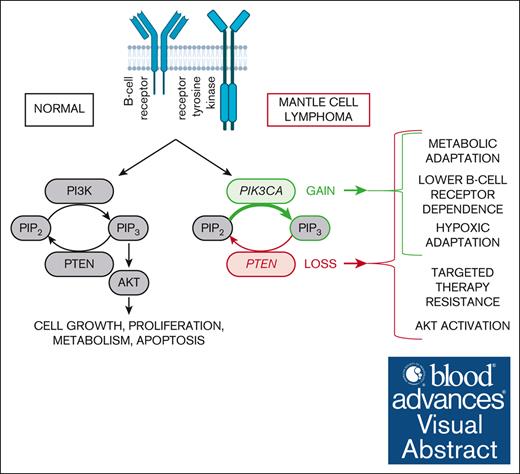
Key Points
PIK3CA gain and PTEN loss decrease the dependence of MCL cells on BCR signaling and antiapoptotic BCL2.
PIK3CA gain and PTEN loss lead to complex metabolic rewiring and increased survival of MCL cells under hypoxia.
Visual Abstract
Besides many other mutations in known cancer driver genes, mantle cell lymphoma (MCL) is characterized by recurrent genetic alterations of important regulators of the phosphoinositol-3-kinase (PI3K) cascade including PIK3CA gains and PTEN losses. To evaluate the biological and functional consequences of these aberrations in MCL, we have introduced transgenic expression of PIK3CA (PIK3CA UP) and performed knockout/knockdown of PTEN gene (PTEN KO/KD) in 5 MCL cell lines. The modified cell lines were tested for associated phenotypes including dependence on upstream B-cell receptor (BCR) signaling (by an additional BCR knockout). PIK3CA overexpression decreased the dependence of the tested MCL on prosurvival signaling from BCR, decreased levels of oxidative phosphorylation, and increased resistance to 2-deoxy-glucose, a glycolysis inhibitor. Unchanged protein kinase B (AKT) phosphorylation status and unchanged sensitivity to a battery of PI3K inhibitors suggested that PIK3CA gain might affect MCL cells in AKT-independent manner. PTEN KO was associated with a more distinct phenotype: AKT hyperphosphorylation and overactivation, increased resistance to multiple inhibitors (most of the tested PI3K inhibitors, Bruton tyrosine kinase inhibitor ibrutinib, and BCL2 inhibitor venetoclax), increased glycolytic rates with resistance to 2-deoxy-glucose, and significantly decreased dependence on prosurvival BCR signaling. Our results suggest that the frequent aberrations of the PI3K pathway may rewire associated signaling with lower dependence on BCR signaling, better metabolic and hypoxic adaptation, and targeted therapy resistance in MCL.
Introduction
Mantle cell lymphoma (MCL) is a B-cell non-Hodgkin lymphoma characterized by a chronically relapsing clinical course.1,2 Besides the canonical translocation t(11;14)(q13;q32) leading to the overexpression of cyclin D1, many other recurrent molecular-cytogenetic events have been reported in patients with newly diagnosed MCL, including lesions of the phosphatidylinositol 3-kinase (PI3K)–protein kinase B (AKT)–mammalian target of rapamycin (mTOR) pathway. These include copy number gains of the P110a catalytic subunit of PI3 kinase (PIK3CA) and loss of phosphatase and tensin homologue (PTEN).3-8
The precise impact of these recurrent aberrations on the biology, survival, and drug resistance of MCL cells remain largely unexplored. Hyperphosphorylation of AKT kinase because of loss of PTEN has been reported in prognostically adverse MCL blastoid variant.5 Increased expression of PIK3CA was associated with MCL progression, whereas increased PI3K-AKT activity was detectable in patients who failed to respond to ibrutinib, an inhibitor of Bruton tyrosine kinase (BTK), a critical kinase involved in B-cell receptor [BCR] signaling).9-11 We have recently demonstrated that PIK3CA copy number gains represent frequent genetic events associated with MCL relapse after failure of standard immunochemotherapy.12 Because of this pathway deregulation, pharmacological targeting of the PI3K-AKT-mTOR cascade has been repeatedly investigated in MCL, either as a single agent or in combination with other targeted agents, having promising results.13-20
In this study, we aimed to investigate the exact consequences of PIK3CA gain and PTEN loss on the biology of MCL cells and on the sensitivity/resistance to PI3K, BTK, AKT, and BCL2 family inhibitors.
Materials and methods
Cell lines and clones
MCL cell lines (MINO, Z138, and JEKO-1) were purchased from German Collection of Microorganisms and Cell Cultures or the American Tissue Culture Collection. UPF1H and UPF19U were derived in our laboratory from leukemized peripheral blood of patients with chemotherapy-resistant MCL. Generation of cells with transgenic PIK3CA (over)expression (PIK3CA UP) or with PTEN knockout (PTEN KO) and cultivation conditions are further described in the supplemental Material.
Cytotoxic agents
Specific PI3K inhibitors, AKT inhibitor, BTK inhibitor (BTKi), and BCL2 family inhibitors are described in the supplemental Material.
