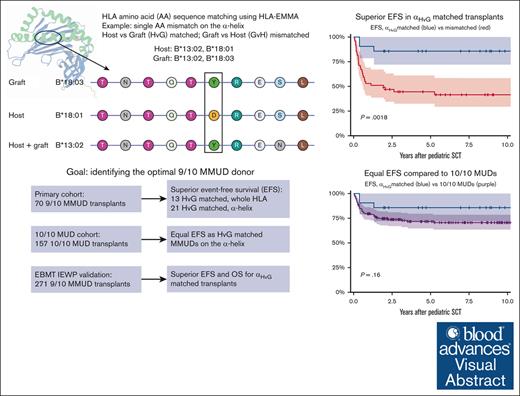
Key Points
Using HLA-EMMA, a subgroup of 9/10 HLA-mismatched transplants with a permissible mismatch can be identified with favorable EFS.
Permissibly mismatched transplants have EFS rates similar to those of 10/10 MUD transplants.
Visual Abstract
Allogeneic hematopoietic stem cell transplantation (HSCT) with mismatched unrelated donors (MMUD) is associated with inferior outcome compared with matched unrelated donors (MUDs). We aimed to identify permissible mismatches using HLA epitope mismatch algorithm, which determines permissibility by analyzing amino acid sequences, in a single-center cohort of 70 pediatric 9/10 MMUD HSCTs and 157 10/10 MUDs for comparison. Amino acid matching was evaluated for the whole HLA protein, the α-helices, and the β-sheets, in both host vs graft (HvG) and graft vs host (GvH) direction. Superior event-free survival (EFS) was found in 13 patients permissibly mismatched in the HvG direction (totalHvG, 92% vs 58% at 1 year; P = .009) and in 21 patients matched on the α-helices (αHvG, 90% vs 53%; P = .002). These rates were similar to EFS rates in patients with 10/10 MUDs (90% vs 80%; P = .60). EFS was not related to β-sheet amino acid matching, nor to matching in the GvH direction. Overall survival (OS) rates trended similarly to those of EFS for amino acid mismatches (totalHvG, 92% vs 74%; P = .075; αHvG, 90% vs 71%; P = .072). These findings were reproduced in an EBMT Registry inborn errors cohort of 271 pediatric 9/10 MMUD HSCTs and 929 10/10 MUD HSCTs, showing a significant effect of αHvG matching on both OS and EFS and similar OS and EFS between αHvG matched MMUDs and 10/10 MUDs. In summary, HvG amino acid matching on the α-helices identifies 9/10 MMUDs with permissible mismatches, which are correlated with favorable transplant outcomes similar to those of matched donors.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) offers an opportunity for treatment of a broad spectrum of malignant and nonmalignant diseases in children. HLA matching is an important factor for overall survival (OS) and event-free survival (EFS) after HSCT.1,2 If a matched family donor is lacking, a 10/10 matched unrelated donor (MUD) is often considered the second best option.3 However, in a substantial part of HSCT candidates, especially those of non-European origin, only HLA-mismatched unrelated donors (MMUD) will be available.4 These HLA mismatches often lead to alloimmune responses including acute and chronic graft-versus-host disease (GVHD) associated with a worse outcome. However, not every HLA mismatch is expected to have the same impact. Therefore, improving the process of donor selection for MMUDs by identifying permissible HLA mismatches could improve clinical outcomes.
HLA mismatches vary in immunogenicity and associated effects on clinical outcome. Some mismatches do not elicit allorecognition and the adverse outcomes associated with it, and are therefore deemed permissible HLA mismatches. Permissible mismatches in the HSCT setting, such as HLA-C∗03:03/03:04 and HLA-DRB1∗14:01/14:54, have been identified by studying their correlation with clinical outcome in cohort studies.5-7 However, if a specific mismatch occurs infrequently, it is difficult to establish its effect on clinical outcome in cohort studies.
Matching algorithms that in silico assess potential immunogenicity of an HLA mismatch have shown promising results, especially in renal transplantation.8,9 In renal transplantation, de novo donor-specific antibody production is more frequent with increasing numbers of molecular HLA mismatches, and strongly impacts graft rejection and survival.10 In HSCT, acute GVHD (aGVHD) is a major complication occurring in over 30% of HSCTs, in which donor T-cell activation by host HLA through the direct pathway of allorecognition is considered to play a key role.11,12 The reverse may also occur, resulting in graft rejection. Therefore, HLA mismatches should be assessed in both host vs graft (HvG) and graft vs host (GvH) direction in HSCT.
Another tool to assess potential immunogenicity in silico is the PIRCHE (Predicted Indirectly ReCognizable HLA Epitopes) algorithm. PIRCHE predicts the number of mismatched peptides derived from patient HLA that could be presented in donor HLA class I (PIRCHE-I score) or HLA class II (PIRCHE-II score), and may predict indirect T-cell allorecognition as a result of a certain HLA mismatch.13 Using the current HSCT module, PIRCHE only predicts reactivity in the GvH direction.
PIRCHE has been used by Stenger et al, who included 20 pediatric patients with a 9/10 MMUD and 85 patients with either a matched sibling donor or an MUD.14 They concluded that a high PIRCHE-I score is related to increased risk of aGVHD. In contrast, HLAMatchmaker, an algorithm that has proven to be relevant in solid organ transplantation, did not predict OS or EFS in large HSCT cohorts.9,15-17
The HLA protein contains a peptide-binding groove flanked by α-helices running in parallel. The β-sheets are positioned directly below the peptide-binding groove.18,19 The α-helices and β-sheets determine not only the peptide repertoire presented by the HLA protein but also direct and indirect allorecognition by T cells.20 Many reported immunogenic amino acid mismatches are part of an α-helix or a β-sheet (eg, the amino acids at position 77 and 156 [α] and 9 and 116 [β]).21-23
HLA epitope mismatch algorithm (HLA-EMMA) is a tool to analyze HLA compatibility on the amino acid level, and was initially developed for analysis of molecular HLA matching in the setting of solid organ transplantation.24 In a recent beta version, new options were added to assess AA mismatches in both the GvH and HvG directions, and to identify which AA mismatches are part of the α-helices or β-sheets.
In a pediatric cohort of MMUD SCT, we investigated whether amino acid matching determined by HLA-EMMA can identify permissible HLA mismatches with favorable clinical outcome in 9/10 MMUD transplants, and compared it with results in 10/10 MUD transplants. A second cohort of pediatric MMUD and MUD transplants for inborn errors was used to validate the findings.
