Key Points
The E592K variant of SF3B1 induces unique RNA missplicing that is nonoverlapping with other MDS-associated variants.
E592K is associated with elevated blasts, lack of RSs, and decreased survival.
Visual Abstract
Among the most common genetic alterations in myelodysplastic syndromes (MDS) are mutations in the spliceosome gene SF3B1. Such mutations induce specific RNA missplicing events, directly promote ring sideroblast (RS) formation, and generally associate with a more favorable prognosis. However, not all SF3B1 mutations are the same, and little is known about how distinct hotspots influence disease. Here, we report that the E592K variant of SF3B1 associates with high-risk disease features in MDS, including a lack of RS, increased myeloblasts, a distinct comutation pattern, and a lack of favorable survival seen with other SF3B1 mutations. Moreover, compared with other hot spot SF3B1 mutations, E592K induces a unique RNA missplicing pattern, retains an interaction with the splicing factor SUGP1, and preserves normal RNA splicing of the sideroblastic anemia genes TMEM14C and ABCB7. These data have implications for our understanding of the functional diversity of spliceosome mutations, as well as the pathobiology, classification, prognosis, and management of SF3B1-mutant MDS.
Introduction
SF3B1 is the most mutated spliceosome gene in myelodysplastic syndromes (MDS), with an overall frequency of >30%.1 The mutations are primarily missense substitutions that induce neomorphic RNA missplicing in thousands of junctions, which in turn alter expression of hundreds of genes in diverse pathways.2 Mutant SF3B1–induced missplicing has been implicated in many MDS phenotypes, including dysfunctional iron metabolism, formation of ring sideroblasts (RSs), activation of innate immune signaling, and promotion of hematopoietic stem cell self-renewal.3-8SF3B1 mutations also contain a prognostic value in MDS, generally associating with more indolent disease progression, though with important exceptions.9-12 In MDS treatment, mutant SF3B1 has been a direct or indirect target of many investigational therapies. Thus, SF3B1 mutations figure prominently in the research of, and clinical practice for, many patients with MDS.
Not well understood is whether and how distinct SF3B1 mutation hotspots differentially affect disease features and/or the RNA missplicing events that drive them. Previously, in an analysis of patients and cell models with SF3B1 exon 14-15 mutations, we found that the K666N variant was enriched in high-risk MDS and produced an asymmetrical lack of missplicing events that are induced by K700E and H662Q mutations.13 Here, we report the results of extending this approach to a larger cohort of patients that included exon 13-16 mutations and additional cell models. This analysis revealed a striking distinctiveness in biological and clinical features of the SF3B1 mutation E592K, which has implications for the understanding and management of SF3B1-mutant MDS.
Materials/subjects and methods
Patients
The mutation-agnostic acquisition of SF3B1 mutations from cases of MDS and acute myeloid leukemia (AML) came from 3 sources: (1) the Sidney Kimmel Cancer Center at Johns Hopkins, the Vanderbilt-Ingram Cancer Center, the MDS and MPN Centre at the Chinese Academy of Medical Sciences, the Munich Leukemia Laboratory, and the Allegheny Health Network Cancer Institute; (2) the Project Genie database14; and (3) extraction from 86 publications (supplemental References). The use of all deidentified patient data was approved by the institutional review boards at the respective institutions. Additional details are in supplemental Methods.
Cells
HEK293T, TF1, and K562 cells were obtained from the ATCC. BC1 human induced pluripotent stem cells (hiPSCs) were a gift from Zack Wang (Johns Hopkins). HEK293T cells were grown in DMEM/10% FBS, K562 in RPMI/20% FBS, TF1 cells in RPMI/20% FBS with 2 ng/mL GM-CSF, and BC1 in mTeSR Plus. STR authentication and mycoplasma testing were done upon receipt and routinely thereafter, with the last testing in February 2022.
hiPSC knockins
Knockins were generated using a 2-step targeting process similar to the CORRECT method of Paquet et al.15 Additional details are in supplemental Methods.
Transcriptome analysis
Total RNA isolation, complementary DNA synthesis, isoform-competitive end point polymerase chain reaction (PCR), and isoform-independent quantitative PCR with SYBR Green were performed as described.13,16 All primer sequences are listed in the supplemental Table 1. RNA sequence (RNA-seq) libraries from TF1 clones were constructed using TruSeq Stranded Total RNA Library Prep. Sequencing was performed on a NovaSeq S1 flow cell. Reads were aligned using STAR.17 Splicing analysis was performed using ASCOT algorithms and gene expression using featureCounts.18,19 Additional details are in supplemental Methods.
Affinity purification of SF3B1-associated proteins
A small-scale protocol was applied to both HEK293T and TF1 cells as previously described, except that for TF1, 10 million cells were used and proteins were eluted with 30 μL (5 μg/μL) 3× FLAG peptide because cells had only 1 affinity tag (FLAG) attached to SF3B1.20
Western blotting
Immunoblotting of proteins following affinity purification of SF3B1 in HEK293T and TF1 cells was performed as previously described.20 Additional details are in supplemental Methods.