Cytogenomic analyses
Copy number alteration analyses of the PIK3CA, PTEN, and TP53 genes were performed either by interphase fluorescence in situ hybridization (FISH) or by array-comparative genomic hybridization/single-nucleotide polymorphism, depending on the availability of material, as part of the routine diagnostic procedure. Detailed protocol description is provided in the supplemental Material.
Generation of cell lines with target genes knockout
Detailed protocol description is provided in the supplemental Material.
Generation of cell lines with target genes overexpression
Detailed protocol description is provided in the supplemental Material.
Cell lines transfection
To introduce all plasmids, we used electroporation according to the manufacturer’s instructions (Neon, ThermoFisher Scientific). A total of 1.2 million cells per sample were used for the 100-μL electroporation system. Cells were washed once by phosphate-buffered saline and resuspended in 120 μL of electroporation R buffer together with plasmid DNA (12 μg for single KO plasmid for green fluorescence protein (GFP) and B-cell receptor knockout (BCR KO); 5 + 5 + 5 μg for 3 plasmids necessary for PTEN KO GFP KI; and 6 + 4 μg for sleeping beauty donor and transposase plasmids, respectively, for stable overexpression) to perform electroporation. Cells were then resuspended in 3 mL of appropriate cell culture media without antibiotics for further experiments or selection. Electroporation conditions for individual cell lines were as follows (voltage, pulse length, number of pulses): Jeko-1 1500 V, 20 ms, 1 pulse; Mino 1300 V, 30 ms, 1 pulse; UPF1H 1500 V, 10 ms, 3 pulses; UPF19U 1500 V, 10 ms, 3 pulses; Z138 1600 V, 20 ms, 1 pulse.
Measurement of AKT activity in living cells
Detailed protocol description is provided in the supplemental Material.
Apoptosis and proliferation assays
Detailed protocol description is provided in the supplemental Material.
Western blotting
Detailed protocol description is provided in the supplemental Material.
Coimmunoprecipitation assay
Detailed protocol description is provided in the supplemental Material.
In vitro assays under hypoxia
The in vitro experiments under hypoxic conditions were implemented in Coy hypoxic box. Detailed protocol description is provided in the supplemental Material.
OCR and ECAR assays
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements were performed using a Seahorse XFe96 analyzer (Agilent Technologies). On the day of the experiment, cells were seeded on XFe96 cell culture microplates coated with Cell-Tak (Corning; 5 × 104 cells per well), in Seahorse base RPMI medium and incubated at 37°C without CO2 for 1 hour. The base medium (180 μL per well) was supplemented with 500-mM pyruvate, 2-mM L-glutamine, 10-mM glucose for OCR measurement and with 2-mM L-glutamine for ECAR measurement. The mito stress test and glycolytic stress test are further described in the supplemental Material.
Statistical analysis
To assess the statistical significance of experiments, unpaired Student t test or ordinary 1-way analysis of variance was used (GraphPad Prism-10). P value <.05 represents statistical significance. Densitometry was carried out using Image-Lab Bio-Rad-6.1 software.
Results
PIK3CA copy number gain and PTEN deletion are common aberrations in patients with MCL
To determine the prevalence of PIK3CA copy number variants and PTEN deletions, we carried out FISH and array-comparative genomic hybridization analyses of 61 consecutive samples obtained from patients with newly diagnosed MCL. Forty-three percent of patients had gain of 1 copy of PIK3CA gene, 7% patients had monoallelic deletion of PTEN, and 1 case had both. There was no obvious correlation between TP53 deletion and PIK3CA gain. Interestingly, PTEN deletion was found exclusively in the context of concurrent TP53 deletion, but the relatively low number of analyzed patients precludes us from drawing any statistically significant correlations (supplemental Table 1).