Materials and methods
Main cohort
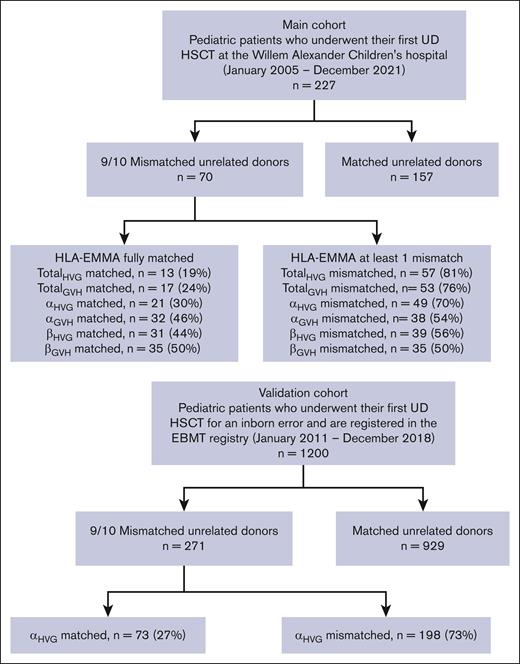
The main study cohort was a retrospective single-center cohort containing pediatric patients, treated with a first HSCT from a 9/10 MMUD with a bone marrow or peripheral blood stem cell graft at the Willem Alexander Children’s hospital (WACH) in Leiden (The Netherlands) between January 2005 and December 2021. A total of 70 patients were included. As a reference group, 157 patients were included with a 10/10 MUD donor, who otherwise met the same inclusion and exclusion criteria (Figure 1).
Flowchart of the inclusion and exclusion of the patient and control groups.
GVHD prophylaxis for mismatched patients was based on methotrexate and a calcineurin inhibitor, most commonly Ciclosporin A. All patients received serotherapy with either alemtuzumab or antithymocyte globulin.
Posttransplant cyclophosphamide (PT-Cy) cohort
Patients receiving cyclophosphamide after 9/10 MMUD transplantation at the WACH were excluded from our main analysis and analyzed as a separate cohort to verify consistency of our main findings in these patients (n = 26, underwent transplantation between December 2017 and June 2023).
Validation cohort
The validation cohort consisted of a cohort extracted from the EBMT Registry, treated with a first HSCT from a 9/10 MMUD with an unmanipulated bone marrow graft between January 2011 and January 2019 for an inborn error of immunity, hematopoiesis, or metabolism. Patients treated at WACH were excluded from this cohort to avoid double inclusion. Patients treated with cyclophosphamide after transplantation (2011-2021; n = 11) were excluded. A total of 271 patients were included. As a reference group, 929 patients with a 10/10 MUD, who otherwise met the same inclusion and exclusion criteria, were included.
All patients and/or patients’ legal guardians have given informed consent for the collection of their data and use for scientific research. The Medical Ethics Committee of the Leiden University Medical Center released a non-WMO waiver and approved the data collection (medical ethical committee reference B17.001/SH/sh).
Study end points
The primary end point was EFS, defined as survival without relapse, subsequent transplant, boost, or autologous reinfusion. Relapse was defined as the recurrence of a malignancy that was previously in remission, and was not considered for nonmalignant indications because these had no uniform irreversible definition of relapse. Secondary end points were OS, the occurrence of Glucksberg grade 2 to 4 aGVHD, neutrophil engraftment, and chimerism. Neutrophil engraftment was defined as a neutrophil count ≥0.5 × 109/L after HSCT.
HLA typing
For the main cohort, patient-donor pairs were DNA typed at the HLA laboratory of the Leiden University Medical Center for HLA-A, -B, -C, -DRB1, and -DQB1. Samples between 2009 and 2019 were typed by Sanger sequencing (GenDx, Genome Diagnostics BV, Utrecht, The Netherlands) with AB 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA). Samples of 2004 to 2009, or those from the 2009 to 2019 period with missing typing for a locus, were retyped to ensure unambiguous second-field HLA typing. These samples and samples from 2019 onward were typed with next-generation sequencing (GenDx) and the MiniSeq System (Illumina, San Diego, CA). Data were analyzed using the SBTengine (GenDx) and the NGSengine software program (GenDx).
For the validation cohort, no information on typing strategy was available. We processed both high-resolution and intermediate-resolution typing, with multiple allele codes (MAC), P- and G-groups, and slash-delimited typing being processed. For intermediate-resolution typing, the most probable HLA allele was assumed to be present, using HaploStats (National Marrow Donor Program) for haplotype frequencies.
Mismatch analyses
HLA-EMMA version 2.0 beta was used to determine the number of amino acid mismatches for every patient-donor pair. This software version has the ability to identify if amino acid mismatches are part of an α-helix or β-sheet, and contains a module specifically designed for (in)compatibility analysis in stem cell transplants. This module performs analyses in both the HvG and GvH direction. We considered a patient as mismatched if there was ≥1 amino acid mismatch, and only considered a patient matched if no mismatching amino acid were present. We analyzed amino acid matching on the whole HLA protein (designated “totalGvH” and “totalHvG”), the α-helices only (designated “αGvH” and “αHvG”), and the β-sheet only (designated “βGvH” and “βHvG”).
To compare against existing matching approaches, we also evaluated amino acid positions on HLA class I that have previously been reported to be potentially immunogenic,21,22 those forming the peptide-binding groove,20 and the PIRCHE-I and PIRCHE-II scores, with cutoffs of 0 and cutoffs as described in the literature where available.25
Statistical analyses
We analyzed survival and engraftment using Kaplan-Meier curves, and tested for differences using a log-rank test. To adjust for relevant covariates, we used Cox proportional hazards regression. Differences in aGVHD were tested using the Fisher exact test. Analysis was done in R 4.3.2 (R Foundation for Statistical Computing), using the packages survival, stats, gtsummary, and mstate.
To analyze chimerism continuously, for each chimerism measurement we determined the lowest level of either peripheral blood mononuclear cells or granulocytes and categorized it as loss of chimerism (≤5% donor cells), mixed chimerism (5%-95% donor cells), and full donor chimerism (>95% donor cells). We then developed a multistate model to determine the percentage of patients with mixed chimerism while accounting for death, relapse, and subsequent transplants, and allowing patients to regain full donor chimerism after having mixed chimerism (supplemental Figure 1).