Results
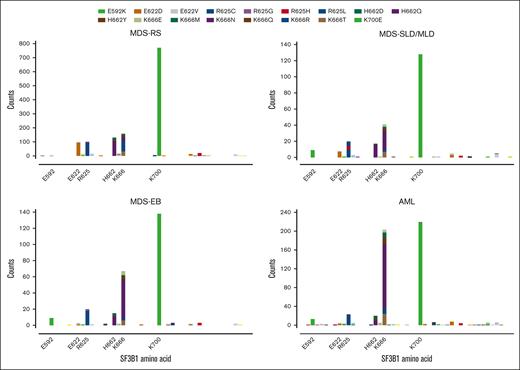
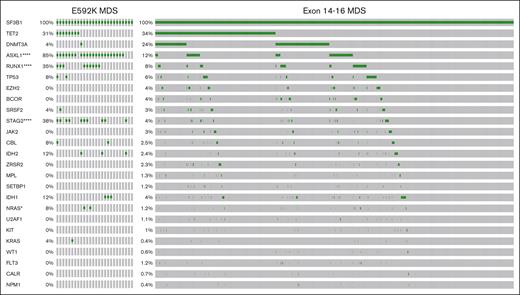
Combining patient data from 5 institutions, publicly available databases, and published literature, we established a data set of 3330 patients with SF3B1-mutant MDS or AML in which exons 13 through 16 had been sequenced. We first determined how SF3B1 mutations were partitioned into World Health Organization (WHO) 2016 classifications, as these data were available for virtually all patients. This distribution showed several asymmetries (Figure 1; supplemental Figure 1). Consistent with our previous report,13 the frequency of K666N was progressively higher in disease types of increasing risk: only 2.1% (28/1360) in MDS-RS vs 9.3% (23/248) in MDS– single-lineage dysplasia/multilineage dysplasia (SLD/MLD), 17.7% (48/272) in MDS-EB, and 25.0% (137/548) in AML (P value compared with MDS-RS = 2.7e-6, 1.5e-19, and 1.6e-51, respectively). With the inclusion of cases in which exon 13 had been sequenced, a disproportionately low frequency in MDS-RS was also revealed for a variant from this exon, E592K: only 0.07% (1/1360) in MDS-RS vs 3.6% in (9/248) in MDS-SLD/MLD, 3.3% (9/272) in MDS-EB, and 2.4% (13/548) in AML (P value compared with MDS-RS = 3.8e-7, 7.6e-7, and 8.4e-7, respectively). Conversely, E622D was decreased in higher-risk disease: 7.0% (95/1360) in MDS-RS vs 2.8% (7/248) in MDS-SLD/MLD (P < .05), 0.4% (1/272) in MDS-EB (P < .0001), and 0.5% (3/548) in AML (P value compared with MDS-RS = 1.0e-2, 6.2e-7, and 2.4e-11, respectively). K666R also decreased: 5.5% (75/1360) in MDS-RS vs 1.2% (3/248) in MDS-SLD/MLD, 1.1% (3/272) in MDS-EB, and 2.2% (12/548) in AML (P value compared with MDS-RS = 1.9e-3, 8.5e-4, and 1.0e-3, respectively). These data confirm and extend the scope of asymmetrical partitioning of distinct SF3B1 hotspots in MDS disease subtypes.
Distribution of exon 13-16 SF3B1 mutations within WHO 2016 classifications of MDS and AML. Amino acid positions are shown along the x-axis, and individual variants are counted along the y-axis according to the legend above the graphs. EB, excess blasts.
Distribution of exon 13-16 SF3B1 mutations within WHO 2016 classifications of MDS and AML. Amino acid positions are shown along the x-axis, and individual variants are counted along the y-axis according to the legend above the graphs. EB, excess blasts.