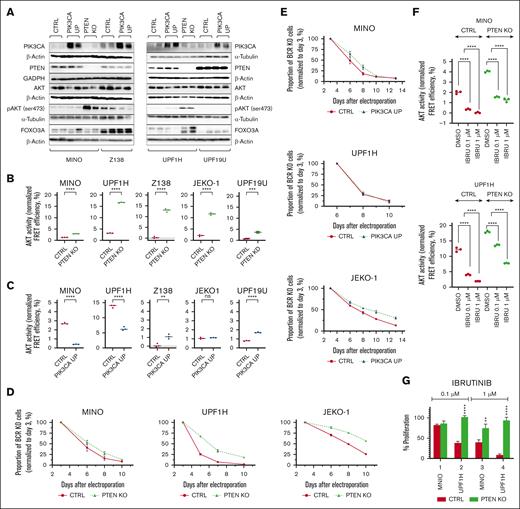
PTEN KO but not PIK3CA overexpression show hyperphosphorylation and overactivation of AKT
Overexpression of PIK3CA in MINO, Z138, UPF1H, UPF19U, and JEKO-1 PIK3CA UP cells and the loss of PTEN expression in UPF1H, MINO, Z138, and JEKO-1 PTEN KO cells were confirmed by western blotting (Figure 1A; supplemental Figure 1). UPF19U and MINO PTEN knockdown (PTEN KD) manifested a decreased expression of PTEN (supplemental Figure 1). Except for JEKO-1, all PTEN KO cells had hyperphosphorylation of the key downstream target of PI3K, the AKT kinase (Figure 1A; supplemental Figure 1). In addition, all PTEN KO/KD cells had increased AKT activity by fluorescence resonance energy transfer (FRET) assay (Figure 1B; supplemental Figure 2). In contrast, transgenic (over)expression of PIK3CA was not associated with consistent changes in AKT phosphorylation or AKT activity (Figure 1C). To determine the extent and potential correlation of PTEN expression and AKT phosphorylation in MCL cells, we conducted western blot profiling of 32 primary MCL samples obtained from patients with newly diagnosed and relapsed MCL. Ten of 32 patient (P) samples (P3, 6, 15, 16, 19, 20, 24, 30, 31, and 32) had higher levels of AKT phosphorylation, all accompanied by lower or no expression of PTEN. Three patients had undetectable PTEN protein (P13, 30, and 31; supplemental Figures 3 and 4A). From these, 2 patients (P13 and 30) had no genetic copy number loss of PTEN gene by standard FISH, suggesting alternative mechanism of loss of PTEN protein expression. Patients with any kind of PTEN genetic copy number loss (P29 and 31) had low or undetectable expression of PTEN protein. Of note, higher levels of AKT phosphorylation were associated with lower expression of PTEN protein (supplemental Figure 4B).
Impact of PTEN loss and PIK3CA gain on AKT activity and BCR prosurvival signaling. Loss of PTEN expression but not PIK3CA (over)expression is associated with hyperphosphorylation and overactivation of AKT. On the contrary, both loss of PTEN and, to a lesser extent, gain of PIK3CA decrease dependance on the prosurvival signaling from BCR. (A) Western blot. Confirmation of the upregulation of PIK3CA in the 4 PIK3CA UP cell lines MINO, Z138, UPF1H, and UPF19U and the loss of PTEN in the PTEN KO clones MINO and UPF1H. Increase of phospho-AKT (ser 473) expression in both PTEN KO cell lines MINO and UPF1H (for each sample, n = 2). (B) AKT activity as measured using genetically encoded FRET-based biosensor. Significantly increased AKT kinase activity in all PTEN KO cell lines compared with respective control cell lines; technical triplicates; (C) FRET assay. Variable effect of PIK3CA (over)expression on AKT activity; technical triplicates. (D) PTEN loss increases survival of MCL cells with knockout of BCR gene (n = 3). (E) Transgenic (over)expression of PIK3CA in MINO and JEKO-1 but not in UPF1H increases survival of MCL cells with knockout of BCR gene (n = 3). (F) AKT activity as measured using genetically encoded FRET-based biosensor. AKT activity in PTEN KO cell lines is higher than that of respective cell lines. In addition, after exposure to BTKi ibrutinib for 3 hours, AKT activity in PTEN KO cell lines remains higher than AKT activity in the respective control cell lines (n = 3). (G) Proliferation assay implemented 72 hours after exposure to ibrutinib (0.1 and 1 μM); the cellular proliferation of the treated cells was normalized to the cellular proliferation of the untreated cells (n = 3); data are represented as mean ± standard deviation (SD); ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates. CTRL, controls; DMSO, dimethyl sulfoxide; ns, not significant.
Impact of PTEN loss and PIK3CA gain on AKT activity and BCR prosurvival signaling. Loss of PTEN expression but not PIK3CA (over)expression is associated with hyperphosphorylation and overactivation of AKT. On the contrary, both loss of PTEN and, to a lesser extent, gain of PIK3CA decrease dependance on the prosurvival signaling from BCR. (A) Western blot. Confirmation of the upregulation of PIK3CA in the 4 PIK3CA UP cell lines MINO, Z138, UPF1H, and UPF19U and the loss of PTEN in the PTEN KO clones MINO and UPF1H. Increase of phospho-AKT (ser 473) expression in both PTEN KO cell lines MINO and UPF1H (for each sample, n = 2). (B) AKT activity as measured using genetically encoded FRET-based biosensor. Significantly increased AKT kinase activity in all PTEN KO cell lines compared with respective control cell lines; technical triplicates; (C) FRET assay. Variable effect of PIK3CA (over)expression on AKT activity; technical triplicates. (D) PTEN loss increases survival of MCL cells with knockout of BCR gene (n = 3). (E) Transgenic (over)expression of PIK3CA in MINO and JEKO-1 but not in UPF1H increases survival of MCL cells with knockout of BCR gene (n = 3). (F) AKT activity as measured using genetically encoded FRET-based biosensor. AKT activity in PTEN KO cell lines is higher than that of respective cell lines. In addition, after exposure to BTKi ibrutinib for 3 hours, AKT activity in PTEN KO cell lines remains higher than AKT activity in the respective control cell lines (n = 3). (G) Proliferation assay implemented 72 hours after exposure to ibrutinib (0.1 and 1 μM); the cellular proliferation of the treated cells was normalized to the cellular proliferation of the untreated cells (n = 3); data are represented as mean ± standard deviation (SD); ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates. CTRL, controls; DMSO, dimethyl sulfoxide; ns, not significant.