Within the validation cohort, we verified the most promising matching strategy as discovered in the main cohort. OS, EFS, and aGVHD rates were evaluated. No multivariate adjustment was performed.
Results
Patient characteristics
A total of 70 MMUD transplants were included in the main cohort. Most patients underwent transplantation for a hematological malignancy (54%), whereas hemoglobinopathies (19%), inborn errors of immunity (17%), and bone marrow failure syndromes (10%) were less common. Most transplants were performed using bone marrow grafts (73%) and myeloablative conditioning (91%). Baseline characteristics were similar between amino acid matching groups (all P values >.1; Table 1). A total of 157 patients with MUD transplants were added for comparison, with a significantly lower donor age, more frequent use of unmanipulated bone marrow as a stem cell source, a trend toward more recent transplants, and otherwise similar characteristics.
Patient characteristics
| Characteristic . | MUD vs MMUD . | P value . | TotalHvG matching . | P value . | αHvG matching . | P value . | |||
|---|---|---|---|---|---|---|---|---|---|
| MUD, n = 157 . | MMUD, n = 70 . | Mismatched, n = 57 . | Matched, n = 13 . | Mismatched, n = 49 . | Matched, n = 21 . | ||||
| Diagnosis | .087 | .24 | .31 | ||||||
| Hematological (pre)malignancy | 67 (43%) | 38 (54%) | 32 (56%) | 6 (46%) | 26 (53%) | 12 (57%) | |||
| Bone marrow failure | 36 (23%) | 7 (10%) | 4 (7.0%) | 3 (23%) | 3 (6.1%) | 4 (19%) | |||
| Hemoglobinopathy | 23 (15%) | 13 (19%) | 12 (21%) | 1 (7.7%) | 11 (22%) | 2 (9.5%) | |||
| Inborn errors of immunity | 31 (20%) | 12 (17%) | 9 (16%) | 3 (23%) | 9 (18%) | 3 (14%) | |||
| Patient age, y | 7.8 (2.6-13.3) | 9.8 (3.4-14.1) | .27 | 8.6 (3.1-13.7) | 13.1 (7.2-14.1) | 0.19 | 8.6 (2.6-13.3) | 12.7 (3.7-14.1) | .20 |
| Year of transplantation | .062 | 1 | .60 | ||||||
| 2005-2011 | 66 (42%) | 39 (56%) | 32 (56%) | 7 (54%) | 26 (53%) | 13 (62%) | |||
| 2012-2021 | 91 (58%) | 31 (44%) | 25 (44%) | 6 (46%) | 23 (47%) | 8 (38%) | |||
| Donor age (cord excluded) | 30 (24-39) | 37 (28-47) | .00022 | 40 (28-47) | 32 (24-45) | .11 | 40 (28-46) | 34 (25-47) | .46 |
| Conditioning strategy | .58 | .31 | .36 | ||||||
| Myeloablative conditioning | 147 (94%) | 64 (91%) | 53 (93%) | 11 (85%) | 46 (94%) | 18 (86%) | |||
| Reduced intensity conditioning | 10 (6.4%) | 6 (8.6%) | 4 (7.0%) | 2 (15%) | 3 (6.1%) | 3 (14%) | |||
| Stem cell source | .0030 | .32 | 1 | ||||||
| BM | 140 (89%) | 51 (73%) | 43 (75%) | 8 (62%) | 36 (73%) | 15 (71%) | |||
| PBSC | 17 (11%) | 19 (27%) | 14 (25%) | 5 (38%) | 13 (27%) | 6 (29%) | |||
| CMV status (patient/donor) | .29 | .44 | .29 | ||||||
| –/– | 57 (37%) | 18 (26%) | 16 (28%) | 2 (15%) | 13 (27%) | 5 (24%) | |||
| –/+ | 19 (12%) | 13 (19%) | 9 (16%) | 4 (31%) | 8 (16%) | 5 (24%) | |||
| +/− | 28 (18%) | 16 (23%) | 12 (21%) | 4 (31%) | 9 (18%) | 7 (33%) | |||
| +/+ | 52 (33%) | 23 (33%) | 20 (35%) | 3 (23%) | 19 (39%) | 4 (19%) | |||
| Unknown | 1 | 0 | |||||||
| Graft manipulation | .0025 | .23 | .63 | ||||||
| Alemtuzumab ex vivo | 0 (0%) | 5 (7.1%) | 3 (5.3%) | 2 (15%) | 3 (6.1%) | 2 (9.5%) | |||
| None | 157 (100%) | 65 (93%) | 54 (95%) | 11 (85%) | 46 (94%) | 19 (90%) | |||
| Characteristic . | MUD vs MMUD . | P value . | TotalHvG matching . | P value . | αHvG matching . | P value . | |||
|---|---|---|---|---|---|---|---|---|---|
| MUD, n = 157 . | MMUD, n = 70 . | Mismatched, n = 57 . | Matched, n = 13 . | Mismatched, n = 49 . | Matched, n = 21 . | ||||
| Diagnosis | .087 | .24 | .31 | ||||||
| Hematological (pre)malignancy | 67 (43%) | 38 (54%) | 32 (56%) | 6 (46%) | 26 (53%) | 12 (57%) | |||
| Bone marrow failure | 36 (23%) | 7 (10%) | 4 (7.0%) | 3 (23%) | 3 (6.1%) | 4 (19%) | |||
| Hemoglobinopathy | 23 (15%) | 13 (19%) | 12 (21%) | 1 (7.7%) | 11 (22%) | 2 (9.5%) | |||
| Inborn errors of immunity | 31 (20%) | 12 (17%) | 9 (16%) | 3 (23%) | 9 (18%) | 3 (14%) | |||
| Patient age, y | 7.8 (2.6-13.3) | 9.8 (3.4-14.1) | .27 | 8.6 (3.1-13.7) | 13.1 (7.2-14.1) | 0.19 | 8.6 (2.6-13.3) | 12.7 (3.7-14.1) | .20 |
| Year of transplantation | .062 | 1 | .60 | ||||||
| 2005-2011 | 66 (42%) | 39 (56%) | 32 (56%) | 7 (54%) | 26 (53%) | 13 (62%) | |||
| 2012-2021 | 91 (58%) | 31 (44%) | 25 (44%) | 6 (46%) | 23 (47%) | 8 (38%) | |||