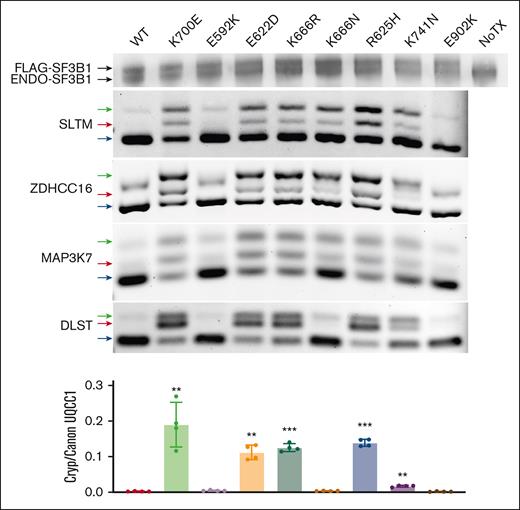
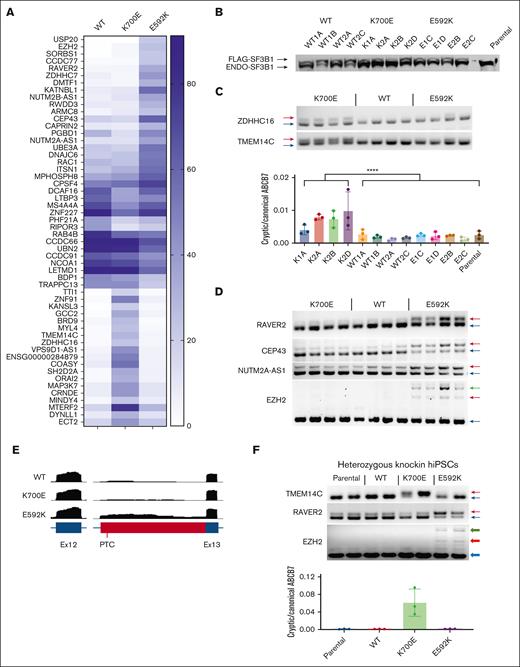
We next examined RNA splicing events produced by those SF3B1 mutations with the most significant asymmetric partitioning. We expressed FLAG–tagged codon-optimized constructs with the E592K, E622D, K666N, and K666R mutations in HEK293 cells, along with wild type SF3B1 and the dominant K700E mutation. For additional comparison, we expressed variants from solid tumors that are rare (R625H, K741N) or absent (E902K) in myeloid malignancies.21 These transfections produced a comparable level of endogenous and exogenous SF3B1 protein (Figure 2). Upon expression of most hot spots, we observed missplicing in junctions known to be affected by SF3B1 mutations, including SLTM, ZDHHC16 and MAP3K7 (Figure 2).2,6 This included MDS mutations K700E, E622D, K666R, and K666N but also solid tumor hotspots R625H and K741N, the last of which produced lower magnitude missplicing as recently reported in the context of uveal melanoma.22 In contrast, these junctions were not misspliced by E902K, which showed its unique missplicing pattern as was noted in TCGA E902K bladder cancer samples (supplemental Figure 2).21 We also found that 2 other mutant SF3B1-associated junctions, in DLST and UQCC1, were misspliced to high magnitude by most mutations but were unaffected by K666N, confirming that the pattern of missplicing by K666N is different from that of other MDS/AML-associated hotspots.13,23 Notably, this pattern occurred with K666N, but not K666R, demonstrating these variants are not functionally equivalent even though they affect the same starting amino acid, a phenomenon also observed with variants in the yeast homolog of SF3B1.24 Finally, conspicuously absent was any missplicing of these events by E592K. Coupled with its near-absence in MDS-RS, this distinct missplicing pattern motivated us to investigate E592K further.
Asymmetric RNA missplicing by distinct SF3B1 mutation hotspots. HEK293T cells were transfected with constructs expressing FLAG-SF3B1 variants. Top row is western blotting with anti-SF3B1 antibody, showing FLAG-SF3B1 and endogenous SF3B1 at similar levels. End point PCR used isoform-competitive primers, with arrows for canonical (blue), cryptic (red), and heteroduplex (green) forms. Cryptic vs canonical UQCC1 was quantified as a ratio between 2 separate isoform-specific quantitative PCRs.
Asymmetric RNA missplicing by distinct SF3B1 mutation hotspots. HEK293T cells were transfected with constructs expressing FLAG-SF3B1 variants. Top row is western blotting with anti-SF3B1 antibody, showing FLAG-SF3B1 and endogenous SF3B1 at similar levels. End point PCR used isoform-competitive primers, with arrows for canonical (blue), cryptic (red), and heteroduplex (green) forms. Cryptic vs canonical UQCC1 was quantified as a ratio between 2 separate isoform-specific quantitative PCRs.
