Key Points
Higher MALT1 level in MM samples correlates with shorter survival of patients with MM.
MALT1 inhibition halts cell growth of MM by inactivation of the B-cell maturation antigen–induced NF-κB pathway and inducing ICD.
Visual Abstract
Because multiple myeloma (MM) poses a formidable therapeutic challenge despite recent progress, exploring novel targets is crucial. Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) emerges as a promising paracaspase with druggable potential, especially unexplored in MM. Our study provided compelling evidence demonstrating a statistically significant elevation of MALT1 expression in human primary MM cells. Moreover, elevated MALT1 expression was associated with a poorer prognosis in MM. Genetic deletion of MALT1 reduced cell growth, colony formation, and tumor growth in vivo. Pharmacological inhibition with 1 μM of a small-molecular MALT1 inhibitor, Mi-2, effectively inhibited cell growth, inducing mitochondria-dependent apoptotic cell death. Mechanistically, MALT1 inhibition disrupted diverse signal transduction pathways, notably impeding nuclear factor κB (NF-κB). Significantly, the inhibition of MALT1 demonstrated a substantial suppression of NF-κB activation by elevating inhibitor of NF-κB, disrupting the nuclear localization of p65 and c-REL. This effect was observed in both the basal state and when stimulated by B-cell maturation antigen, highlighting the pivotal role of MALT1 inhibition in influencing MM cell survival. It was noteworthy that Mi-2 induces properties associated with immunogenic cell death (ICD), as evidenced by increased calreticulin, adenosine triphosphate release, and high-mobility group protein B1 upregulation, consequently triggering ICD-associated immune activation and enhancing CD8+ T-cell cytotoxicity in vitro. In conclusion, our research highlights MALT1 as a promising druggable target for therapeutic interventions in MM, providing insights into its molecular mechanisms in MM progression.
Introduction
Multiple myeloma (MM) is a neoplastic plasma cell disorder characterized by abnormal proliferation of plasma cells in the bone marrow (BM).1 The dysregulated progression of MM cells is a complicated process with a lot of genetic and molecular aberrations in vital genes or signaling pathways.2 Despite recent advances in diagnostic and treatment approaches, MM remains incurable, with a significant proportion of patients experiencing relapse. Therefore, elucidation of novel molecular mechanisms and introduction of therapeutic vulnerabilities of MM are of utmost importance.
Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) serves as a paracaspase originally identified because of its involvement in chromosomal translocations observed in MALT lymphoma cells. In the context of MALT lymphoma, MALT1 functions as a pivotal player in the oncogenic process, in which its aberrant activation contributes to the dysregulation of cellular signaling pathways, and it is ubiquitously expressed in several cell types.3-5 MALT1 is a paracaspase that controls antigen receptor–induced nuclear factor κB (NF-κB) signaling as a scaffolding protein or an arginine-specific protease.6,7 Specifically, MALT1 acts as an adaptor protein, forming a signaling complex known as CARD11-BCL10-MALT1 (CBM) in conjunction with caspase recruitment domain family member 11 (CARMA1/CARD11) and BCL10. This complex, in turn, regulates coupled upstream signaling events of the canonical NF-κB via recruitment of E3 ubiquitin ligase, tumor necrosis factor receptor–associated factor 6.8,9 The dysregulated NF-κB signaling induced by MALT1 in MALT lymphoma cells promotes oncogenic processes, ultimately contributing to the development and progression of the disease. In addition to its adaptor function, MALT1 as a paracaspase significantly affects the NF-κB response through the cleavage of TNF alpha induced protein 3 (TNFAIP3/A20), RelB, RANBP2-type and C3HC4-type zinc finger containing 1 (RBCK1/HOIL-1), and CYLD, which adversely affect the NF-κB pathway in lymphocytes.10-13 Except for lymphomas, the dysregulation of MALT1 in human malignancies has been extensively studied and elucidated in several hematological cancers.14-18 Nevertheless, the paracaspase MALT1 has barely been reported in relation to MM in previous studies.
Activation of NF-κB in MM cells is closely linked to the regulation of genes governing cell survival, proliferation, and resistance to apoptosis. Factors within the BM microenvironment, including insulin-like growth factor, proliferation-inducing ligand (APRIL), and B-cell activating factor (BAFF), can directly or indirectly stimulate NF-κB activation in plasma cells and MM cells.19-21 Sustained NF-κB activation in MM cells contributes to uncontrolled proliferation, enhanced survival mechanisms, and resistance to antimyeloma therapies. Consequently, targeting the NF-κB signaling pathway emerges as a promising therapeutic strategy to disrupt these critical cellular processes and combat MM effectively.
B-cell maturation antigen (BCMA) is a critical transmembrane glycoprotein expressed on MM plasma cells and acts as a receptor for ligands such as BAFF and APRIL in the BM microenvironment.22-24 The interaction between BCMA and these ligands activates the NF-κB pathway, contributing to the survival and uncontrolled proliferation of malignant plasma cells. Recognizing the significance of BCMA-induced NF-κB activation is vital for devising targeted therapies to disrupt these pathways and advance novel strategies for managing MM progression and improving treatment outcomes.
With the aim of extending upon existing knowledge, in this study we investigated the molecular functions and effects of MALT1 in MM. We found that MALT1 expression was high in primary MM cells and a panel of MM cell lines. Genetic deletion through RNA interference or clustered regularly interspaced short palindromic repeats–associated protein 9-mediated knockdown suppressed colony formation and inhibited cell growth in vitro and in vivo. Pharmacological targeting with the small-molecular inhibitor Mi-2 resulted in suppressed cell growth, colony formation, and osteoclast formation, induced apoptotic death, and triggered immunogenic cell death. Mechanistically, MALT1 inhibition led to increased IκB levels, disrupting p65 and c-REL nuclear localization in MM cells. Importantly, MALT1 depletion blocked BCMA-dependent NF-κB activation in MM cells. Our findings highlight MALT1's role in regulating NF-κB activation in MM, including BCMA-induced activation, suggesting a pivotal role in MM development and progression. This insight provides a potential therapeutic target to disrupt BCMA-mediated NF-κB activation, offering a novel strategy to impede MM cell survival and growth and potentially enhancing the efficacy of existing MM treatment approaches.
Materials and methods
Cells culture
The human myeloma cell lines RPMI8226, IM9, H929, U266, OPM2, MM1R, and MM1S were cultured in RPMI-1640 medium (KeyGEN BioTECH). The human BM stromal cell line HS-5 was cultured in high glucose DMEM medium (KeyGEN BioTECH). All culture media was supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (VICMED). CD138+ or CD138− cells from patents with MM were isolated from the patients’ BM aspirates. Primary normal hematopoietic cells were isolated from healthy individuals who generously provided peripheral blood stem cells for allogeneic hematopoietic stem cell transplantation after being mobilized with granulocyte colony-stimulating factor.