| Donor age (cord excluded) | 30 (24-39) | 37 (28-47) | .00022 | 40 (28-47) | 32 (24-45) | .11 | 40 (28-46) | 34 (25-47) | .46 |
| Conditioning strategy | .58 | .31 | .36 | ||||||
| Myeloablative conditioning | 147 (94%) | 64 (91%) | 53 (93%) | 11 (85%) | 46 (94%) | 18 (86%) | |||
| Reduced intensity conditioning | 10 (6.4%) | 6 (8.6%) | 4 (7.0%) | 2 (15%) | 3 (6.1%) | 3 (14%) | |||
| Stem cell source | .0030 | .32 | 1 | ||||||
| BM | 140 (89%) | 51 (73%) | 43 (75%) | 8 (62%) | 36 (73%) | 15 (71%) | |||
| PBSC | 17 (11%) | 19 (27%) | 14 (25%) | 5 (38%) | 13 (27%) | 6 (29%) | |||
| CMV status (patient/donor) | .29 | .44 | .29 | ||||||
| –/– | 57 (37%) | 18 (26%) | 16 (28%) | 2 (15%) | 13 (27%) | 5 (24%) | |||
| –/+ | 19 (12%) | 13 (19%) | 9 (16%) | 4 (31%) | 8 (16%) | 5 (24%) | |||
| +/− | 28 (18%) | 16 (23%) | 12 (21%) | 4 (31%) | 9 (18%) | 7 (33%) | |||
| +/+ | 52 (33%) | 23 (33%) | 20 (35%) | 3 (23%) | 19 (39%) | 4 (19%) | |||
| Unknown | 1 | 0 | |||||||
| Graft manipulation | .0025 | .23 | .63 | ||||||
| Alemtuzumab ex vivo | 0 (0%) | 5 (7.1%) | 3 (5.3%) | 2 (15%) | 3 (6.1%) | 2 (9.5%) | |||
| None | 157 (100%) | 65 (93%) | 54 (95%) | 11 (85%) | 46 (94%) | 19 (90%) | |||
The data presented as n (%), with median (interquartile range) and used Fisher exact test for categorical data and Wilcoxon rank sum test for continuous data.
BM, bone marrow; CMV, cytomegalovirus; PBSC, peripheral blood stem cell.
HLA-EMMA scores in the HvG direction are associated with improved EFS
HLA-EMMA classified 13 patients (19%) as totalHvG matched and 17 (24%) as totalGvH matched; 21 patients (30%) were αHvG matched and 32 (45%) were αGvH matched, whereas 31 (44%) were βHvG matched and 35 (50%) βGvH matched (Figure 1).
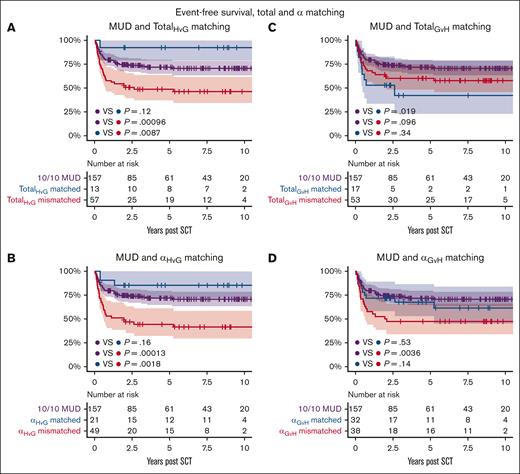
EFS was 64% at 1 year and 57% at 5 years for patients receiving an MMUD transplant. Events included death in 11, relapse of malignancy in 10, and a subsequent transplantation due to graft failure in 10 cases. amino acid matching in the HvG direction strongly impacted EFS, with a 5-year EFS of 92% for totalHvG matched and 49% for totalHvG mismatched patients (P = .0087). In the GvH direction, 5-year EFS was similar for matched and mismatched transplants (42% for totalGvH matched vs 60% for totalGvH mismatched; P = .34; Table 2; Figure 2).
OS and EFS split by different matching categories
| . | 1-y OS . | 2-y OS . | P value . | 1-y EFS . | 2-y EFS . | P value . |
|---|---|---|---|---|---|---|
| TotalHvGmatching | ||||||
| mismatched (n = 57) | 74% | 67% | .075 | 58% | 53% | .0087 |
| matched (n = 13) | 92% | 92% | 92% | 92% | ||
| TotalGvHmatching | ||||||
| mismatched (n = 53) | 79% | 75% | .21 | 68% | 62% | .34 |
| matched (n = 17) | 71% | 59% | 53% | 53% | ||
| αHvGmatching | ||||||
| mismatched (n = 49) | 71% | 65% | .072 | 53% | 49% | .0018 |
| matched (n = 21) | 90% | 86% | 90% | 86% | ||
| αGvHmatching | ||||||
| mismatched (n = 38) | 74% | 68% | .49 | 58% | 50% | .14 |
| matched (n = 32) | 81% | 75% | 72% | 72% | ||
| βHvGmatching | ||||||
| mismatched (n = 39) | 77% | 72% | .91 | 62% | 54% | .15 |
| matched (n = 31) | 77% | 71% | 68% | 68% | ||
| βGvHmatching | ||||||
| mismatched (n = 35) | 77% | 71% | .97 | 60% | 51% | .49 |
| matched (n = 35) | 77% | 71% | 69% | 69% | ||
| 10/10 MUD (n = 157) | 86% | 84% | 80% | 75% |
| . | 1-y OS . | 2-y OS . | P value . | 1-y EFS . | 2-y EFS . | P value . |
|---|---|---|---|---|---|---|
| TotalHvGmatching | ||||||
| mismatched (n = 57) | 74% | 67% | .075 | 58% | 53% | .0087 |
| matched (n = 13) | 92% | 92% | 92% | 92% | ||
| TotalGvHmatching | ||||||
| mismatched (n = 53) | 79% | 75% | .21 | 68% | 62% | .34 |
| matched (n = 17) | 71% | 59% | 53% | 53% | ||
| αHvGmatching | ||||||
| mismatched (n = 49) | 71% | 65% | .072 | 53% | 49% | .0018 |
| matched (n = 21) | 90% | 86% | 90% | 86% | ||
| αGvHmatching | ||||||
| mismatched (n = 38) | 74% | 68% | .49 | 58% | 50% | .14 |
| matched (n = 32) | 81% | 75% | 72% | 72% | ||
| βHvGmatching | ||||||
| mismatched (n = 39) | 77% | 72% | .91 | 62% | 54% | .15 |
| matched (n = 31) | 77% | 71% | 68% | 68% | ||
| βGvHmatching | ||||||
| mismatched (n = 35) | 77% | 71% | .97 | 60% | 51% | .49 |
| matched (n = 35) | 77% | 71% | 69% | 69% | ||
| 10/10 MUD (n = 157) | 86% | 84% | 80% | 75% |
Survival percentages are extracted from Kaplan-Meier curves, with P values corresponding to the log-rank test for differences between these curves.