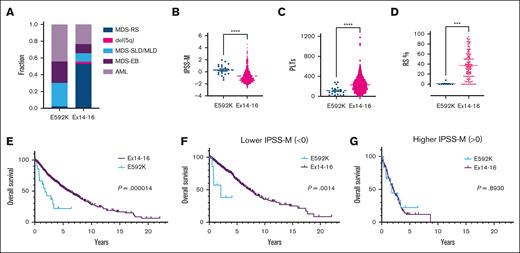
We characterized the clinical features of E592K patients in more detail. The hot spot agnostic collection of all patients with exon 13-16 sequencing had produced 40 cases of E592K, comprising 1.2% (40/3300) of SF3B1-mutant MDS/AML in our data set and 0.25% (8/3323) of all MDS in the IPSS-M cohort.10 By specifically seeking out additional cases from multiple institutions, we gathered a total of 51 patients with E592K-mutated myeloid neoplasms, 45 of whom were with MDS or AML (supplemental Table 2). This expanded E592K cohort also showed asymmetry in 2016 WHO classifications, and these patients had higher IPSS-M scores, IPSS-R scores, and lower platelets than cases with exon 14-16 mutations (Figure 3A-C; supplemental Figure 3A). Hemoglobin and WBC were not significantly different (supplemental Figure 3B-C). E592K cases also had a notable lack of RS (supplemental Figure 3D), with only 1 instance of low-blast E592K MDS reporting any RS, at 8%, therein being the only E592K patient to meet WHO 2016 criteria for MDS-RS (>5% RS if SF3B1 mutation present). By contrast, for low-blast MDS with exon 14-16 mutations in which exact RS percentages were available, the average was 37%, consistent with the known pathological and mechanistic links between mutant SF3B1 and RS in MDS (Figure 3D).5,25 E592K MDS and AML also had a notable comutation profile (Figure 4; supplemental Figures 4-7). Like exon 14-16 mutations, E592K co-occurred rarely with mutations in other splicing factors (SRSF2, U2AF1, ZRSR2), a pattern consistent with pathogenicity for spliceosome mutations. However, a striking distinction was the nearly ubiquitous comutation of ASXL1 (85%) in E592K MDS, compared with only 12% in exon 14-16 MDS (P = 1.1e-16, q = 2.4e-15). High ASXL1 comutation characterized both low and high blast E592K cases, suggesting this relationship occurs early in disease evolution (supplemental Figures 4-6). E592K MDS patients also had increased comutations of RUNX1 (35% vs 8%) and STAG2 (38% vs 4%) (P = 2.1e-10 and 5.0e-8; q = 2.2e-9 and 3.5e-7, respectively). Additionally, there was a noteworthy trend toward mutual exclusivity between E592K and DNMT3A comutations (4%) in MDS, despite DNMT3A being the second-most commonly comutated gene (24%) in exon 14-16 MDS (P = .017 but q = 0.074). In fact, the 1 DNMT3A mutation in E592K MDS was a VAF of 2.3% in a sample in which the VAFs for E592K and ASXL1 mutations were 20%, raising the possibility that the DNMT3A mutation was in a separate clone with wild type SF3B1. When all MDS and AML cases were considered together, the mutual exclusivity between E592K and DNMT3A mutations was significant (P = 1.8e-3, q = 8.8e-3; supplemental Figure 8). Finally, both overall survival and leukemia-free survival were markedly shorter in E592K MDS patients (Figure 3E; supplemental Figure 3E). Binary stratification into IPSS-M higher (score > 0) or lower (score < 0) categories showed that although outcomes were similarly poor for E592K and exon 14-16 patients in the higher group, E592K had significantly worse survival in the lower group (Figure 3F-G; supplemental Figure 3F-G). Analogous stratification using the IPSS-R yielded similar results (supplemental Figure 8D-E). For additional context, we compared survival of our data set to non–SF3B1-mutant patients from the cohort that trained the IPSS-M (supplemental Figure 8). Like E592K, non–SF3B1-mutant MDS had lower survival than exon 14-16, although binary stratification by the IPSS-M—and to a lesser extent the IPSS-R—separated the lower- and higher-risk non–SF3B1-mutant patients better than E592K. Taken together, these data demonstrate that SF3B1-mutant MDS patients with E592K have distinct and high-risk clinical features.
Clinical parameters of MDS patients with the E592K variant of SF3B1. Compared with cases with exon 14-16 mutations, patients with E592K have (A) higher fractions of MDS-SLD/MLD, MDS-EB, and AML, (B) higher IPSS-M, (C) lower platelets, (D) nearly-absent RS, and (E) lower overall survival which is retained in (F) lower IPSS-M score groups. Outcomes are similarly poor for E592K and exon 14-16 patients in (G) higher IPSS-M score groups.
Clinical parameters of MDS patients with the E592K variant of SF3B1. Compared with cases with exon 14-16 mutations, patients with E592K have (A) higher fractions of MDS-SLD/MLD, MDS-EB, and AML, (B) higher IPSS-M, (C) lower platelets, (D) nearly-absent RS, and (E) lower overall survival which is retained in (F) lower IPSS-M score groups. Outcomes are similarly poor for E592K and exon 14-16 patients in (G) higher IPSS-M score groups.
Comutation landscape of MDS with E592K or exon 14-16 SF3B1 mutations. All cases of SF3B1-mutant MDS that sequenced at least the 24 additional genes shown were included. Fisher exact test P values and Benjamini linear multiple testing q values for FDR < 0.05 were as follows: ∗∗∗∗ASXL1 = 1.13E-16 and 2.37E-15; ∗∗∗∗RUNX1 = 2.06E-10 and 2.16E-09; ∗∗∗∗STAG2 = 5.03E-08 and 3.52E-07; and ∗NRAS = 4.61E-03 and 2.42E-02.
Comutation landscape of MDS with E592K or exon 14-16 SF3B1 mutations. All cases of SF3B1-mutant MDS that sequenced at least the 24 additional genes shown were included. Fisher exact test P values and Benjamini linear multiple testing q values for FDR < 0.05 were as follows: ∗∗∗∗ASXL1 = 1.13E-16 and 2.37E-15; ∗∗∗∗RUNX1 = 2.06E-10 and 2.16E-09; ∗∗∗∗STAG2 = 5.03E-08 and 3.52E-07; and ∗NRAS = 4.61E-03 and 2.42E-02.