Antibodies and chemicals
The following primary antibodies were used in this study: MALT1, Bcl2, Bax, P21 (Santa Cruz), cleaved-caspase 9, cleaved-caspase 3, Poly(ADP-ribose) polymerase 1 (PARP), P27, p-P65, P65, phospho-IκB (p-IκB), IκB, p-NF-κB, NF-κB, c-REL, phospho-PERK (p-PERK), eukaryotic translation initiation factor 2 alpha kinase 3 (EIF2AK3/PERK), TNF receptor superfamily member 17 (TNFRSF17/BCMA), BCL10, CARD11, phospho-eIF2α (p-eIF2α), and eukaryotic translation initiation factor 2α (eIF2α) antibodies (Cell Signaling Technology); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin, and histone-H3 (Proteintech); and Flag (Sigma-Aldrich). Mi-2 was purchased from MedChemExpress. Human macrophage colony-stimulating factor and human receptor activator of the NF-κB ligand were purchased from R&D Systems. Cytokines including human vascular endothelial growth factor (VEGF), human insulin-like growth factor I (IGF-I), and human interleukin-6 (IL-6) were purchased from PeproTech. Pan-caspase inhibitor Z-VAD-FMK was purchased from Proteintech. Phytohemagglutinin was purchased from Sigma-Aldrich.
Cell viability, proliferation, and apoptosis assay
Cell Counting Kit-8 (CCK8, VICMED) was used to measure cell viability in accordance with the manufacturer’s instruction. Apoptotic cell death was detected by flow cytometry using Fluorescein isothiocyanate Annexin V Apoptosis Detection Kit I (BD Biosciences).
EdU assays for cell cycle detection
5-Ethynyl-2'-deoxyuridine (EdU) incorporation assay was performed to investigate DNA synthesis using the keyFlour647 Click-iT EdU flow cytometry kit (KeyGEN). Briefly, cells treated with or without Mi-2 were collected and incubated with 10 μM EdU for 2 hours. The cells were then fixed and washed. Subsequently, Click-iT reaction solution was added for 30 minutes, followed by incubation with DAPI (4’,6-diamidino-2-phenylindole, 1 μg/mL) for 15 minutes. Flow cytometry was used to analyze the results.
Colony-formation assay
Clonogeneic growth of indicated cells was evaluated by seeding 800 cells into a 6-well plate in 2 mL methylcellulose semisolid medium (RPMI-1640 containing 0.9% methylcellulose and 10% fetal bovine serum). After 14 days, colonies were stained with 0.4% (weight-to-volume ratio) crystal violet in 20% (volume-to-volume ratio) methanol for 30 minutes, and the number of colonies was counted.
Immunofluorescence (IF)
Cells were seeded onto glass coverslips and treated as indicated in the figure legends. After harvesting and washing, the cells were fixed, permeabilized, and blocked. Subsequently, they were incubated with P65 or c-REL antibody and corresponding fluorescent secondary antibodies. Next, 1 μg/mL DAPI (Beyotime) was added to stain the nuclei. After rinsing with phosphate-buffered saline, the coverslips were mounted onto glass slides and imaged using confocal laser scanning microscopy (LSM880, Zeiss).
Caspase activity assay
Cells treated with or without Mi-2 were harvested and Caspase-Glo Assay Systems (Promega) was used to measure caspase-9 and caspase-3/7 activity. Briefly, 100 μL Caspase-Glo Reagent was added to each well of a white-walled 96-well plate containing 100 μL of blank and samples. The contents of wells were gently mixed and incubated at room temperature for 30 minutes. Then, the luminescence was measured by a GloMax Luminometer as directed by the luminometer manufacturer.
RNA-sequencing analysis
RNA-sequencing libraries were constructed with Illumina NovaSeq reagent kit using 1 μg of total RNA. Briefly, total RNA was extracted using TRIzol reagent (Invitrogen). Then messenger RNA (mRNA) was isolated and, randomly fragmented into 300–base pair small fragments and complementary DNA was synthesized by reverse transcription polymerase chain reaction. Next, the synthesized complementary DNA was ligated to the adapter for further purification and sorted to obtain the final libraries. After quantification by Qubit 4.0, the paired-end RNA-sequencing library was sequenced with the Illumina NovaSeq 6000 sequencer (2 × 150-base pair read length). All process steps were performed by Shanghai Majorbio Bio-pharm Technology Co, Ltd. Gene set enrichment analysis (GSEA) was performed via computational platform from the Broad institute with Hallmark gene signature and curated gene sets.
Immunogenic cell death (ICD)–related molecules analysis
Cells were treated with Mi-2 for 24 hours and then harvested for detection of ICD-related indicators. Briefly, for the analysis of calreticulin (CRT) exposure, the cells were stained with Alexa Fluor647–conjugated anticalreticulin antibody (Abcam) and propidium iodide (PI) (eBioscience) on ice for 30 minutes and detected by flow cytometry; analysis of fluorescence intensity was assessed on viable (propidium iodide-negative) cells. For adenosine triphosphate (ATP) release, ATP Assay Kit (Beyotime) was used for quantitative analysis of ATP in accordance with the manufacturer’s instructions. For heat shock proteins 70/90 (HSP70/90) secretion assays, cells were stained with anti-HSP70/90 primary antibody and CoraLite488-conjugated goat anti-rabbit immunoglobulin G (Proteintech). The HSP70/90 expression on cell surface was measured by flow cytometry. For high-mobility group box 1 (HMGB1) secretion assay, human HMGB1 enzyme-linked immunosorbent assay kit (Elabscience) was used in accordance with the manufacturer’s protocol.
Dendritic cell (DC) culture and maturation assay
Monocyte-derived DCs, generated from peripheral blood mononuclear cells (PBMCs) from healthy donors, were cultured in a 100-mm dish for 1 hour. The suspended cells were removed, and the adherent cells were cultured in RPMI-1640 complete medium supplemented with 100 ng/mL granulocyte-macrophage colony-stimulating factor (PrepoTech,) and 100 ng/mL IL-4 (PrepoTech) for 5 days to generate immature DCs. Immature DCs were then cultured alone or with untreated or Mi-2–pretreated MM cells for 24 hours and analyzed by flow cytometry using the antibodies against: CD45-APC, CD11c-BV421, CD80-FITC, and CD86-PE (BioLegend), and live/dead dye (Invitrogen). Dead cells were excluded by live/dead staining positivity, and CD80 and CD86 expression was evaluated on CD11c+ cells.
T-cell isolation and T-cell–mediated cytotoxicity assay
T cells were isolated from PBMCs using the EasySep Human T-Cell Isolation Kit (STEMCELL Technologies) in accordance with the manufacturer’s instructions, and frozen until immature DCs were generated. For T-cell–mediated cytotoxicity assay, MM cells were treated with Mi-2 for 24 hours and labeled with carboxyfluorescein diacetate duccinimidyl ester (CFSE) (BD Biosciences). Then, the cells were cocultured with immature DCs and T cells at the indicated ratios for further 48 hours. Percentage of live MM cells was determined using flow cytometry.
