumor types using the TCGA database. Statistical analysis was performed using a 1-way ANOVA followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (B) HERC1 expression was analyzed in a panel of 1296 different cell lines. Statistical analysis was performed as described in panel A. (C) Kaplan-Meier analysis of the BEAT-AML data set based on HERC1 expression (n = 132). (D) Kaplan-Meier analysis of the BEAT-AML data set based on HERC1 expression (n = 132). (E-F) Similar to panels C-D respectively, except that DCK was analyzed instead. Statistical analysis was performed using the Mantel-Cox test. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast adenocarcinoma; CESC, cervical squamous cell carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OPSCC, oropharyngeal squamous cell carcinoma; OV, ovarian cancer; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
HERC1 expression has prognostic potential in human AML. (A) HERC1 expression was analyzed in a total of 34 different tumor types using the TCGA database. Statistical analysis was performed using a 1-way ANOVA followed by Holm-Šídák multiple comparisons test. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (B) HERC1 expression was analyzed in a panel of 1296 different cell lines. Statistical analysis was performed as described in panel A. (C) Kaplan-Meier analysis of the BEAT-AML data set based on HERC1 expression (n = 132). (D) Kaplan-Meier analysis of the BEAT-AML data set based on HERC1 expression (n = 132). (E-F) Similar to panels C-D respectively, except that DCK was analyzed instead. Statistical analysis was performed using the Mantel-Cox test. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast adenocarcinoma; CESC, cervical squamous cell carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OPSCC, oropharyngeal squamous cell carcinoma; OV, ovarian cancer; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Discussion
Comprehensive CRISPR-based functional genomics screens have been powerful in identifying in vitro and in vivo genetic vulnerabilities in AML cells with therapeutic potential.18,44-48 In this study, we employed an in vitro CRISPR screening approach to identify genes that regulate murine AML cell susceptibility to Ara-C cytotoxicity. Systematic approaches have been employed previously to map the landscape of modulators of Ara-C,9,10,49 which identified loss of DCK as a major mechanism of resistance to Ara-C treatment. Similar findings were observed in our CRISPR-based approach in which targeting of Dck provided resistance to Ara-C in both MA and HM AML cell lines. Previous work has also shown that loss of SAMHD1 activity potentiated the cytotoxicity of Ara-C in AML cells,11 which was confirmed in our CRISPR screens, thereby validating our approach in murine AML cells.
Aside from these well-established modulators of Ara-C response, our screen identified that depleting the E3 Ub ligase HERC1 increased the susceptibility of murine and human AML cells to Ara-C–induced apoptosis. Furthermore, we observed that depleting Herc1 sensitized AML cells to other nucleoside analogs, such as Gem and Flu, which are also processed by DCK.7 We showed that Herc1-depleted MA cells are hypersensitive to Ara-C treatment in vivo and that HERC1 depletion in the human U937 cell line enhanced the apoptotic effect of Ara-C. In human AML, we found that the HERC1 and DCK proteins were inversely correlated in terms of abundance in KMT2A-translocated and RAS-mutated cell lines and that higher HERC1 expression was predictive of a poor prognosis in 2 large AML patient cohorts (BEAT-AML and TCGA), although this effect was not found in all cohorts. These findings suggest that HERC1 contributes to nucleoside metabolism in AML, which may be context or oncogene specific. Additional characterization of nucleotide levels or fluxes in HERC1-depleted cells would be informative of its regulatory role.