Loss of PTEN expression and gain of PIK3CA both decrease dependence of MCL cells on prosurvival signaling from BCR
We have clearly documented before that BCR signaling critically contributes to PI3K/AKT signaling activation in lymphomas. The degree of PI3K/AKT activation is largely dependent on BCR surface abundance.21 Therefore, we tested whether PI3K/AKT hyperactivation (triggered by PTEN KO or PIK3CA overexpression) can decrease the dependency of MCL cells on upstream BCR-mediated PI3K/AKT activation. We have performed BCR knockout in 3 PTEN KO cell line variants (MINO, UPF1H, and JEKO-1 PTEN/BCR KO subclones) and in 3 PIK3CA UP cell line variants (MINO, UPF1H, and JEKO-1 PIK3CA UP/BCR KO subclones). Loss of PTEN partially decreased the dependence on the prosurvival signaling from the BCR (Figure 1D). Partial PTEN KO–mediated BCR KO rescue was detectable especially in cell lines with large increase of AKT activity after PTEN KO (Figure 1B) and was consisted with presumed “chronic active” type of BCR signaling in MCL that activates multiple signaling pathways than just PI3K/AKT.22 Curiously, in 2 of 3 tested PIK3CA UP/BCR KO subclones (MINO and JEKO-1 but not UPF1H), gain of PIK3CA also decreased the dependence on BCR prosurvival signaling, although to a lower extent in comparison with PTEN/BCR KO subclones (Figure 1E). As for the remaining MCL cell lines used in this study, Z138 cells are BCR independent (supplemental Figure 3), and UPF19U cells do not express BCR. Additional analysis in MINO and JEKO-1 cell lines showed that PTEN KO supported growth specifically in BCR KO cells and did not affect (JEKO-1) or slightly decreased (MINO) the growth of parental cell lines with intact BCR (supplemental Figure 6). PI3KCA overexpression increased the growth of MINO cells, more so in BCR-negative variant, and did not affect the cell growth of parental and BCR KO JEKO-1 cells (supplemental Figure 6). PTEN KD in MINO cell line did not rescue the cells from BCR KO (supplemental Figure 7).
Importantly, after BTK inhibition by ibrutinib, PTEN KO cells showed lower dependence on BCR-mediated AKT activation compared with parental cell lines (as detected by genetically encoded FRET-based AKT activity biosensor; Figure 1F; supplemental Figure 8), which was to a lesser extend detectable also in MINO PTEN KD cells (supplemental Figure 9).23 This implies that the loss of PTEN overcomes ibrutinib-mediated decrease of AKT activity, which should translate into BTK inhibition resistance. Indeed, proliferation assays confirmed that PTEN KO cells were more resistant to ibrutinib (Figure 1G). On the contrary, PIK3CA overexpression did not consistently increase AKT activity and caused a paradoxical AKT activity decrease in several cell lines (Figure 1C). In accordance with this, AKT activity decrease after ibrutinib treatment was not rescued by PIK3CA overexpression (supplemental Figure 10), and no changes in the sensitivity to ibrutinib were detected (supplemental Figure 11).
Loss of PTEN and gain of PIK3CA affect metabolic activity and survival of MCL cells under hypoxia
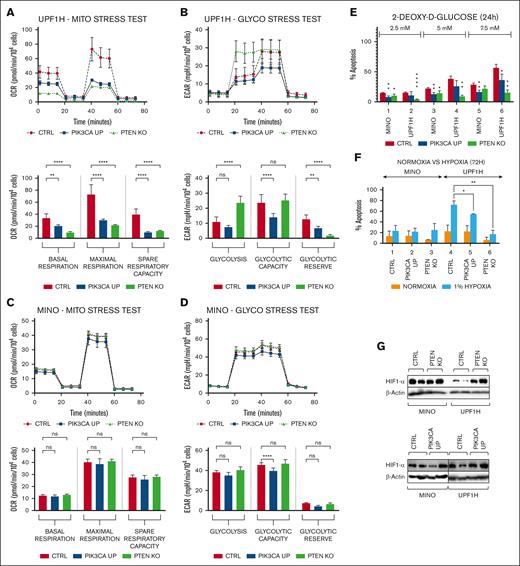
The PI3K-AKT signaling pathway plays a pivotal role in the regulation of cell energy-metabolic pathways. Therefore, we used Seahorse analyzer to assess the potential impact of PIK3CA gain or PTEN loss on metabolic reprogramming of MCL. In UPF1H cells, both PTEN KO and PIK3CA overexpression decreased basal as well as maximal respiration (Figure 2A). Interestingly, these 2 genetic alterations had different effects on glycolysis in UPF1H cells. PTEN KO massively increased basal glycolysis (with almost no further glycolytic reserve left). Basal glycolysis was not affected by PIK3CA overexpression; however, PIK3CA overexpression decreased overall glycolytic capacity and glycolytic reserve (Figure 2B). In MINO cells, PIK3CA overexpression had a similar effect as in UPF1H (decreased maximal respiration and decreased glycolytic parameters); however, it was much less pronounced. PTEN KO did not affect respiration or glycolysis in UPF1H cells (Figure 2C-D). Substantial decrease of mitochondrial function, accompanied by increased basal glycolysis in PTEN KO cells, suggest increased glycolysis to oxidative phosphorylation (ox/phos) ratio in PTEN KO as well as PIK3CA UP UPF1H cells, which was not clearly detectable in MINO cells.