Because our initial splicing analysis merely showed the absence of known SF3B1-mutant events in E592K cells, we next sought missplicing events specifically induced by this variant. To do so, we stably expressed the WT, K700E, and E592K mutations through lentiviral delivery in TF1 cells, isolated multiple independent clones for each genotype, and performed RNA-seq to quantify percent spliced in (PSI) values for all splice junctions (Figure 5A). K700E produced a characteristic pattern of increased expression of junctions using alternative 3’ acceptors, with high magnitude missplicing of genes like MAP3K7 and ZDHCC16, as expected. In contrast, the E592K mutation produced a fundamentally different pattern of missplicing, with its affected genes nonoverlapping with those misspliced by K700E, and vice versa. Western blotting showed that exogenous FLAG–tagged SF3B1 was equal to or less than the endogenous SF3B1 form, not overexpressed above it (Figure 5B). We then validated several missplicing events with end point and quantitative PCR assays. For K700E-specific junctions, this included missplicing of ZDHHC16, TMEM14C, and ABCB7 (Figure 5B-C). For E592K-specific junctions, this included RAVER2, CEP43, NUTM2A-AS1, and EZH2 (Figure 5D). Interestingly, the EZH2 missplicing event was an alternate acceptor in intron 12, creating a premature termination codon predicted to activate nonsense-mediated decay (Figure 5E). We further validated the hot spot specificity of these events in 2 other overexpression contexts: transiently transfected HEK293T and stably transduced K562 cells (supplemental Figure 9). To determine the effect of precisely heterozygous expression of these variants from their natural genetic elements, we also queried hot spot–specific missplicing in hiPSCs that we engineered with heterozygous knockins of K700E, E592K, and R702R (a single nucleotide polymorphism in the human population) (Figure 5E-F; supplemental Figure 10). In all cases, K700E- and E592K–dependent missplicing events were present and distinct. Of note, missplicing of TMEM14C and ABCB7 were recently shown to drive RS formation in iPSCs derived from SF3B1-mutant MDS.5 Both genes were clearly misspliced by K700E, but not by E592K, consistent with the lack of sideroblastic anemia in E592K patients. Together, these data show that the E592K variant of SF3B1 has a unique pattern of RNA missplicing.
E592K induces unique RNA missplicing events. TF1 cells transduced with different SF3B1 variants were analyzed by RNA-seq, with (A) highest-scoring ΔPSIs shown. (B) Western blot with anti-SF3B1 antibody. (C) End point PCR/quantitative PCR validation of K700E–specific missplicing events. (D) Validation of E592K-specific events. (E) RNA-seq reads from TF1 cells showing the cryptic event in EZH2. (F) Orthogonal validation of missplicing in heterozygous SF3B1 knockin hiPSCs. PTC, premature termination codon.
E592K induces unique RNA missplicing events. TF1 cells transduced with different SF3B1 variants were analyzed by RNA-seq, with (A) highest-scoring ΔPSIs shown. (B) Western blot with anti-SF3B1 antibody. (C) End point PCR/quantitative PCR validation of K700E–specific missplicing events. (D) Validation of E592K-specific events. (E) RNA-seq reads from TF1 cells showing the cryptic event in EZH2. (F) Orthogonal validation of missplicing in heterozygous SF3B1 knockin hiPSCs. PTC, premature termination codon.
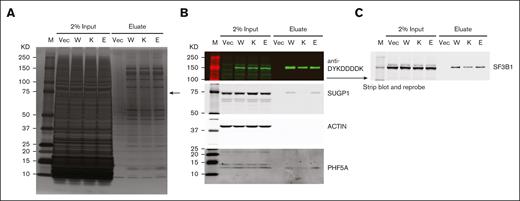
In addition to shared RNA missplicing events, the most well-studied SF3B1 hot spot mutations also share a specific biochemical defect: disruption of the interaction between SF3B1 and SUGP1.20 This disruption is not incidental to missplicing but directly mediates it; inactivation of SUGP1 recapitulates SF3B1-mutant missplicing and overexpression of SUGP1 partially rescues it.20 We therefore, asked whether E592K, with its nonoverlapping missplicing events, might preserve the interaction of SF3B1 with SUGP1. Indeed, when His6-FLAG-SF3B1 variants were introduced into HEK293T cells and affinity purified using anti-DYKDDDDK (FLAG) antibody and cobalt beads, the association of His6-FLAG-SF3B1 with endogenous SUGP1 was disrupted by K700E but not by wild type or E592K SF3B1 (Figure 6). We then asked whether E592K might instead disrupt the interaction between SF3B1 and PHF5A, given that E592 is in HEAT repeat 3 at the interface with PHF5A.26 However, this interaction was preserved by each His6-FLAG-SF3B1 variant (Figure 6). We also observed these effects in a second cell context, TF1 cells expressing FLAG-SF3B1 variants (supplemental Figure 11). Together, these data indicate that, consistent with its induction of unique RNA missplicing events, the E592K variant does not participate in the disruption of the SF3B1-SUGP1 interaction that drives the cryptic splicing of other SF3B1 mutations.