MM xenograft mouse model
NOD.Cg-PrkdcscidIl2rgtm1Wjl (NSG) mice (6 to 8 weeks old, male) were purchased from Beijing Vital River Laboratories. The animal experimental protocols were approved by the ethics committee of Xuzhou Medical University. A total of 1 × 106 MALT1-scramble cells and MALT1 knockout (KO) MM cells in 100 μL phosphate-buffered saline were intravenously injected into NSG mice, to examine the survival of the mice. A total of 5 × 106 IM9 cells or 5 × 106 RPMI8226 cells were subcutaneously injected into the NSG mice to establish a human MM xenograft model. When the tumor became measurable (100 mm3), the mice were treated on a 5-days-per-week schedule with vehicle or Mi-2. The mice were euthanized when the tumor volume reached 2000 mm3.
Statistical analysis
The variables of different experimental groups were analyzed using GraphPad Prism and presented as mean ± standard deviation. The experiments were routinely performed in triplicate unless specified differently. For comparisons between 2 groups, an unpaired Student t test was used. For comparison of multiple groups, 1-way analysis of variance was used. P value of < .05 was considered statistically significant. The survival of mice was analyzed by Kaplan-Meier survival curves.
The animal experimental protocols were approved by the ethics committee of Xuzhou Medical University.
Results
MALT1 is highly expressed in MM cells and is critical for cell growth and viability
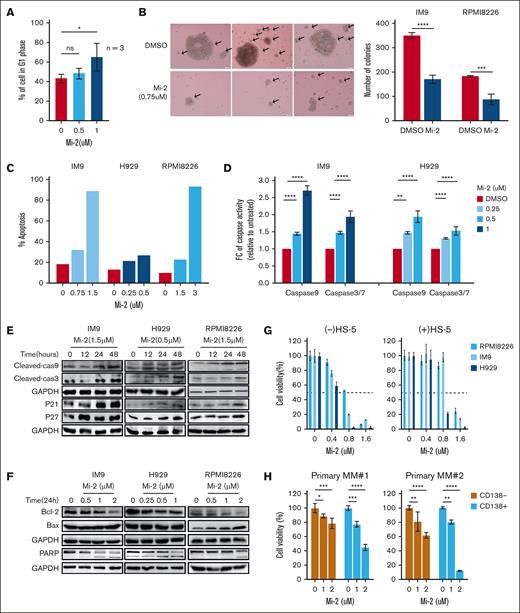
We first evaluated the protein and mRNA expression levels of MALT1 in a panel of MM cell lines and primary MM cells. MALT1 expression was significantly elevated in the MM cell lines and primary MM cells compared with CD138− BM mononuclear cells (Figure 1A; supplemental Figure 1A-B). To assess the functional impact of MALT1 on MM cell growth, we used clustered regularly interspaced short palindromic repeats-associated protein 9 to generate MALT1 KO cell lines, which resulted in significantly impaired cell growth and colony formation (Figure 1B; supplemental Figure 1C). Although MM cells rely on the surrounding BM microenvironment for survival, MALT1 KO also robustly halted cell proliferation in the presence of cytokines (IL-6, VEGF, or IGF) or HS-5 conditional medium (Figure 1C; supplemental Figure 1D). Cell growth inhibition was also confirmed by short-hairpin RNA targeting MALT1 (supplemental Figure 1E). Furthermore, the impact of MALT1 on tumor growth in vivo was evaluated. MALT1-depleted cells and scramble cells were injected intravenously into NSG mice, and the survival was significantly improved (Figure 1D). Compared with the scramble group, the volume of tumors was decreased in the MALT1 KO group in the subcutaneous tumor model (supplemental Figure 1F).
MALT1 is highly expressed in MM cells. (A) Whole-cell lysates of human myeloma cell lines, 2 CD138+ primary MM cells, and 2 CD138− primary BM mononuclear cells (BMMCs) were subjected to WB analysis and probed with indicated antibodies, with GAPDH as a loading control. (B) IM9 and U266 cell lines were transduced with single guide RNA (sgRNA) targeting MALT1. WB analysis of MALT1 in MALT1-KO cells (labeled as KO) compared with MALT1-scramble cells (labeled as Scr). Cell viability was measured daily by CCK-8 assay. Data are shown as optical density (OD) value at 450 nm. (C) Scramble and MALT1-KO cells were cultured with RPMI-1640 medium, HS-5 conditional medium, VEGF (100 ng/mL), insulin-like growth factor (IGF; 100 ng/mL), and IL-6 (25 ng/mL). Cell viability was measured by CCK8 assay at 24, 48, or 72 hours, and OD value at 450 nm is shown. (D) NSG mice were inoculated intravenously with scramble cells (n = 6) and MALT-KO IM9 cells (n = 6). The survival of mice was analyzed by Kaplan-Meier survival curves; hazard ratio (HR), 0.3759; 95% confidence interval (CI), 0.11-1.33. (E) The mRNA level of MALT1 in BMMCs from patients with MM or normal healthy donors (NR) was measured by quantitative reverse transcription polymerase chain reaction assay (qRT-PCR). (F) The dot blot shows the mRNA expression level of MALT1 in MM and other tumors. Data were obtained from Depmap. (G) Correlation of MALT1 with overall survival was investigated in the MMRF CoMMpass trial (772 newly diagnosed patients). No significance (ns), P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
MALT1 is highly expressed in MM cells. (A) Whole-cell lysates of human myeloma cell lines, 2 CD138+ primary MM cells, and 2 CD138− primary BM mononuclear cells (BMMCs) were subjected to WB analysis and probed with indicated antibodies, with GAPDH as a loading control. (B) IM9 and U266 cell lines were transduced with single guide RNA (sgRNA) targeting MALT1. WB analysis of MALT1 in MALT1-KO cells (labeled as KO) compared with MALT1-scramble cells (labeled as Scr). Cell viability was measured daily by CCK-8 assay. Data are shown as optical density (OD) value at 450 nm. (C) Scramble and MALT1-KO cells were cultured with RPMI-1640 medium, HS-5 conditional medium, VEGF (100 ng/mL), insulin-like growth factor (IGF; 100 ng/mL), and IL-6 (25 ng/mL). Cell viability was measured by CCK8 assay at 24, 48, or 72 hours, and OD value at 450 nm is shown. (D) NSG mice were inoculated intravenously with scramble cells (n = 6) and MALT-KO IM9 cells (n = 6). The survival of mice was analyzed by Kaplan-Meier survival curves; hazard ratio (HR), 0.3759; 95% confidence interval (CI), 0.11-1.33. (E) The mRNA level of MALT1 in BMMCs from patients with MM or normal healthy donors (NR) was measured by quantitative reverse transcription polymerase chain reaction assay (qRT-PCR). (F) The dot blot shows the mRNA expression level of MALT1 in MM and other tumors. Data were obtained from Depmap. (G) Correlation of MALT1 with overall survival was investigated in the MMRF CoMMpass trial (772 newly diagnosed patients). No significance (ns), P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
Next, we tried to better understand the significance of MALT1 gene in the clinical setting of MM. Therefore, we performed quantitative reverse transcription polymerase chain reaction analysis of MALT1 mRNA expression in BM mononuclear cells from patients with MM, which revealed elevated MALT1 expression (Figure 1E). We also analyzed the gene expression of MALT1 in a public data set (GSE39754) and found that MALT1 expression was higher than that in normal plasma cells (supplemental Figure 1G). The gene expression level of MALT1 from Depmap data was also higher in MM cells than in other tumors (Figure 1F). Notably, we analyzed MM samples from the Multiple Myeloma Research Foundation (MMRF) CoMMpass (772 newly diagnosed patients) and found that the patients with high MALT1 expression had a poorer prognosis (P < .05; Figure 1G). We further analyzed the expression levels of MALT1 within different subgroups with CoMMpass data. However, there were no significant differences in MALT1 expression levels across the various cytogenetic subgroups (supplemental Figure 1H). These findings suggest that MALT1 may serve as a potential drug target across all stages of MM.