PTEN KO and PIK3CA UP cell lines have changed the activity of key energy-metabolic pathways and have increased survival under hypoxia. (A-D) Mito stress tests and glycol stress tests performed using Seahorse analyzer. (A) Decrease in the basal respiration, maximal respiration, and spare respiratory capacity in UPF1H PIK3CA UP and PTEN KO cells compared with UPF1H unmodified cell line; measured by OCR. (B) Increase in basal glycolysis in UPF1H PTEN KO cells, decrease of glycolytic capacity in UPF1H PIK3CA UP cells, and decrease of glycolytic reserve in both tested variants; measured by ECAR. (C) No significant changes of mitochondrial function in MINO PIK3CA UP modified cells compared with MINO CTRL. (D) Slight decrease of glycolysis parameters in in MINO PIK3CA UP modified cells compared with MINO CTRL. (E) Number of apoptotic cells 24 hours after exposure to the inhibitor of glycolysis 2-DG (2.5, 5, and 7.5 mM); apoptosis of the treated cells was normalized to the apoptosis of the untreated cells (n = 3). (F) Increased survival of PTEN KO and PIK3CA UP cell lines after 72-hour exposure to 1% hypoxia; apoptosis of the cells cultured for 72 hours under 1% hypoxia (as well as apoptosis of the cells cultured for 72 hours in parallel under normoxia) was normalized to the apoptosis of the cells before placement into hypoxia (n = 3). (G) Western blot analysis of HIF1-alpha in PTEN KO and PIK3CA UP cell lines compared with respective control cell lines (for each sample, n = 2); data are represented by means ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates. CTRl, control; ns, not significant.
PTEN KO and PIK3CA UP cell lines have changed the activity of key energy-metabolic pathways and have increased survival under hypoxia. (A-D) Mito stress tests and glycol stress tests performed using Seahorse analyzer. (A) Decrease in the basal respiration, maximal respiration, and spare respiratory capacity in UPF1H PIK3CA UP and PTEN KO cells compared with UPF1H unmodified cell line; measured by OCR. (B) Increase in basal glycolysis in UPF1H PTEN KO cells, decrease of glycolytic capacity in UPF1H PIK3CA UP cells, and decrease of glycolytic reserve in both tested variants; measured by ECAR. (C) No significant changes of mitochondrial function in MINO PIK3CA UP modified cells compared with MINO CTRL. (D) Slight decrease of glycolysis parameters in in MINO PIK3CA UP modified cells compared with MINO CTRL. (E) Number of apoptotic cells 24 hours after exposure to the inhibitor of glycolysis 2-DG (2.5, 5, and 7.5 mM); apoptosis of the treated cells was normalized to the apoptosis of the untreated cells (n = 3). (F) Increased survival of PTEN KO and PIK3CA UP cell lines after 72-hour exposure to 1% hypoxia; apoptosis of the cells cultured for 72 hours under 1% hypoxia (as well as apoptosis of the cells cultured for 72 hours in parallel under normoxia) was normalized to the apoptosis of the cells before placement into hypoxia (n = 3). (G) Western blot analysis of HIF1-alpha in PTEN KO and PIK3CA UP cell lines compared with respective control cell lines (for each sample, n = 2); data are represented by means ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates. CTRl, control; ns, not significant.
Functionally, both PTEN KO and PIK3CA UP UPF1H and MINO cell lines were generally more resistant to 2-deoxy-glucose (2-DG), an inhibitor of glycolysis (Figure 2E). Similar to metabolic changes, this phenotype was more prominent in UPF1H cells. MINO PTEN KD did not show increased resistance to 2-DG compared with parental cell line (supplemental Figure 12).
Increased glycolytic rate is characteristic for malignant cells growing under hypoxia. Therefore, we tested the survival of modified cell lines under 3-day exposure to hypoxia (1% oxygen). UPF1H PTEN KO and PIK3CA UP cells were both more resistant to long-term exposure to 1% hypoxia (Figure 2F). UPF1H PTEN KO cells and, to a lesser extent, UPF1H PIK3CA UP cells had higher levels of hypoxia induced factor 1 (HIF1)-alpha protein by western blotting than the parental UPF1H cells (Figure 2G; supplemental Figure 13). In contrast, MINO cells were generally resistant to low tensions of oxygen, and therefore, no significant differences were observed between PTEN KO or PIK3CA UP modified cells and the parental MINO cells regarding differences in growth under hypoxia and HIF1-alpha upregulation (Figure 2F-G; supplemental Figure 13).