The E592K variant preserves association of SF3B1 with SUGP1. HEK293T cells were transfected with His6-FLAG-SF3B1 variants and subjected to affinity purification with anti-DYKDDDDK (FLAG) antibody. (A) Silver-stained protein gel, with arrow pointing to the size of SUGP1, which is decreased in K700E but not E592K eluate. (B) Western blot showing decreased SUGP1 in K700E, but not E592K, eluate. PHF5A is present with all SF3B1 variants. (C) Reprobing with anti-SF3B1 shows native and His6-FLAG–tagged protein levels. E, E592K; K, K700E; KD, kilodaltons; M, marker; Vec, vector only; W, wild type SF3B1.
The E592K variant preserves association of SF3B1 with SUGP1. HEK293T cells were transfected with His6-FLAG-SF3B1 variants and subjected to affinity purification with anti-DYKDDDDK (FLAG) antibody. (A) Silver-stained protein gel, with arrow pointing to the size of SUGP1, which is decreased in K700E but not E592K eluate. (B) Western blot showing decreased SUGP1 in K700E, but not E592K, eluate. PHF5A is present with all SF3B1 variants. (C) Reprobing with anti-SF3B1 shows native and His6-FLAG–tagged protein levels. E, E592K; K, K700E; KD, kilodaltons; M, marker; Vec, vector only; W, wild type SF3B1.
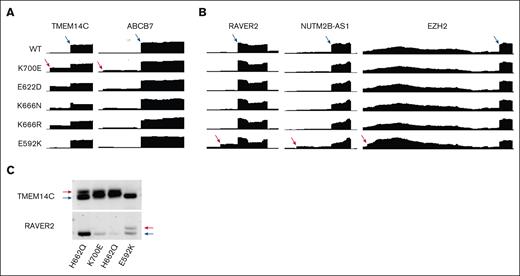
Finally, we identified and analyzed RNA-seq from 2 patients with E592K mutation, compared with other SF3B1 mutations, from the recent Munich Leukemia Laboratory cohort of spliceosome-mutant myeloid malignancy patients.27 Inspection of the junctions validated in our cell models also demonstrated the same specificity of missplicing in these primary patient samples, when compared with other SF3B1 mutations (Figure 7A). In addition, end point PCR validation of TMEM14C and RAVER2 from a third primary E592K sample at a separate institution demonstrated the same hot spot specificity of RNA missplicing (Figure 7B), validating the findings from our cell models.
E592K exhibits unique RNA missplicing in primary MDS samples. RNA-seq read distribution in the MLL cohort shows that E592K exhibits (A) canonical TMEM14C and ABCB7 missplicing and (B) cryptic RAVER2, NUTM2B-AS1, and EZH2 missplicing. (C) Distinct TMEM14C and RAVER2 missplicing was validated in marrow CD34+ cells from an independent patient by end point PCR.
E592K exhibits unique RNA missplicing in primary MDS samples. RNA-seq read distribution in the MLL cohort shows that E592K exhibits (A) canonical TMEM14C and ABCB7 missplicing and (B) cryptic RAVER2, NUTM2B-AS1, and EZH2 missplicing. (C) Distinct TMEM14C and RAVER2 missplicing was validated in marrow CD34+ cells from an independent patient by end point PCR.
Discussion
Here, we show the E592K variant of SF3B1 produces unique RNA missplicing and is associated with high-risk MDS. These results have several implications. First, the distinctiveness of E592K informs the pathobiology of SF3B1-mutant MDS. Clough et al elegantly showed that RS is formed by TMEM14C and ABCB7 missplicing in MDS-derived iPSCs with the G742D variant of SF3B1.5 These missplicing events have been seen with other hotspots, including K700E, which we corroborated here.2,28 Cells with E592K, in contrast, preserved canonical splicing of these genes, and cases of E592K MDS lacked RS. Other events that have been implicated in SF3B1-mutant MDS pathobiology include innate immune activation by missplicing of MAP3K7 and IRAK4, enhanced self-renewal by MECOM missplicing, impaired erythropoiesis by COASY missplicing, and hepcidin suppression by ERFE missplicing; all these genes were also canonically spliced in E592K cells (supplemental Figure 12).3,4,6-8 A distinct E592K pathobiology is also suggested by its high co-occurrence with ASXL1/RUNX1/STAG2 mutations and mutual exclusivity with DNMT3A mutations, a starkly different pattern than that of other SF3B1 hot spots. Like most comutation patterns in cancer, it is unclear whether these co-occurrences are driven by synergism or permissiveness and whether the mutual exclusivity is driven by antagonism or redundancy. Interestingly, the same pattern of co-occurrence (ASXL1/RUNX1/STAG2) and mutual exclusivity (DNMT3A) occurs with loss-of-function EZH2 mutations in MDS (supplemental Figure 13).10,14 It is therefore intriguing that, E592K missplicing produced a frameshifted EZH2 transcript in our studies here, and it is tempting to speculate this may create “mutant EZH2-ness” in E592K cells, an area for future investigation. If so, mutual exclusivity due to redundancy might be expected between mutant EZH2 and E592K. While there were indeed no EZH2 mutations in the 51 E592K patients here, the low overall frequency (4%) of EZH2 in SF3B1-mutant myeloid malignancies means a larger cohort would be needed for significance testing of this particular pair. Nonetheless, the distinctiveness of E592K here shows that certain MDS phenotypes associated with SF3B1 mutation, including sideroblastic anemia, are not inevitable pathobiological outcomes of RNA missplicing by all SF3B1 variants.