MALT1 inhibition stalls cell growth of MM cells ex vivo and in vivo
Selective pharmacological targeting of MALT1 paracaspase by the small-molecule inhibitor Mi-2 has demonstrated therapeutic potential in a wide range of human tumors, including glioblastoma, breast cancer, and osteosarcoma.25-27 Thus, we assessed the impact of Mi-2 on MM cell growth, and revealed a robust, dose-dependent cell death promotion after Mi-2 treatment (supplemental Figure 2A). In addition, Mi-2 strongly induced G0-G1 phase cell cycle arrest, hindered colony formation, and significantly decreased the volume and number of colonies (Figure 2A-B; supplemental Figure 2B). Moreover, Mi-2 induced substantial apoptotic cell death, evidenced by increased activities of caspases-3/7 and caspase-9 (Figure 2C-D; supplemental Figure 2C) and also confirmed by upregulation of cleaved-caspase 3, caspase-9, PARP, p21, and p27, and the elevated Bax/Bcl2 ratio through western blot (WB) assay (Figure 2E-F). To address whether Mi-2 induced cytotoxicity completely through apoptosis, we used the caspase inhibitor Z-VAD-FMK to overcome the effect of Mi-2. We showed that Z-VAD-FMK overcame almost half of apoptotic cell death induced by Mi-2 (supplemental Figure 2D). Additionally, we found that Mi-2 could not induce more cytotoxicity in the MALT1-KO cell lines (supplemental Figure 2E). Thus, MALT1 is the target of Mi-2 in MM, and Mi-2 induces cell death through a combination of cell cycle arrest and mitochondria-dependent apoptosis in MM cells in vitro.
MALT1 inhibition inhibits cell growth of MM cells ex vivo and in vivo. (A) RPMI8226, H929, and IM9 cells were treated with Mi-2 (0.5 and 1 μM) for 48 hours. Cell cycle was evaluated by flow-cytometric analysis after EdU incorporation. Percentages of cells in G1 phase were calculated and compared among the groups. (B) IM9 and RPMI8226 cells were treated with Mi-2 (0.75 μM). Colony formation assay was performed to evaluate the clonogeneic ability after Mi-2 treatment. The representative figure of IM9 cells is shown (left), and the number of colonies is shown in bar graph (right). (C) IM9, H929, and RPMI8226 cells were treated with different concentration of Mi-2 for 48 hours, and apoptotic cell death was analyzed by flow-cytometric analysis staining with Annexin V/PI. The bar graph shows the ratio of apoptotic death cell. (D) IM9 and H929 cells were treated with indicated concentrations of Mi-2. The caspase-9 and caspase-3/7 activity was assessed by a luminescent assay after MI-2 treatment. The bar graph shows the fold change (FC) of caspase activity compared with dimethyl sulfoxide (DMSO). (E-F) RPMI8226, H929, and IM9 cells were treated with Mi-2 under the conditions indicated. WB assay was used to evaluate the expression of cleaved-caspase 9, cleaved-caspase 3, PARP, Bcl2, Bax, p21, and p27 proteins. GAPDH was used as a loading control. (G) RPMI8226, H929, and IM9 cells were treated with indicated concentrations of Mi-2 in the absence or presence of HS-5 conditional medium for 48 hours. Cell viability was measured by CCK-8 assay. (H) CD138+ primary MM cells from patients with MM were cultured with the indicated concentrations of Mi-2 for 48 hours. Cell viability was assessed by CCK-8 assay. (I) Primary normal hematopoietic cells were treated with the indicated concentrations of Mi-2 for 48 hours. Cell viability was assessed by CCK-8 assay. (J) NSG mice injected subcutaneously with RPMI8226 cells were treated with Mi-2 (placebo, 25 mg/kg, 5 days per week) intraperitoneally for 4 weeks. The survival of mice was analyzed by Kaplan-Meier survival curves. HR, 0.2785; 95% CI, 0.08-0.98. ns, P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
MALT1 inhibition inhibits cell growth of MM cells ex vivo and in vivo. (A) RPMI8226, H929, and IM9 cells were treated with Mi-2 (0.5 and 1 μM) for 48 hours. Cell cycle was evaluated by flow-cytometric analysis after EdU incorporation. Percentages of cells in G1 phase were calculated and compared among the groups. (B) IM9 and RPMI8226 cells were treated with Mi-2 (0.75 μM). Colony formation assay was performed to evaluate the clonogeneic ability after Mi-2 treatment. The representative figure of IM9 cells is shown (left), and the number of colonies is shown in bar graph (right). (C) IM9, H929, and RPMI8226 cells were treated with different concentration of Mi-2 for 48 hours, and apoptotic cell death was analyzed by flow-cytometric analysis staining with Annexin V/PI. The bar graph shows the ratio of apoptotic death cell. (D) IM9 and H929 cells were treated with indicated concentrations of Mi-2. The caspase-9 and caspase-3/7 activity was assessed by a luminescent assay after MI-2 treatment. The bar graph shows the fold change (FC) of caspase activity compared with dimethyl sulfoxide (DMSO). (E-F) RPMI8226, H929, and IM9 cells were treated with Mi-2 under the conditions indicated. WB assay was used to evaluate the expression of cleaved-caspase 9, cleaved-caspase 3, PARP, Bcl2, Bax, p21, and p27 proteins. GAPDH was used as a loading control. (G) RPMI8226, H929, and IM9 cells were treated with indicated concentrations of Mi-2 in the absence or presence of HS-5 conditional medium for 48 hours. Cell viability was measured by CCK-8 assay. (H) CD138+ primary MM cells from patients with MM were cultured with the indicated concentrations of Mi-2 for 48 hours. Cell viability was assessed by CCK-8 assay. (I) Primary normal hematopoietic cells were treated with the indicated concentrations of Mi-2 for 48 hours. Cell viability was assessed by CCK-8 assay. (J) NSG mice injected subcutaneously with RPMI8226 cells were treated with Mi-2 (placebo, 25 mg/kg, 5 days per week) intraperitoneally for 4 weeks. The survival of mice was analyzed by Kaplan-Meier survival curves. HR, 0.2785; 95% CI, 0.08-0.98. ns, P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
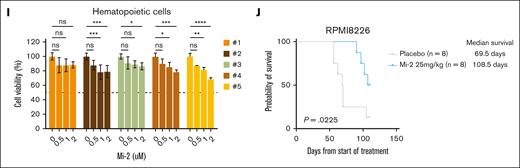
Based on the superior impact of Mi-2 on MM cells, we were interested in whether Mi-2 also affects cell growth in the presence of the BM microenvironment. We evaluated the cell growth of MM in the absence or presence of HS-5 conditional medium with Mi-2 treatment, and showed that Mi-2 still significantly impaired cell viability of MM cells in the BM microenvironment (Figure 2G). Next, we assessed whether Mi-2 affected primary MM cells, and showed that Mi-2 exerted strong cytotoxicity against primary CD138+ MM cells (Figure 2H). Importantly, Mi-2 was not effective in hematopoietic cells and healthy donor PBMCs activated with phytohemagglutinin (Figure 2I; supplemental Figure 3A). The effect of Mi-2 in vitro prompted us to assess the efficacy of Mi-2 in an in vivo model of MM. IM9 or RPMI 8226 cells were subcutaneously injected in NSG mice and treated with placebo or Mi-2. Compared with the placebo group, intraperitoneal injection of Mi-2 for 4 weeks significantly increased overall survival in both models without weight loss (Figure 2J; supplemental Figure 3B-C).