MCL PIK3CA UP and PTEN KO cell lines have changed sensitivity to BH3 mimetics
Given the important antiapoptotic role of the PI3K/AKT pathway and the clinical potential of BH3 mimetics, we also tested the cytotoxic efficacy of selected BH3 mimetics in the context of PTEN KO and PIK3CA overexpression. To investigate whether partial loss of PTEN affects sensitivity to BH3 mimetics in MCL, we tested the same panel of BCL2 family inhibitors on the MINO PTEN KD cell line.
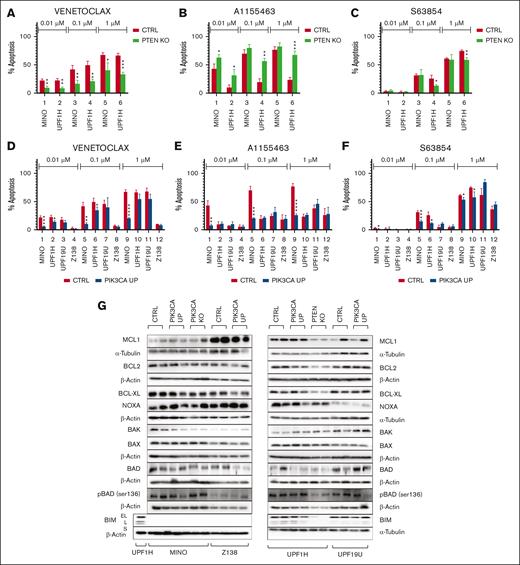
Although all tested PTEN KO cells were more resistant to the BCL2 inhibitor venetoclax (VEN), their sensitivity to the BCL-XL inhibitor A1155463 was increased (Figure 3A-C). MINO PTEN KD cells were also more resistant to VEN and manifested no change in their sensitivity to A1155463 (supplemental Figure 14A-B). Two of 4 tested PIK3CA UP cells (UPF1H and MINO) were more resistant to VEN and S63845, a MCL1 inhibitor (Figure 3D,F).
PIK3CA UP and PTEN KO leads to different sensitivities to BH3 mimetics. (A-F) Number of apoptotic cells 24 hours after exposure to the BH3 mimetics venetoclax (0.01, 0.1, and 1 μM), S63845 (0.01, 0.1, and 1 μM), and A1155463 (0.01, 0.1, and 1 μM). Apoptosis of the treated cells was normalized to the apoptosis of the untreated cells (n = 3). (G) Western blot analysis of selected BCL2 family proteins in PIK3CA UP and PTEN KO cell lines compared with the respective control MCL cell lines (for each sample, n = 2); data are represented by means ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates.
PIK3CA UP and PTEN KO leads to different sensitivities to BH3 mimetics. (A-F) Number of apoptotic cells 24 hours after exposure to the BH3 mimetics venetoclax (0.01, 0.1, and 1 μM), S63845 (0.01, 0.1, and 1 μM), and A1155463 (0.01, 0.1, and 1 μM). Apoptosis of the treated cells was normalized to the apoptosis of the untreated cells (n = 3). (G) Western blot analysis of selected BCL2 family proteins in PIK3CA UP and PTEN KO cell lines compared with the respective control MCL cell lines (for each sample, n = 2); data are represented by means ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates.
To decipher the molecular mechanisms underlying the observed changes in the sensitivity to the tested BH3 mimetics, we implemented western blot analysis to determine the expression of key BCL2 family proteins and immunoprecipitated BCL2 and BCL-XL to identify their respective binding partners.
UPF1H PTEN KO cells had significantly decreased the expression of proapoptotic BCL2L11/BIM (MINO cells have biallelic deletion of BIM). MINO PTEN KO cells expressed more phospho-BAD (Figure 3D). Immunoprecipitation experiments demonstrated decreased amount of proapoptotic BIM protein on BCL-XL and BCL2 in UPF1H PTEN KO cells. On the contrary, there was a decreased BAX interaction with BCL2 in MINO PTEN KO cells (supplemental Figure 15).
PTEN KO and, to a lesser degree, PIK3CA overexpression mediated resistance to a panel of PI3K inhibitors
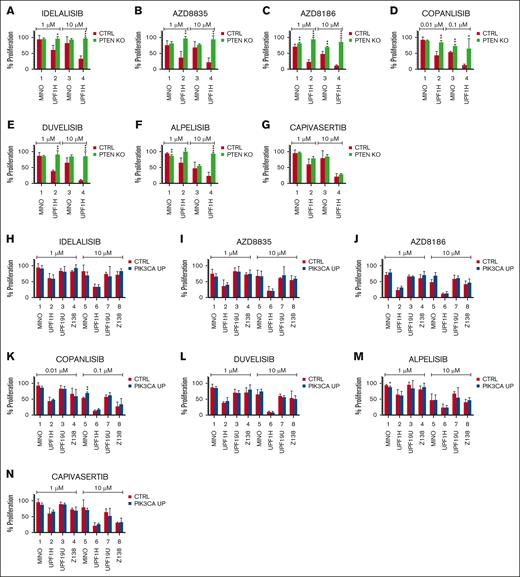
To explore the therapeutic consequences of PTEN loss and PIK3CA gain, we implemented a cytotoxic screen of the established PIK3CA UP and PTEN KO/KD cells using a panel of selected pathway inhibitors including PI3K inhibitors (idelalisib, duvelisib, AZD8186, AZD8835, alpelisib, and copanlisib) and an AKT inhibitor capivasertib. For the functional assays, we used 2 PTEN KO (UPF1H and MINO), 1 PTEN KD (MINO), and 4 PIK3CA UP cell lines variants (UPF1H, MINO, UPF19U, and Z138). Loss of PTEN correlated with increased resistance to PI3K inhibitors (Figure 4A-F). Although UPF1H PTEN KO cells were more resistant to all tested PI3K inhibitors, MINO PTEN KO cells were more resistant to AZD8186 and copanlisib. MINO PTEN KD were more resistant to copanlisib. Both PTEN KO cell lines and MINO PTEN KD retained sensitivity to AKT inhibitor capivasertib (Figure 4G; supplemental Figure 16). With few exceptions, gain of PIK3CA was not associated with significant changes in the sensitivity to PI3K inhibitors or capivasertib (Figure 4H-N).