Our findings reinforce the importance of ubiquitylated/deubiquitylated events in the pathobiology of AML and its response to different stress conditions.50 Apart from HERC1, we identified several members of the SAGA de-ubiquitylating module (SAGA-DUBm) as regulators of Ara-C cytotoxicity, including ATXN7L3 and USP22. A previous study revealed that inactivating the SAGA-DUBm sensitized AML cells to double-negative T cell–mediated cytotoxicity,51 suggesting a potential role for the SAGA-DUBm as resistance marker for different regimens used in the treatment of patients with AML. Numerous members of the Ub-proteasome system have been found to play crucial roles in AML biology, particularly in the context of disease development and progression. Recently, 2 separate studies have shed light on how resistance to Ara-C in AML is mediated at the proteasomal level through distinct cellular pathways. One group revealed that the dNTP hydrolase SAMHD1 is under the control of 2 factors, namely non-POU domain–containing octamer binding (NONO) and the DDB1-DCAF1 E3 ligase.52 This regulation leads to increased stability of SAMHD1, which leads to the development of resistance to Ara-C in AML cells.52 In another investigation, the DUB USP7 was found to play a pivotal role in AML chemoresistance by modulating the levels of the CHK1 protein.53 This modulation enables AML cells to promote the progression of replication forks and to adapt to replication stress, thus contributing to resistance against Ara-C treatment.53
Our approach further highlights the central role of nucleotide metabolism in predicting the outcome of Ara-C treatment in AML cells. Both mutated54 and alternatively spliced55-57 forms of DCK have been linked to the development of resistance in patients with AML. Our study delineated an additional layer of regulation of murine Dck, namely at the posttranscriptional level by the E3 Ub ligase Herc1, which modulates the abundance of this critical enzyme. Our data suggest that a complex network of regulatory processes controls DCK levels, thereby influencing the response of AML cells to Ara-C, and that modulation of HERC1 levels in AML cells may represent a strategy to potentiate Ara-C response.
Acknowledgments
The authors are grateful for assistance from the Genomics Platform at Institute for Research in Immunology and Cancer, Université de Montréal, Montréal, Canada, for RNA sequencing analysis. Mass spectrometry analysis was performed by the Proteomics and Molecular Analysis Platform at the Research Institute of the McGill University Health Centre, Montréal, Canada.
M.J. is a recipient of a Cole Foundation doctoral scholarship and Lady Davis Institute/TD Bank Studentship Award. Work in the F.E.M. laboratory was supported by a transition grant from the Cole Foundation and an internal operating fund from the Sir Mortimer B. Davis Foundation of the Jewish General Hospital. Work in the F.E.M. and A.O. laboratories was jointly supported by a CRS operating grant (OG-940771), transition grants from the Cole Foundation and internal operating funds from the Sir Mortimer B. Davis Foundation of the Jewish General Hospital. F.E.M. is a Junior 2 clinical research scholar at the Fonds de Recherche du Québec – Santé. A.O. is the Canada Research Chair (Tier 2) in Genome Stability and Hematological Malignancies.
Authorship
Contributions: M.J. designed and completed most of the experiments presented in the manuscript, analyzed all data, and helped with the writing of the manuscript under the supervision of F.E.M. and A.O; W.W.L.P. performed the genome-wide screen and helped to prepare the libraries for next-generation sequencing; M.J. analyzed the genome-wide screen. C.G.-L. assisted with the western blot experiments and cell viability assays; G.G.V., C.C.-D., and A.I., under the supervision of M.J., helped in the generation of the reagents for the experiments presented in this study; B.S.-A. assisted with in vivo experiments and mouse colony management; Y.W. and A.K.J. performed the correlation analysis using RNA and proteome data sets from patients with acute myeloid leukemia; Y.D. helped with the analysis of RNA sequencing data under the supervision of J.S.; and M.J., A.O., and F.E.M. conceived the study, designed the research, provided supervision, and wrote the manuscript with input from all the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for G.G.V. is Paraza Pharma Inc.
The current affiliation for M.J. is Paraza Pharma Inc.
The current affiliation for W.W.L.P. is Strand Therapeutics Inc.
Correspondence: Francois E. Mercier, Lady Davis Institute, Segal Cancer Centre Jewish General Hospital, Montréal, QC, H3T 1E2, Canada; email: francois.mercier@mcgill.ca; and Alexandre Orthwein, Department of Radiation Oncology, Winship Cancer Center, Emory University, Atlanta, GA 30322; email: alexandre.orthwein@emory.edu.
References
Author notes
The RNA sequencing and proteomics data have been deposited in the Gene Expression Omnibus database (accession number GSE273667) and Proteomics Identifications database (accession number PXD054402), respectively.
The full-text version of this article contains a data supplement.