We measured the secretion levels of VEGF or IL-6 from MM and HS-5 cells. As shown in supplemental Figure 3D, the secretion level of VEGF but not IL-6 (data not shown) was decreased in MM cells after Mi-2 treatment alone or cocultured with HS-5 cells. The in vitro osteoclast formation of healthy donor PBMCs was significantly impaired, with limited evidence of tartrate resistant acid phosphatase (TRAP)-positive multinuclear osteoclasts (OCs) when Mi-2 was used at the concentration of 0.25 μM (supplemental Figure 3E). These results indicate that Mi-2 significantly inhibits MM cell growth ex vivo and in vivo, and partially impairs the BM microenvironment.
MALT1 inhibition halts the activation of the NF-κB signaling pathway
To gain insight into the molecular implications of MALT1-mediated cell growth in MM, we conducted global transcriptional profiling and an unbiased GSEA in MM cells treated with Mi-2 (1 μM, 24 hours) and in MM cells with MALT1 depletion. The GSEA revealed a significant negative enrichment in NF-κB pathways after MALT1 inhibition in both the Mi-2 treated and MALT1-depletion groups (Figure 3A-B; supplemental Figure 4A-B).
MALT1 inhibition blocks the activation of the NF-κB pathway. (A) H929 cells were treated with Mi-2 for 24 hours, and H929 cells transduced with MALT1-depletion or scramble vector were subjected to RNA-sequencing and GSEA. The enriched pathways are shown in bubble graph (false discovery rate [FDR] < 0.25; absolute value of normalized enrichment score [|NES|]> 1.4). (B) Enrichment plots of GSEA revealing significantly negative enrichments for the NF-κB pathway after MALT1 inhibition. (C) Scramble and MALT1-KO cells (IM9, RPMI8226, and U266) were subjected to WB analysis and probed with p-p65 and p65 antibodies, with GAPDH as a loading control. (D) IF analysis of -c-REL protein in scramble and MALT1-KO cells (RPMI8226 and U266). Nuclei were stained with DAPI (1 μg/mL). Bar = 10 μM. (E) WB analysis of p-IκB and IκB in scramble and MALT1-KO IM9 cells, with GAPDH as a loading control. (F) IM9 cells were treated with Mi-2 (1.5 μM) for different times (0, 0.5, 1, 2, 4, and 8 hours) and p-NF-κB was examined by WB assay. (G) IM9 cells were treated with indicated concentrations of Mi-2 for 24 hours in the absence or presence of PMA (10 ng/mL). The nuclear proteins c-REL and p65 were measured by WB assay. Histone H3 was used as a control. (H) IF analysis of p65 and c-REL protein in IM9 cells treated with Mi-2 or DMSO. Nuclei were stained with DAPI (1 μg/mL).
MALT1 inhibition blocks the activation of the NF-κB pathway. (A) H929 cells were treated with Mi-2 for 24 hours, and H929 cells transduced with MALT1-depletion or scramble vector were subjected to RNA-sequencing and GSEA. The enriched pathways are shown in bubble graph (false discovery rate [FDR] < 0.25; absolute value of normalized enrichment score [|NES|]> 1.4). (B) Enrichment plots of GSEA revealing significantly negative enrichments for the NF-κB pathway after MALT1 inhibition. (C) Scramble and MALT1-KO cells (IM9, RPMI8226, and U266) were subjected to WB analysis and probed with p-p65 and p65 antibodies, with GAPDH as a loading control. (D) IF analysis of -c-REL protein in scramble and MALT1-KO cells (RPMI8226 and U266). Nuclei were stained with DAPI (1 μg/mL). Bar = 10 μM. (E) WB analysis of p-IκB and IκB in scramble and MALT1-KO IM9 cells, with GAPDH as a loading control. (F) IM9 cells were treated with Mi-2 (1.5 μM) for different times (0, 0.5, 1, 2, 4, and 8 hours) and p-NF-κB was examined by WB assay. (G) IM9 cells were treated with indicated concentrations of Mi-2 for 24 hours in the absence or presence of PMA (10 ng/mL). The nuclear proteins c-REL and p65 were measured by WB assay. Histone H3 was used as a control. (H) IF analysis of p65 and c-REL protein in IM9 cells treated with Mi-2 or DMSO. Nuclei were stained with DAPI (1 μg/mL).
Building upon the GSEA, we aimed to further examine the activation of the NF-κB pathway in MM cells after MALT1 inhibition. As anticipated, the activation of NF-κB was impeded after MALT1 depletion, leading to decreased phosphorylation of p65 and reduction in nuclear c-REL (Figure 3C-D), as well as increased expression level of IκB (Figure 3E). Similarly, Mi-2 treatment significantly blocked the phosphorylation of NF-κB in a short time–dependent manner (Figure 3F). Western blots for p65 and c-REL in nuclear extracts of MM cells treated with Mi-2 (at concentrations of 1.5 and 3 μM) in the absence or presence of phorbol 12-myristate 13-acetate (PMA) demonstrated a clear reduction in nuclear c-REL and p65 after Mi-2 treatment (Figure 3G). To further validate the inactivation of the NF-κB signaling pathway, we used an IF assay to monitor the cellular localization of p65 and c-REL. We found that p65 and c-REL were also less localization in the nucleus after Mi-2 treatment (Figure 3H). Taken together, the results confirmed that MALT1 inhibition blocks the activation of the NF-κB pathway in MM cells by interrupting the nuclear translocation of p65 and c-REL.