PTEN KO and, to a lesser degree, PIK3CA overexpression mediated resistance to a panel of PI3K inhibitors. (A-N) Proliferation assays implemented 72 hours after exposure to the indicated agents: idelalisib (1 and 10 μM), duvelisib (1 and 10 μM), AZD8186 (1 and 10 μM), AZD8835 (1 and 10 μM), alpelisib (1 and 10 μM) (F), copanlisib (0.01 and 0.1 μM), and capivasertib (1 and 10 μM); the cellular proliferation of the treated cells was normalized to the cellular proliferation of the untreated cells (n = 3); data are represented as means ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates. CTRL, control.
PTEN KO and, to a lesser degree, PIK3CA overexpression mediated resistance to a panel of PI3K inhibitors. (A-N) Proliferation assays implemented 72 hours after exposure to the indicated agents: idelalisib (1 and 10 μM), duvelisib (1 and 10 μM), AZD8186 (1 and 10 μM), AZD8835 (1 and 10 μM), alpelisib (1 and 10 μM) (F), copanlisib (0.01 and 0.1 μM), and capivasertib (1 and 10 μM); the cellular proliferation of the treated cells was normalized to the cellular proliferation of the untreated cells (n = 3); data are represented as means ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; (N) represents the number of biological replicates. CTRL, control.
Discussion
Aberrant overactivation of the PI3K-AKT signaling pathway has been repeatedly reported in MCL and was associated with blastoid morphology and resistance to BTKis.5,7,9,10,24,25 In this study, we confirmed that gain of PIK3CA and deletion of PTEN are recurrent aberrations in patients with newly diagnosed MCL. Several studies demonstrated that PTEN is a haploinsufficient tumor suppressor gene.26-28 We also demonstrated that patients with any kind of genetic copy number loss of PTEN (as determined by locus specific FISH) had lower PTEN protein than patients with intact PTEN gene. In addition, low or even undetectable PTEN protein was observed in several MCL samples obtained from patients with intact PTEN gene. Our results thus confirm observations from previous studies that PTEN can be downregulated or completely silenced either by PTEN gene promoter hypermethylation or by diverse posttranscriptional and posttranslational modifications of PTEN messenger RNA or PTEN protein.29-32 Interestingly, the most frequent type of aberration was a gain of just 1 copy of PIK3CA gene found in 43% of patients. We may speculate that fine-tuning of the PI3K-AKT pathway, rather than massive uncontrolled overactivation, is needed for accurate benefits from its activation in MCL cells. This is in line with previous reports by Shojaee et al who demonstrated that both the blockage and massive overactivation of PI3K-AKT is detrimental for malignant cells.33 Importantly, FISH analysis was performed only in patients with bone marrow involvement (≥20% infiltration). Therefore, frequency of PIK3CA gains in patients with low or no bone marrow infiltration remains unknown. Frequencies of PIK3CA gains and PTEN losses at the first or subsequent disease relapses also remain elusive. However, our recent whole-exome sequencing–based study on clonal evolution of 25 patients with MCL at diagnosis and at first relapse (after failure of standard immunochemotherapy) revealed that PIK3CA gain is one of the most frequently identified copy number variant associated with MCL relapse.12
Surprisingly, transgenic (over)expression of PIK3CA was not associated with measurable changes in the phosphorylation or activity levels of AKT kinase or increased resistance to a wide spectrum of tested PI3K inhibitors or the BTKi ibrutinib in the majority of tested MCL cell lines. On the contrary, most of the tested MCL cell lines with PIK3CA overexpression were less sensitive to BH3 mimetics. Molecular mechanisms underlying the observed increase in resistance to BH3 mimetics, however, remain elusive. Importantly, PIK3CA gain variably decreased the dependence of the tested MCL cell lines on prosurvival signaling from the BCR, decreased the levels of ox/phos, and increased resistance to 2-DG. Generally, these changes may suggest that PIK3CA gain contributes to increased survival of MCL cells, especially under hypoxia. Even though precise molecular mechanisms that mediate these observations remain largely elusive, unchanged activity and phosphorylation status of AKT suggest that consequences of PIK3CA gain are mediated independently of AKT. This is supported by a study in endometrial carcinoma. Holst et al showed that endometrial tumors with PIK3CA amplification were associated with aggressive behavior; however, had decreased levels of activated AKT at the same time.34 AKT-independent cancer-promoting effect was reported also for PIK3CA mutations in breast cancer.35