Probability of EFS between MUDs, amino acid matched and mismatched transplants for AA matching approaches. The AA matching approaches are: (A) TotalHvG matching, (B) αHvG matching, (C) TotalGvH matching, and (D) αGvH matching. SCT, stem cell transplantation.
Probability of EFS between MUDs, amino acid matched and mismatched transplants for AA matching approaches. The AA matching approaches are: (A) TotalHvG matching, (B) αHvG matching, (C) TotalGvH matching, and (D) αGvH matching. SCT, stem cell transplantation.
Matching on the α-helices resulted in a 5-year EFS of 86% of the 21 αHvG matched patients vs 45% of the 49 mismatched patients (P = .0018). No difference was observed in the GvH direction (68% for αGvH matched and 47% for mismatched patients; P = .14). Matching on the β-sheets did not impact 5-year EFS in either the HvG direction (68% vs 48% for βHvG matched vs mismatched; P = .15) or the GvH direction (62% vs 51% for βGvH matched vs mismatched; P = .49). OS showed a trend similar to EFS but did not reach statistical significance in any matching group (Table 2; supplemental Figure 2).
These favorable EFS rates for totalHvG and αHvG remained significant in bivariate analysis with the diagnosis. Both totalHvG and αHvG matching were still strongly associated with EFS (hazard ratio [HR], 0.18; P = .034 and HR, 0.18; P = .0048, respectively), whereas they were not significantly associated with OS (HR, 0.21; P = .13 and HR, 0.33; P = .079; respectively). In bivariate analysis including year of transplantation, we observed an even stronger effect of amino acid matching, both for OS (totalHvG [HR, 0.19; P = .11]; αHvG [HR, 0.29; P = .048]) and EFS (totalHvG [HR, 0.11; P = .030]; αHvG [HR, 0.16; P = .0029]; Table 3), as year of transplantation appeared significantly and independently associated with OS and EFS in our center.
Bivariate Cox proportional hazards analyses for diagnosis and amino acid matching, and year and amino acid matching, for both OS and EFS
| . | Variables . | HR (95% CI) . | P value . |
|---|---|---|---|
| OS | TotalHvG matched | 0.21 (0.03-1.55) | .13 |
| Hematological (pre)malignancy | 1.91 (0.73-4.97)) | .19 | |
| αHvG matched | 0.33 (0.10-1.13) | .079 | |
| Hematological (pre)malignancy | 2.10 (0.81-5.48) | .13 | |
| TotalHvG matched | 0.19 (0.03-1.42) | .11 | |
| Year of transplantation >2011 | 0.26 (0.09-0.79) | .017 | |
| αHvG matched | 0.29 (0.08-0.99) | .048 | |
| Year of transplantation >2011 | 0.24 (0.08-0.71) | .010 | |
| EFS | TotalHvG matched | 0.18 (0.05-0.59) | .034 |
| Hematological (pre)malignancy | 1.55 (0.74-3.24) | .24 | |
| αHvG matched | 0.18 (0.05-0.59) | .0048 | |
| Hematological (pre)malignancy | 1.75 (0.84-3.66) | .14 | |
| TotalHvG matched | 0.11 (0.01-0.80) | .030 | |
| Year of transplantation >2011 | 0.45 (0.21-0.97) | .042 | |
| αHvG matched | 0.16 (0.05-0.54) | .0029 | |
| Year of transplantation >2011 | 0.39 (0.08-0.71) | .017 |
| . | Variables . | HR (95% CI) . | P value . |
|---|---|---|---|
| OS | TotalHvG matched | 0.21 (0.03-1.55) | .13 |
| Hematological (pre)malignancy | 1.91 (0.73-4.97)) | .19 | |
| αHvG matched | 0.33 (0.10-1.13) | .079 | |
| Hematological (pre)malignancy | 2.10 (0.81-5.48) | .13 | |
| TotalHvG matched | 0.19 (0.03-1.42) | .11 | |
| Year of transplantation >2011 | 0.26 (0.09-0.79) | .017 | |
| αHvG matched | 0.29 (0.08-0.99) | .048 | |
| Year of transplantation >2011 | 0.24 (0.08-0.71) | .010 | |
| EFS | TotalHvG matched | 0.18 (0.05-0.59) | .034 |
| Hematological (pre)malignancy | 1.55 (0.74-3.24) | .24 | |
| αHvG matched | 0.18 (0.05-0.59) | .0048 | |
| Hematological (pre)malignancy | 1.75 (0.84-3.66) | .14 | |
| TotalHvG matched | 0.11 (0.01-0.80) | .030 | |
| Year of transplantation >2011 | 0.45 (0.21-0.97) | .042 | |
| αHvG matched | 0.16 (0.05-0.54) | .0029 | |
| Year of transplantation >2011 | 0.39 (0.08-0.71) | .017 |
We analyzed a separate cohort including 23 patients receiving a 9/10 MMUD HSCT and treated with PT-Cy, wherein all patients had nonmalignant disease (supplemental Table 1). In this cohort, 7 patients were αHvG matched, and none of them experienced any events (OS and EFS = 100%), whereas 6 had a follow-up longer than 1 year. However, in the 16 αHvG mismatched patients, 1-year EFS was 67% (P = .10) and OS was 87% (P = .36). Because of the limited size of this cohort, no further analysis was done.