Mi-2 blocks the BCMA-dependent activation of NF-κB
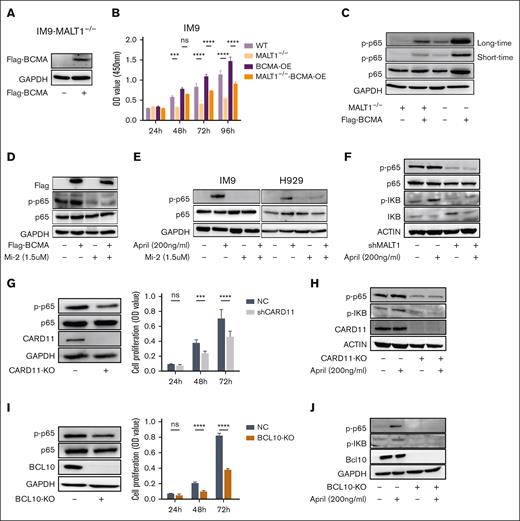
It is widely recognized that APRIL/BCMA promotes MM progression through the activation of the NF-κB pathway. However, uncertainty persists as to whether targeting MALT1 or the CBM complex can effectively halt the activation of NF-κB through APRIL/BCMA induction. Herein, we used a strategy of overexpressing the BCMA gene in MALT1-KO MM cells (Figure 4A). Notably, the overexpression of BCMA significantly promoted cell proliferation in control MM cells but failed to support cell growth in MALT1-KO cells (Figure 4B). Accordingly, the exogenous expression of BCMA led to an increased phosphorylation level of p65, subsequently activating NF-κB. Nevertheless, MALT1 KO markedly reduced the phosphorylation of p65 in MM cells bearing overexpressed BCMA (Figure 4C). Consistently, impaired NF-κB signal was found in MM cells bearing BCMA overexpression after Mi-2 treatment (Figure 4D). Furthermore, Mi-2 treatment reduced the phosphorylation level of p65 induced by APRIL (Figure 4E). Similar results were obtained in MM cells with MALT1 inhibition via short-hairpin RNA silencing (Figure 4F).
The CBM complex impedes the BCMA-induced activation of NF-κB. (A-C) Flag-BCMA protein was overexpressed in MALT1-KO IM9 cells. BCMA expression was detected by WB assay (A). Cell viability of these cells was detected for 4 consecutive days by CCK-8 assay (B). WB analysis of p65 and p-p65 in cells treated as indicated in the panel. GAPDH was used as control (C). (D) BCMA-overexpressing cells were treated with Mi-2 for 4 hours, and p-p65 was detected by WB assay. GAPDH was used as control. (E) IM9 or H929 cells were treated with APRIL (200 ng/mL) in absence or presence of Mi-2 (1.5 μM) for 4 hours, p-p65 was detected by WB assay. (F) RPMI8226 cells with shMALT1 and scramble were cultured with APRIL (200 ng/mL) for 4 hours, and levels of p-p65 and p-IκB level were detected by WB assay. (G-H) IM9 cells were transduced with sgRNA targeting CARD11. The protein expression level of CARD11 was detected by WB (left), and cell viability of these cells was assessed by CCK-8 assay (right) (G). p-p65 and p-IκB were detected by WB assay in absence or presence of APRIL (200 ng/mL) (H). I-J. IM9 cells were transduced with sgRNA targeting BCL10. The protein expression level of BCL10 was detected by WB (left), and cell viability of these cells was assessed by CCK8 assay (right) (I). p-p65 and p-IκB were detected by WB assay in absence or presence of APRIL (200 ng/mL) (J). ns, P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
The CBM complex impedes the BCMA-induced activation of NF-κB. (A-C) Flag-BCMA protein was overexpressed in MALT1-KO IM9 cells. BCMA expression was detected by WB assay (A). Cell viability of these cells was detected for 4 consecutive days by CCK-8 assay (B). WB analysis of p65 and p-p65 in cells treated as indicated in the panel. GAPDH was used as control (C). (D) BCMA-overexpressing cells were treated with Mi-2 for 4 hours, and p-p65 was detected by WB assay. GAPDH was used as control. (E) IM9 or H929 cells were treated with APRIL (200 ng/mL) in absence or presence of Mi-2 (1.5 μM) for 4 hours, p-p65 was detected by WB assay. (F) RPMI8226 cells with shMALT1 and scramble were cultured with APRIL (200 ng/mL) for 4 hours, and levels of p-p65 and p-IκB level were detected by WB assay. (G-H) IM9 cells were transduced with sgRNA targeting CARD11. The protein expression level of CARD11 was detected by WB (left), and cell viability of these cells was assessed by CCK-8 assay (right) (G). p-p65 and p-IκB were detected by WB assay in absence or presence of APRIL (200 ng/mL) (H). I-J. IM9 cells were transduced with sgRNA targeting BCL10. The protein expression level of BCL10 was detected by WB (left), and cell viability of these cells was assessed by CCK8 assay (right) (I). p-p65 and p-IκB were detected by WB assay in absence or presence of APRIL (200 ng/mL) (J). ns, P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
To further validate the role of the CBM complex in the BCMA-dependent activation of the NF-κB signaling cascade in MM cells, we generated MM cells with CARD11 or BCL10 deletion. The deletion of either CARD11 or BCL10 significantly reduced the phosphorylation of p65 and blocked cell proliferation (Figure 4G,I). Importantly, any disruption of the CBM complex components clearly impeded the BCMA-dependent NF-κB signal transduction cascades in MM cells (Figure 4H,J).
Mi-2 induces ICD-associated immune activation via endoplasmic reticulum (ER) stress in vitro
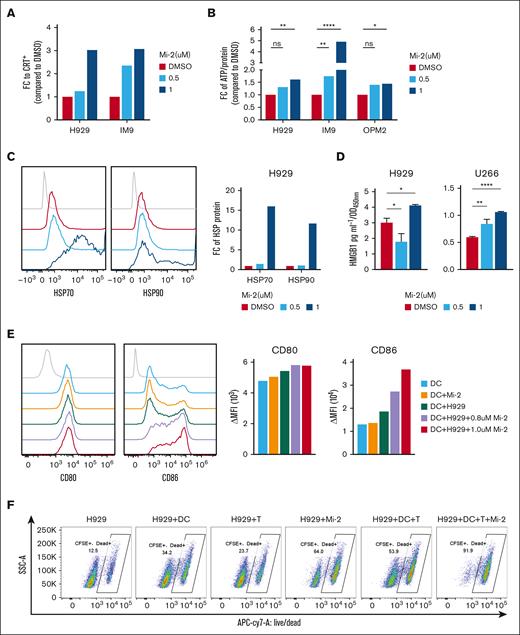
It is intriguing that type I interferon pathways were identified as positively enriched pathways in MALT1-depleted MM cell lines (supplemental Figure 4C). Given the essential role of interferon response and inflammatory chemokines in achieving optimal therapeutic efficacy of agents inducing ICD,28-30 our interest was piqued. Thus, we aimed to ascertain whether Mi-2 induces key indicators of ICD, including the cell surface exposure of CRT and the release of damage-associated molecular patterns, such as ATP, HSP70/90, or HMGB1. As illustrated in Figure 5A, Mi-2 treatment resulted in a significant increase in CRT expression, as determined by flow cytometry analysis, compared with the control. Consistent with the elevated CRT levels, the release of other damage-associated molecular patterns, including ATP, HSP70/90, and HMGB1, also showed a substantial increase in MM cells at 24 hours after Mi-2 treatment (Figure 5B-D). ICD has the capability to induce the antigen-presenting function of DCs, subsequently activating a cascade of immune responses, including cytokine secretion and T-cell activation. To investigate whether Mi-2 treatment induces DC maturation and activation, we cultured DCs isolated from PBMCs and cocultured them with MM cells in the absence or presence of Mi-2 pretreatment. Flow cytometry analysis revealed that the expression of DC maturation and activation markers (CD80 and CD86) increased after Mi-2 treatment (Figure 5E), especially CD86 expression. These results strongly indicate that coculturing with Mi-2–treated MM cells significantly enhances DC maturation and activation.