In contrast to PIK3CA (over)expression, knockout of PTEN (which mimics complete loss of PTEN protein/function) was associated with a distinct phenotype, including hyperphosphorylation and overactivation of AKT kinase, increased resistance to most of the tested PI3K inhibitors, resistance to the BTKi ibrutinib and BCL2 inhibitor VEN, markedly decreased dependence on prosurvival BCR signaling, and metabolic reprogramming of MCL cells with increased glycolytic rate and increased resistance to 2-DG. Importantly, sensitivity to AKT inhibitor capivasertib remained unchanged, which is consistent with data published thus far.36 Interestingly, sensitivity to BCL-XL inhibitor A1155463 was increased. The observations from the western blot analyses (ie, downregulation of BCL2 and BIM proteins in UPF1H PTEN KO and downregulation of BAK1 in MINO PTEN KO) and immunoprecipitation experiments (ie, decreased interaction of BIM with BCL2 in UPF1H PTEN KO and decreased amount of BAX on BCL2 in MINO PTEN KO) may at least partially explain increased resistance of PTEN KO cell lines to VEN. However, molecular mechanisms underlying the observed increased sensitivity to BCL-XL inhibition remain elusive.
Of note, knockdown of PTEN (which mimics partial loss of PTEN protein/function) was associated with slight but significant increase in AKT activity, lower AKT activity reduction in response to ibrutinib, and increased resistance to copanlisib, ibrutinib, VEN, and S63854. This is important because our data showed that even partial loss of PTEN protein negatively correlated with sensitivity to BTKis. PTEN might thus represent an important predictive factor of sensitivity to BTKi. However, this correlation must be confirmed on primary MCL samples before and after failure of BTKi therapy.
We proved that both aberrations (ie, gain of PIK3CA and loss of PTEN) might lead to metabolic reprogramming of MCL cells, which results in significant changes in the rates of glycolysis and ox/phos, increased resistance to 2-DG, and increased survival under hypoxia. Previous studies demonstrated that hypoxia adaptation was associated with allelic loss or inactivation of PTEN.37,38 Conversely, loss of PTEN facilitated HIF1–mediated gene expression.39-41 Metabolic reprogramming belongs to hallmarks of cancer.42-44 Overactivation of PI3K pathway promotes glycolysis in an AKT-dependent and -independent manner. One example of AKT-independent activation of glycolysis is the effect of the above-mentioned PIK3CA mutation.45,46 Together with the observed decreased dependence on the prosurvival signaling from BCR, we propose that both PTEN loss and PIK3CA gain foster survival of MCL cells under hypoxia.
Conclusion
Our results suggest that frequent aberrations of the PI3K pathway observed in MCL may rewire associated signaling with lower dependence on BCR signaling, better metabolic and hypoxic adaptation, and targeted therapy resistance.
Acknowledgments
The authors thank all the patients who generously donated samples for this study. The authors acknowledge Imaging Methods Core Facility at the Biotechnology Center in Vestec supported by the Ministry of Education, Youth and Sports of the Czech Republic (LM2023050 Czech-BioImaging), for their support in this work. Graphical abstract was created with BioRender.com.
The manuscript was supported by Ministry of Health of the Czech Republic (grant AZV NU21-03-00386, all rights reserved), National Institute for Cancer Research- Programme EXCELES (ID project number LX22NPO5102), funded by the European Union, Next Generation EU, MH CZ DRO VFN 64165, and the Czech Science Foundation (20-01969Y). This work was also supported by a grant from The Leukemia & Lymphoma Society (grant ID MCL 7005-24), a COOPERATIO Program, research area “Hematooncology,” and Charles University graduate students research program SVV 260634/2023, and SVV 260637.
Authorship
Contribution: P.K. and O.H. designed the research, analyzed data, and supervised the preparation of the manuscript; N.B. performed most of the research and wrote the manuscript; J.S., K.K., A.R., and V.H. derived genetically modified cell lines, performed measurements of AKT activity, and performed experiments with the BCR KO; L.A. and D.S. implemented Seahorse analyses; K.S., A.B., and Z.Z. performed FISH analyses; L.D. and A.D. performed hypoxic experiments; and M.T. and R.E.D. analyzed data and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pavel Klener, Institute of Pathological Physiology, First Faculty of Medicine, Charles University, U Nemocnice 5, Praha 2, 128 53 Prague, Czech Republic; email: pavel.klener2@lf1.cuni.cz.
References
Author notes
Data are available upon reasonable request from the corresponding author, Pavel Klener (pavel.klener2@lf1.cuni.cz).
The full-text version of this article contains a data supplement.