Secondary end points not associated with HLA-EMMA scores
In the main cohort, 13 patients (19%) experienced grade 2 to 4 aGVHD. We found no significant association between aGVHD and any categorization of amino acid mismatches. However, of the 13 patients without any amino acid mismatch in the HvG direction, none experienced grade 2 to 4 aGVHD, and for those without any amino acid mismatches on the α-helices, only 1 out of 21 patients experienced aGVHD. The overall rate of grade 2 to 4 aGVHD was similar between MMUDs (19%) and MUDs (21%; Table 4). Neutrophil engraftment was reached in nearly all patients, and was not influenced by amino acid matching (supplemental Figure 3).
Occurrence of aGVHD in the different amino acid matching categories and in 10/10 MUDs, both for the main and validation cohort
| . | Grade 0-1 aGVHD . | Grade 2-4 aGVHD . | P value aGVHD . |
|---|---|---|---|
| TotalHvGmatching | .11 | ||
| mismatched (n = 57) | 44 (77%) | 13 (23%) | |
| matched (n = 13) | 13 (100%) | 0 (0%) | |
| TotalGvHmatching | .28 | ||
| mismatched (n = 53) | 45 (85%) | 8 (15%) | |
| matched (n = 17) | 12 (71%) | 5 (29%) | |
| αHvGmatching | .09 | ||
| mismatched (n = 49) | 37 (76%) | 12 (24%) | |
| matched (n = 21) | 20 (95%) | 1 (5%) | |
| αGvHmatching | .76 | ||
| mismatched (n = 38) | 30 (79%) | 8 (21%) | |
| matched (n = 32) | 27 (84%) | 5 (16%) | |
| βHvGmatching | .12 | ||
| mismatched (n = 39) | 29 (74%) | 10 (26%) | |
| matched (n = 31) | 28 (90%) | 3 (10%) | |
| βGvHmatching | 1 | ||
| mismatched (n = 35) | 28 (80%) | 7 (20%) | |
| matched (n = 35) | 29 (83%) | 6 (17%) | |
| 10/10 matched controls (n = 157) | 123 (78%) | 34 (22%) | |
| Validation cohort, MUD vs MMUD | .22 | ||
| 10/10 MUD (n = 894, 35 missing) | 621 (69%) | 273 (31%) | |
| 9/10 MMUD (n = 257, 14 missing) | 168 (65%) | 89 (35%) |
| . | Grade 0-1 aGVHD . | Grade 2-4 aGVHD . | P value aGVHD . |
|---|---|---|---|
| TotalHvGmatching | .11 | ||
| mismatched (n = 57) | 44 (77%) | 13 (23%) | |
| matched (n = 13) | 13 (100%) | 0 (0%) | |
| TotalGvHmatching | .28 | ||
| mismatched (n = 53) | 45 (85%) | 8 (15%) | |
| matched (n = 17) | 12 (71%) | 5 (29%) | |
| αHvGmatching | .09 | ||
| mismatched (n = 49) | 37 (76%) | 12 (24%) | |
| matched (n = 21) | 20 (95%) | 1 (5%) | |
| αGvHmatching | .76 | ||
| mismatched (n = 38) | 30 (79%) | 8 (21%) | |
| matched (n = 32) | 27 (84%) | 5 (16%) | |
| βHvGmatching | .12 | ||
| mismatched (n = 39) | 29 (74%) | 10 (26%) | |
| matched (n = 31) | 28 (90%) | 3 (10%) | |
| βGvHmatching | 1 | ||
| mismatched (n = 35) | 28 (80%) | 7 (20%) | |
| matched (n = 35) | 29 (83%) | 6 (17%) | |
| 10/10 matched controls (n = 157) | 123 (78%) | 34 (22%) | |
| Validation cohort, MUD vs MMUD | .22 | ||
| 10/10 MUD (n = 894, 35 missing) | 621 (69%) | 273 (31%) | |
| 9/10 MMUD (n = 257, 14 missing) | 168 (65%) | 89 (35%) |
Mixed chimerism was detectable in 14% of patients at 1 year and 11% at 5 years. Although EFS was influenced by amino acid matching, the percentage of patients with mixed chimerism was not impacted, with mixed chimerism occurring in 23% of totalHvG matched patients at 1 year and 15% at 5 years, whereas these rates were 12% and 10% for totalHvG mismatched patients at 1 and 5 years, respectively. Comparable results were observed for αHvG matching and for matching in the GvH direction (Figure 3).
Multistate model describing the occurrence of mixed chimerism split by amino acid matching, while allowing patients to recover from mixed chimerism and accounting for competing events. Subanalyses for (A) TotalHvG matched (B) TotalHvG mismatched, (C) αHvG matched, (D) αHvG mismatched, (E) TotalGvH matched, (F) TotalGvH mismatched, (G) αGvH matched, and (H) αGvH mismatched patients.
Multistate model describing the occurrence of mixed chimerism split by amino acid matching, while allowing patients to recover from mixed chimerism and accounting for competing events. Subanalyses for (A) TotalHvG matched (B) TotalHvG mismatched, (C) αHvG matched, (D) αHvG mismatched, (E) TotalGvH matched, (F) TotalGvH mismatched, (G) αGvH matched, and (H) αGvH mismatched patients.
PIRCHE and potentially immunogenic amino acid positions do not identify permissible mismatches in MMUD transplants
To determine whether alternative approaches could determine permissible mismatches in the setting of MMUD transplants, we also investigated the PIRCHE scores, amino acid positions reported to potentially be immunogenic in previous literature,20 and amino acid positions involved in the HLA-I peptide–binding groove as defined by Garrett et al,26 with most of these positions being within an α-helix. Both PIRCHE scores and mismatches on potentially immunogenic amino acid positions were not associated with differences in OS, EFS, or aGVHD. Amino acid matching on the peptide-binding groove in the HvG direction was associated with improved EFS (P = .008) and aGVHD (P = .044; supplemental Table 2).
Amino acid matched MMUD transplants showed OS and EFS comparable to those of MUD transplants
When comparing against transplants with an MUD, MMUD transplants with a totalHvG mismatch had inferior OS and EFS (P < .001 for both), whereas those with a totalHvG match had similar OS and EFS (P = .12 and P = .16, respectively). Similarly, when considering only mismatches on the α-helices, αHvG mismatched transplants had inferior OS and EFS (P = .004; P < .001), whereas αHvG matched transplants had similar OS and EFS compared with MUD transplants (P = .60; P = .16; supplemental Figure 4).