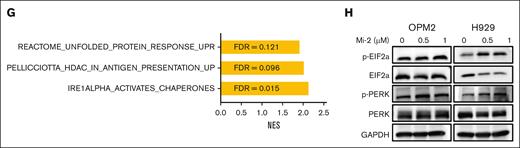
Mi-2 induces the immunogenic cell death via ER stress. (A-D) MM cells were treated with indicated concentrations of Mi-2 for 24 hours. The CRT expression was measured by flow-cytometric analysis, the bar graph shows the FC of CRT’s mean fluorescence intensity (MFI) compared with that of DMSO (A). The ATP released was assessed by ATP assay kit, the bar graph shows the FC of ATP compared with that of DMSO (B). The HSP70 and HSP90 protein exposure were measured by flow-cytometric analysis (left), the bar graph shows the FC of HSP proteins’ MFI compared with that of DMSO (right) (C). The HMGB1 secretion was assessed by enzyme-linked immunosorbent assay analysis, the bar graph shows the relative HMGB1 concentration in groups normalized to OD value at 450 nm (D). (E) DCs were cocultured with MM cells pretreated with DMSO or Mi-2 for 24 hours, and the expression of CD80 and CD86 were detected by flow cytometry (left). The bar graph shows the MFI of CD80 or CD86 (right). (F) The ration of dead cells of the following groups was determined by live/dead staining: H929, H929/DCs, H929/CD8+ T cells, 1 μM Mi-2–treated H929, H929/DCs/CD8+ T cells, and 1 μM Mi-2–treated H929/DCs/CD8+ T cells. The representative scatter plots are shown. (G) The bar graph shows that positive enriched pathways after Mi-2 treatment. FDR < 0.25. (H) Whole-cell lysates from OPM2 and H929 treated with the indicated concentration of Mi-2 for 24 hours were subjected to WB analysis and probed with indicated antibodies, with GAPDH as a loading control. ns, P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
Mi-2 induces the immunogenic cell death via ER stress. (A-D) MM cells were treated with indicated concentrations of Mi-2 for 24 hours. The CRT expression was measured by flow-cytometric analysis, the bar graph shows the FC of CRT’s mean fluorescence intensity (MFI) compared with that of DMSO (A). The ATP released was assessed by ATP assay kit, the bar graph shows the FC of ATP compared with that of DMSO (B). The HSP70 and HSP90 protein exposure were measured by flow-cytometric analysis (left), the bar graph shows the FC of HSP proteins’ MFI compared with that of DMSO (right) (C). The HMGB1 secretion was assessed by enzyme-linked immunosorbent assay analysis, the bar graph shows the relative HMGB1 concentration in groups normalized to OD value at 450 nm (D). (E) DCs were cocultured with MM cells pretreated with DMSO or Mi-2 for 24 hours, and the expression of CD80 and CD86 were detected by flow cytometry (left). The bar graph shows the MFI of CD80 or CD86 (right). (F) The ration of dead cells of the following groups was determined by live/dead staining: H929, H929/DCs, H929/CD8+ T cells, 1 μM Mi-2–treated H929, H929/DCs/CD8+ T cells, and 1 μM Mi-2–treated H929/DCs/CD8+ T cells. The representative scatter plots are shown. (G) The bar graph shows that positive enriched pathways after Mi-2 treatment. FDR < 0.25. (H) Whole-cell lysates from OPM2 and H929 treated with the indicated concentration of Mi-2 for 24 hours were subjected to WB analysis and probed with indicated antibodies, with GAPDH as a loading control. ns, P > .05; ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
To further investigate whether Mi-2 treatment enhances DC-mediated anti-MM response, H929 cells treated with Mi-2 were cocultured with DCs and CD8+ T cells, and cell death was measured by live/dead staining. The pretreatment with Mi-2 significantly enhanced DC-mediated T-cell cytotoxicity, as almost all the cells were dead (91.9%; Figure 5F). Notably, the GSEA revealed positive enrichment of unfolded protein response, endoplasmic reticulum to nucleus signalling 1 (IRE1α)-activated chaperone, and antigen presentation up pathways in MM cells after Mi-2 treatment (Figure 5G). This suggests that Mi-2 induces an imbalance in protein homeostasis, leading to ER stress. Furthermore, we investigated the ER transmembrane sensors PERK and its substrate eIF2α. The results indicated enhanced phosphorylation levels of PERK and eIF2α after Mi-2 treatment (Figure 5H).
Discussion
Despite the progress in advancing therapeutic approaches for MM, relapse and treatment resistance remain significant challenges. Hence, the identification of novel therapeutic targets remains a crucial long-term pursuit. Extensive research has elucidated the molecular functions of MALT1 in various hematological malignancies and solid tumors.31 As the first-in-man MALT1 targeting clinical trial begins enrolling more patients, the potential of MALT1 as a promising clinical target is evident. Consequently, our study aimed to explore the molecular characteristics and oncogenic implications of MALT1 in MM cells, with the goal of incorporating MALT1 as a novel target in MM therapeutic strategies. Our findings highlight the essential role of MALT1 in MM cell proliferation and survival, demonstrating that the MALT1 inhibitor Mi-2 can suppress tumor growth in vitro and in vivo, significantly extending the survival of xenografted mouse models. These results underscore the vulnerability of MM cells to MALT1 inhibition.
NF-κB transcription factors are pivotal in MM proliferation and survival. APRIL and BAFF are 2 of the main survival factors that can directly activate the NF-κB pathway in MM.32,33 Sequestration of APRIL or BAFF has been reported to prevent the extrinsic NF-κB activation and reduce the tumor burden.34 Hence, blocking intrinsic components of the NF-κB pathway should augment the inhibition of extrinsic signaling. Our study revealed that both ectopic KO and pharmacological inhibition of MALT1 diminished NF-κB activation by preventing nuclear localization of p65, including the BCMA-dependent NF-κB activation. Intriguingly, CARD11 or BCL10 KO also reduces the BCMA-dependent activation of NF-κB. These results suggest that BCMA-induced NF-κB activation requires MALT1 as a paracaspase, along with MALT1 as an adaptor forming the CBM complex. Beyond disrupting the axis of BCMA/CBM/NF-κB, MI-2 also impaired the formation of OCs. OCs, known to stimulate MM growth and bone lesions, produce a high level of APRIL.35,36 Thus, MALT1 inhibition not only impairs the intrinsic components of the NF-κB pathway but also limits extrinsic factors influencing of NF-κB.