Second, there are implications for MDS classification. Recently, both the International Consensus Classification (ICC) and the fifth edition of the WHO classification of myeloid neoplasms created nosologic entities for SF3B1-mutant MDS.29,30 These are based on the International Working Group findings that SF3B1 mutation, low blasts, and lack of certain co-occurring genetic aberrations defined an indolent MDS with shared pathobiology of idiosyncratic RNA missplicing that was more homogenous than the MDS-RS classification.31 This was due, in part, to exclusion of SF3B1-unmutated MDS-RS which had greater myeloid/megakaryocyte dysplasia, more TP53 comutations, and poorer outcomes. Both the ICC and WHO criteria require SF3B1 mutation, low blasts, cytopenias, dysplasia, and lack of multihit TP53/del(5q)/-7/complex cytogenetics; the ICC also requires SF3B1 VAF>10% and lack of RUNX1/del(7q)/abn3q26.2 and the WHO allows for >15% RS to substitute for SF3B1 mutation. As we have seen here, E592K patients do not fit these groups: they have unique RNA missplicing, higher blasts, lack of RS, increased RUNX1 mutations, and poorer prognosis. Although E592K is, overall, an infrequent mutation, these differences do suggest that E592K could be considered an additional exclusion criterion for these entities. They also suggest consideration of specific hot spot mutations in future classification efforts. If so, this would be the precise mutation, not just the affected amino acid, as we demonstrated different patterns of disease partitioning and missplicing between K666R and K666N.
Third, our data have implications for the prognostication of E592K patients. Along with low blasts, shallow cytopenias, and certain cytogenetic abnormalities such as del(11q), SF3B1 mutations have been consistently associated with better MDS outcomes in multiple independent data sets.1,9-12 Accordingly, new MDS prognosis scoring tools incorporating mutational data have generally weighted SF3B1 mutations favorably, although there is important context-dependence that overrides this favorability, such as increased blasts and aberrations such as del(5q), RUNX1 mutation, and others.10-12 However, given the much larger patient cohorts that would be required, prognostication tools do not yet separately weight the myriad individual variants of mutated genes (SF3B1 or otherwise) into their scores. In our cohort, the group defined by higher IPSS-M risk categories included E592K and exon 14-16 patients with similarly poor outcomes, whereas the group composed of lower risk categories captured patients with better outcomes for exon 14-16, but not E592K, disease (Figure 3E-G). Although the number of E592K patients in these groups are still modest overall, these data nonetheless suggest caution in regarding any E592K patients as lower risk. For example, patients 9 and 1 progressed from MDS to AML at 6 and 9 months, respectively, despite lower-risk scores from both the IPSS-M and SEX-GSS (supplemental Figure 14). Furthermore, with the caveat that E592K is uncommon and may represent a special case, our findings suggest consideration of incorporating specific hot spot mutations in future prognostication tools.