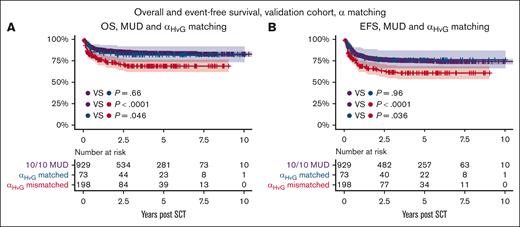
αHvG matched transplants had favorable OS and EFS in the validation cohort
The main findings were validated against an EBMT cohort of pediatric patients who underwent transplantation for an inborn error, with 271 9/10 MMUD transplants and 929 10/10 MUD transplants. A total of 73 patients (27%) were αHvG matched, with superior 5-year EFS (76% vs 61%; P = .036) and OS (82% vs 69%; P = .046). OS and EFS for αHvG matched transplants were similar to those of 10/10 MUD transplants (P = .96 and P = .66, respectively), whereas αHvG mismatched transplants had inferior OS and EFS (P < .0001 for both; Figure 4). The rate of grade 2 to 4 aGVHD was not statistically significantly different between 9/10 MMUD and 10/10 MUD transplants (35% vs 31%; P = .22; Table 4), and was not analyzed further.
Discussion
In our pediatric HSCT main cohort, we found that HLA-EMMA identified permissible HLA mismatches in 30% of 9/10 MMUD transplants. These transplants were αHvG matched, meaning that in the HvG direction all amino acids on the α-helices were matched. They had similar OS and EFS compared with MUD transplants, and significantly superior EFS compared with those with an αHvG mismatch.
This finding was fully consistent between the main and validation cohort, although these cohorts differed substantially regarding disease (all diseases vs inborn errors), HLA typing (high vs intermediate resolution), and aGVHD rate (19% vs 35%). Within the validation cohort, the disparity in EFS and OS between 10/10 MUDs and 9/10 MMUDs was smaller than in the main cohort, also leading to a smaller window of effect of αHvG matching. This might reflect the intermediate HLA typing for many patients and donors in the validation cohort. It has been shown that higher-resolution HLA typing can identify a substantial number of additional mismatches that still impact outcome.27
Comparable outcomes were observed with totalHvG matching, in which all amino acids on the HLA protein are matched. However, this limited the number of patients being permissibly matched (19 % for totalHvG matching vs 30% for αHvG matching). Therefore, by matching on the α-helices, a large group of 9/10 MMUDs can be identified that could lead to equal clinical outcome as 10/10 MUDs.
HLA-EMMA considers both HLA proteins per locus of donor and of patient to identify matched amino acids. For example, if a patient has an HLA-B∗18:03 and an HLA-B∗13:02, and the donor has an HLA-B∗18:01 and an HLA-B∗13:02, HLA-EMMA identifies this combination of HLA proteins as matched in the HvG direction. If no HLA-B∗13:02 is present in patient and donor, HLA-EMMA identifies this combination of HLA-B∗18 proteins as bidirectionally mismatched. This example illustrates that both the matched and the mismatched allele are used by HLA-EMMA to determine if a specific amino acid difference is considered permissible and thus matched (supplemental Table 3).
In our cohort, HvG matching on the amino acid level resulted in superior EFS compared with HvG mismatching, in contrast to GvH (mis)matching. The events that define EFS included relapse, second transplants due to poor graft functioning and graft failure, and mortality. For each of these events, host immunity against the graft could be a key driver. At the same time, HvG mismatches do not carry the potential benefit of a graft-versus-leukemia effect because they likely do not contribute to allorecognition of host malignant cells by the graft. However, we caution against extrapolation of our results to an adult patient population given the contradictory outcomes in different patient cohorts.28,29
The contribution of HvG vs GvH mismatches to clinical outcome is not clear from previous studies. It has been described that HvG matched cord blood transplants have superior nonrelapse mortality in children.30,31 A study on bone marrow and peripheral blood stem cell grafts found that unidirectionally GvH matched transplants resulted in more favorable outcome than unidirectionally HvG matched transplants. However, most of the patients were adult, and there was no subgroup analysis for pediatric patients.28 In addition, in a Japanese registry cohort, unidirectionally HvG mismatched and GvH mismatched unrelated donor (UD) transplants had equally favorable survival, whereas bidirectionally mismatched UD transplants had worse survival.29
In our center, donors with a unidirectional GvH match were preferred for donor selection when otherwise equally valid donors were available. This preference is reflected in the number of patients having a unidirectional match in the GvH direction vs the number of patients having a unidirectional match in the HvG direction (12 vs 5; supplemental Table 4). This selection bias implies that a donor selection strategy based on αHvG matching may result in a higher percentage of αHvG matched transplants than the 30% achieved in our cohort.
Alternative matching approaches such as PIRCHE, peptide-binding motif matching, and HLAMatchmaker have identified a correlation between GvH mismatches and an increased risk of aGVHD.14,15,25 These strategies all depend on different working mechanisms and hypothesized biology (Table 5). However, we did not observe an effect on aGVHD for PIRCHE or amino acid matching. In both the main and the validation cohort, there was no significant difference in incidence of aGVHD between MUDs and MMUDs, indicating that the effect of a single HLA mismatch in the evaluated loci, if present, is minor. This may be different for adult patients given that there is strong evidence in adult cohorts that mismatches lead to increased risk of aGVHD, which may be abrogated by identifying permissible mismatches.