For long-term control of tumor growth, effective cancer therapies should not only diminish tumor mass but also provoke a protective anticancer immune response. Chemotherapeutic agents such as oxaliplatin, taxanes, cyclophosphamide, and bortezomib are acknowledged for their contribution to the antitumor effect via indirect, immune-dependent mechanisms.30,37,38 Notably, we observed an intriguing phenomenon in which Mi-2 treatment induced changes in the levels of markers associated with ICD, such as an increase in CRT, ATP release, and upregulation of HMGB1, and enhanced DC maturation and activation.39 ICD is characterized by an autophagic response that facilitates the release of ATP during the blebbing phase of apoptosis or necrotic demise, as well as ER stress response. Our data showed that Mi-2 treatment induced ER stress and impaired the imbalance in protein homeostasis. The alterations in these immunogenic molecules suggest that after MALT1 inhibition, MM cells emit “find me” and “eat me” signals, potentially attracting immune cells to kill MM cells. Additionally, the ER stress response exposes CRT on the cell surface. Although further in vivo experiments are needed to validate these findings, they already hint at the possibility that combining MALT1 targeting may enhance the efficacy of immunotherapeutic strategies for clinical MM.
As the use of MALT1 inhibitors in preclinical research increases, potential limitations in signal transduction targeted therapies may emerge. A recent study has shown that MALT1 inhibition promotes increased mechanistic target of rapamycin complex 1 activity and phosphorylation of S6K1 and S6 as a potential feedback mechanism. Combination of MALT1 and mechanistic target of rapamycin complex 1 inhibition has shown enhanced therapeutic efficacy.40 Consistent with this, our CCK-8 assay evaluated the combination effect index of MALT1 inhibition with various clinical drugs for MM, such as bortezomib, melphalan, or dexamethasone, revealing that only dexamethasone combined with MALT1 inhibitor, exerted a stronger synergistic effect on MM cells in the absence or presence of HS-5 conditional medium, compared with other drugs (supplemental Figure 5). This could be attributed to dexamethasone’s capability to inhibit mechanistic target of rapamycin signaling.41 This finding proposes a promising clinical strategy for patients with MM that involves a combination of MALT1 inhibitor and dexamethasone.
This study marks a pioneering investigation into the role of MALT1 in the progression of MM, aiming to unravel its clinical significance. Our findings highlight the central involvement of MALT1 in driving cell proliferation, both in vitro and in vivo. This positions the paracaspase MALT1 as a promising and previously unexplored therapeutic target for MM. Notably, our research brings to the forefront the crucial significance of MALT1 within the BCMA/CBM/NF-κB signaling axis in the context of MM. These revelations not only advance our understanding of MM pathogenesis but also present a compelling avenue for the development of novel clinical strategies with significant potential in the management of MM.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81870163 and 82260042), and Major Basic Research Project of the Natural Science Foundation of the Jiangsu Higher Education Institutions (21KJA320005 and 19KJA560001).
Authorship
Contribution: Y.Y. and M.N. designed the experiments, wrote the manuscript, and prepared presentation of the data; M.Y. and M.S. performed the experiments, analyzed the results, and created the figures; W.L., Y.Z., J.L., and Y.S. helped with the experiments; C.L. helped with the data analysis; and Z.L. and K.X. oversaw the interpretation of data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kailin Xu, Blood Disease Institute, Xuzhou Medical University, 84 West Huaihai Rd, Xuzhou, China 221001; email: lihmd@163.com; Mingshan Niu, Blood Disease Institute, Xuzhou Medical University, 84 West Huaihai Rd, Xuzhou, China 221001; email: msniu24@126.com; and Yao Yao, Blood Disease Institute, Xuzhou Medical University, 84 West Huaihai Rd, Xuzhou, China 221001; email: yaoy412@126.com.
References
Author notes
Y.Y., M.Y., and M.S. are joint first authors.
The data reported in this article have been deposited in the GEO (Gene Expression Omnibus) database “Halting multiple myeloma with MALT1 inhibition: suppressing BCMA-induced NF-κB and inducing immunogenic cell death” (accession number: GSE266548).
All data generated or analyzed during this study are included in this published article.
The full-text version of this article contains a data supplement.


![MALT1 inhibition blocks the activation of the NF-κB pathway. (A) H929 cells were treated with Mi-2 for 24 hours, and H929 cells transduced with MALT1-depletion or scramble vector were subjected to RNA-sequencing and GSEA. The enriched pathways are shown in bubble graph (false discovery rate [FDR] < 0.25; absolute value of normalized enrichment score [|NES|]> 1.4). (B) Enrichment plots of GSEA revealing significantly negative enrichments for the NF-κB pathway after MALT1 inhibition. (C) Scramble and MALT1-KO cells (IM9, RPMI8226, and U266) were subjected to WB analysis and probed with p-p65 and p65 antibodies, with GAPDH as a loading control. (D) IF analysis of -c-REL protein in scramble and MALT1-KO cells (RPMI8226 and U266). Nuclei were stained with DAPI (1 μg/mL). Bar = 10 μM. (E) WB analysis of p-IκB and IκB in scramble and MALT1-KO IM9 cells, with GAPDH as a loading control. (F) IM9 cells were treated with Mi-2 (1.5 μM) for different times (0, 0.5, 1, 2, 4, and 8 hours) and p-NF-κB was examined by WB assay. (G) IM9 cells were treated with indicated concentrations of Mi-2 for 24 hours in the absence or presence of PMA (10 ng/mL). The nuclear proteins c-REL and p65 were measured by WB assay. Histone H3 was used as a control. (H) IF analysis of p65 and c-REL protein in IM9 cells treated with Mi-2 or DMSO. Nuclei were stained with DAPI (1 μg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/15/10.1182_bloodadvances.2023012394/2/m_blooda_adv-2023-012394-gr3.jpeg?Expires=1764955684&Signature=OVjVBmXdoq4UPvOMZ0buEjwnjLMFAJezu8YsHH-uNSpnu2iQeWcPnoZNI2YJVJiGndpWJZjKOcaB0~clpFeLWXynpHqrzN~uOr5jx9UB2nPA8plGeTvHiYGYqsRLGbn-f7WQotyjETcpeOMeZvD3kOI9vEZq~kieUmM9CqyZmhCEf84kIGhSzmsNMqfxwaUH~Zki6fi9tK4YyfqdZFkUF5YI8DUDoKTCIB3FcyOgcoUGWQ9UTCDFmXfbdGD3Kff2ZMPV~Q4qOJNGFpoNAVk~iqH48g9M-niMj-vM5eNf-YJVHdVhddbv7rSwgBJD~vOn1GvdnIKBwdGitHmStqKhMg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)