Fourth, these results have implications for investigational therapies that target the missplicing defects of SF3B1-mutant cells. Gene vector therapies that leverage missplicing to deploy therapeutic cargo against spliceosome-mutant cells are promising, but such vectors would need to be not only gene-specific (ie, SF3B1 vs SRSF2) but, in the case of E592K, hot spot-specific due to its nonoverlapping missplicing.32,33 Antisense oligonucleotide therapy aimed at rescuing BRD9 missplicing showed activity against SF3B1-mutant tumors in vivo, but such therapy would not apply to E592K, which does not missplice BRD9 (supplemental Figure 10).34 It is also possible that synthetic lethality approaches of further disrupting the spliceosome itself in SF3B1-mutant cells could affect E592K cells differently.35
Finally, these findings add to our understanding of the functional diversity of spliceosome mutations. After these mutations were first discovered as conspicuously enriched in myeloid neoplasms in a mutually exclusive manner, efforts have sought unifying downstream functional effects that might explain this occurrence pattern.36 This has revealed shared phenotypes, such as convergent disruption of certain pathways or genes (eg, mutant SRSF2 missplices EZH2 but at a different junction than we observed here for SF3B1 E592K), creation of genotoxic R-loops, and sensitization to further disruption of the spliceosome.6,35,37-41 At the same time, bulk and then single-cell analyses have shown that spliceosome mutations are not always mutually exclusive, with multiple mutations sometimes selected for in the same cells.23 This indicates that different spliceosome mutations can confer different, even complementary, advantages to cancer cells, and the mutual exclusivity that does occur may be more from the splicing toxicity of combining certain mutations than from functional redundancy.23 Consistent with this finding, studies have shown direct functional differences between spliceosome mutations: the missplicing events induced by mutations in different spliceosome genes (ie, SRSF2 vs U2AF1, etc) are nonoverlapping, the 2 dominant hot spot mutations in U2AF1 missplice different genes, and some less common SRSF2 and U2AF1 variants only partially or “dually” recapitulate the RNA missplicing of hot spot mutations.2,40,42-44 For SF3B1 mutations, their asymmetric partitioning among cancer types (ie, K700E/H662Q in MDS, R625C/R625H in uveal melanoma, G642D in CLL, etc) first suggested the mutations may have functional differences.36,45,46 Later, Seiler et al noted different missplicing events in TCGA RNA-seq from bladder tumors with the E902K variant, events that we experimentally confirmed here.21 For other SF3B1 hotspots, studies have described differences of degree in RNA missplicing events, including K700E vs R625C/R625H in primary samples, R625H vs K741Q vs others in isogenic cells, and K666N vs K700E/H662Q in primary samples and isogenic cells.13,22,23,47 To these, we add E592K, whose missplicing events are differences of kind, nonoverlapping with those of other MDS-associated hotspots. We also find distinct biochemistry for E592K, with preservation of the interaction between SUGP1 and SF3B1, and future work should include screening for altered interactions between E592K and other spliceosome proteins. Although it remains possible that the important underlying oncogenic effects of E592K are phenotypes still shared with other SF3B1 hotspots, the distinctiveness of E592K suggests to us that different SF3B1 variants can promote different kinds of leukemia by inducing different RNA missplicing events. Because these differences can impact our understanding and management of patients with myeloid neoplasms, additional studies of differences between other spliceosome variants are warranted.
Acknowledgments
This work was supported by grants from the National Heart Lung Blood Institute of the National Institutes of Health (NIH) (HL159306), Edward P. Evans Foundation, Leukemia & Lymphoma Society, Maryland Stem Cell Research Fund, Gabrielle’s Angel Foundation for Cancer Research, Emerson Collective, and AbbVie (W.B.D.), NIH R35 GM118136 (J.L.M.), Lady Tata Memorial Trust (3218), and the Barts Charity (G-002167) (C.P. and K.R.-P). D.H.W. is supported by an Advanced Clinician Scientist Fellowship from Cancer Research UK, a grant from the The Oglesby Charitable Trust, and the University of Manchester’s Sybil Mary Pilkington Leukaemia Research Fellowship.
Authorship
Contribution: A.E.D. and W.B.D. conceived the study; I.Y.C., J.P.L., J.Z., E.H., J.L.M., K.R.-P., A.E.D., and W.B.D. designed the research; I.Y.C., J.P.L., J.Z., E.H., P.T., C.P., B.D., and W.B.D. performed the experiments; J.L.M., K.R.-P., and W.B.D. supervised the experiments; I.Y.C., W.W., R.E.B., C.P., B.L., D.H.W., K.B., M.O., E.B., X.L., T.H., A.K., S.F., C.D.G., T.J., A.E.D., and W.B.D. collected the data; I.Y.C., J.P.L., J.Z., W.W., J.L.M., K.R.-P., A.E.D., and W.B.D. analyzed the data; I.Y.C., J.P.L., J.Z., K.R.-P., and W.B.D. prepared the figures; I.Y.C. and W.B.D. wrote the manuscript; and all authors critically reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: T.J. received institutional research support from CTI Biopharma, Kartos Therapeutics, Incyte, and Bristol Myers Squibb, and has been a participant on the advisory boards with Bristol Myers Squibb, Incyte, AbbVie, CTI Biopharma, Kite, Cogent Biosciences, Blueprint Medicine, Telios Pharma, Protagonist Therapeutics, Galapagos, TScan Therapeutics, Karyopharm, and MorphoSys. S.F. has consulted for and received honoraria from Blueprint Medicine, CTI Biopharma, Sobi, Bristol Myers Squibb, AbbVie, Gilead, GlaxoSmithKline, Incyte, Janssen, Jazz, Karyopharm, Novartis, PharmaEssentia, Sanofi, Servier, Stemline, Taiho, and Takeda. The remaining authors declare no competing financial interests.
Correspondence: William Brian Dalton, The Johns Hopkins Sidney Kimmel Comprehensive Cancer Center, Cancer Research Building Room 2M07, 1650 Orleans St, Baltimore, MD 21231; email: wdalton2@jhmi.edu.
References
Author notes
Raw FASTQ files are deposited in the NCBI Sequence Read Archive under accession number PRJNA1107944.
The full-text version of this article contains a data supplement.