Overview of different permissible matching strategies
| Name . | Strategy . | Possible biology . |
|---|---|---|
| HLAMatchmaker (Duquesnoy et al16,17) | Identification of amino acid triplets present in HLA that are mismatched at known accessible/polymorphic positions. Special attention to triplets confirmed to have been binding sites for donor-specific antibodies. Interlocus comparisons are performed. | Triplets accessible on the cell surface (eplets/epitopes) may lead to donor-specific antibody formation and solid organ transplant rejection. |
| PIRCHE (Stenger et al14) | Identification of mismatching peptide fragments derived from HLA, which can also be presented in HLA class I (PIRCHE-I) or class II (PIRCHE-II). | Presentation of HLA-derived nonself peptides may lead to indirect allorecognition and both aGVHD and rejection. |
| PBM matching (Crivello et al32) | Identification of mismatching peptide binding motif (PBM) categories, where PBM categories have been defined using hierarchical clustering on the peptide repertoire presented. | Differences in the PBM lead to a different peptide repertoire presented, against which T cells have not been tolerized. Distinct PBM clusters can be identified with a similar presented peptide repertoire. If these clusters match, there may be a reduction in indirect allorecognition. |
| HLA-EMMA (the present article) | Identification of single amino acids that are mismatched on specific positions within the HLA molecule. Special attention to α-helices and β-sheets. Interlocus comparisons are optional, but not applied in this study. | Alterations in the HLA molecule may lead to both changes in the peptide repertoire presented and the way the T-cell receptor docks to the HLA molecule and recognizes the presented peptide in the context of HLA. This may lead to both direct and indirect allorecognition. The α-helices are particularly important because they can directly affect both. |
| Name . | Strategy . | Possible biology . |
|---|---|---|
| HLAMatchmaker (Duquesnoy et al16,17) | Identification of amino acid triplets present in HLA that are mismatched at known accessible/polymorphic positions. Special attention to triplets confirmed to have been binding sites for donor-specific antibodies. Interlocus comparisons are performed. | Triplets accessible on the cell surface (eplets/epitopes) may lead to donor-specific antibody formation and solid organ transplant rejection. |
| PIRCHE (Stenger et al14) | Identification of mismatching peptide fragments derived from HLA, which can also be presented in HLA class I (PIRCHE-I) or class II (PIRCHE-II). | Presentation of HLA-derived nonself peptides may lead to indirect allorecognition and both aGVHD and rejection. |
| PBM matching (Crivello et al32) | Identification of mismatching peptide binding motif (PBM) categories, where PBM categories have been defined using hierarchical clustering on the peptide repertoire presented. | Differences in the PBM lead to a different peptide repertoire presented, against which T cells have not been tolerized. Distinct PBM clusters can be identified with a similar presented peptide repertoire. If these clusters match, there may be a reduction in indirect allorecognition. |
| HLA-EMMA (the present article) | Identification of single amino acids that are mismatched on specific positions within the HLA molecule. Special attention to α-helices and β-sheets. Interlocus comparisons are optional, but not applied in this study. | Alterations in the HLA molecule may lead to both changes in the peptide repertoire presented and the way the T-cell receptor docks to the HLA molecule and recognizes the presented peptide in the context of HLA. This may lead to both direct and indirect allorecognition. The α-helices are particularly important because they can directly affect both. |
The finding that HvG amino acid mismatches on the α-helices are particularly important is consistent with previous literature, and plausible given the role of the α-helix in the HLA protein in peptide presentation and T-cell recognition.18,19,21 For HLA class I, we observed a similar effect with αHvG matching on binding groove positions, which substantially overlap with the α-helix positions.26
Out of the 21 αHvG matched patients, 13 were also totalHvG matched, whereas 8 had at least 1 HvG amino acid mismatch outside of the α-helices. Investigating survival in a larger cohort of patients with amino acid mismatches outside the α-helices is required to confirm that superior EFS is linked to having no mismatches on the α-helices.
Our main cohort comprised pediatric patients with both malignant and nonmalignant diseases. We performed bivariate analysis to check for confounding by disease category, and found that HvG amino acid matching still predicted survival. After correcting for year of transplantation, the effect of HvG amino acid matching appeared even more pronounced. The effect of year of transplantation is unclear given that no major changes were made in our transplant regimen besides the introduction of PT-Cy, which was analyzed separately. However, many small changes were made, such as optimization of serotherapy exposure, introduction of new second-line therapies for steroid-refractory aGVHD, and less use of busulfan but more use of treosulfan in conditioning regimens. We chose not to perform more extensive multivariate analysis because relevant transplant characteristics were homogeneously distributed between amino acid matching groups, indicating that there was no risk of confounding.
Conflicting results have been presented on whether MMUD outcomes still differ from those of MUDs when PT-Cy is used.33,34 Out of 23 patients treated with PT-Cy, none of the 7 αHvG matched patients experienced any event, whereas a third of the αHvG mismatched patients experienced 1 before 1 year. Within the EBMT Registry, only 11 transplants could be identified that received PT-Cy with a 9/10 MMUD for an inborn error between 2011 and 2021. This indicates that there may still be a role for identifying permissible mismatches in the context of PT-Cy, possibly using HLA-EMMA, but future studies are needed to draw conclusions.
In our study, we focused on the HLA-A, -B, -C, -DRB1, and -DQB1 loci and the comparison with 10/10 MUD donors, as matching on 10 alleles was the most common matching strategy in this study cohort. However, it would be of interest to investigate other HLA loci such as DPB1 and DQA1, and explore how HLA-EMMA performs in the context of locus-specific scores and adjustments such as HLA-DPB1 T-cell epitope matching, DQA1-DQB1 heterodimer formation, and HLA-B leader matching, which have shown promising results in other studies.35-37
In conclusion, we have demonstrated that 9/10 MMUD transplants with a HvG permissible mismatch on the α-helix, as identified by HLA-EMMA version 2.0 beta, had OS and EFS comparable to those of 10/10 MUD transplants, both in a single-center and an EBMT Registry cohort. Similar results could not be obtained with other matching algorithms. Our study indicates that adding αHvG matching allows for expansion of the donor pool with permissibly HLA matched donors, without adversely affecting clinical outcome. Future studies should confirm that prospectively identified permissibly matched (MMUD) donors maintain outcomes similar to those of fully MUDs.
Acknowledgment
This study was funded internally.
Authorship
Contribution: E.G.J.v.A., S.H., A.B.M., D.L.R., A.C.L., and A.A.v.B. designed the study; E.G.J.v.A. and F.H. performed analyses and drafted the manuscript; A.C.L. and A.A.v.B. supervised the study and drafted the manuscript; and S.H., A.B.M., D.L.R., C.S.M.K., F.H.J.C., M.H.A., and B.N. reviewed and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Erik G. J. von Asmuth, Willem Alexander Children’s Hospital, Leiden University Medical Center, Albinusdreef 2, J6-P, Leiden 2333ZA, The Netherlands; email: e.g.j.von_asmuth@lumc.nl.
References
Author notes
E.G.J.v.A. and F.H. contributed equally to this study. A.C.L. and A.A.v.B. contributed equally to this study.
Data are available on request from the corresponding author, Erik G. J. von Asmuth (e.g.j.von_asmuth@lumc.nl).
The full-text version of this article contains a data supplement.