Key Points
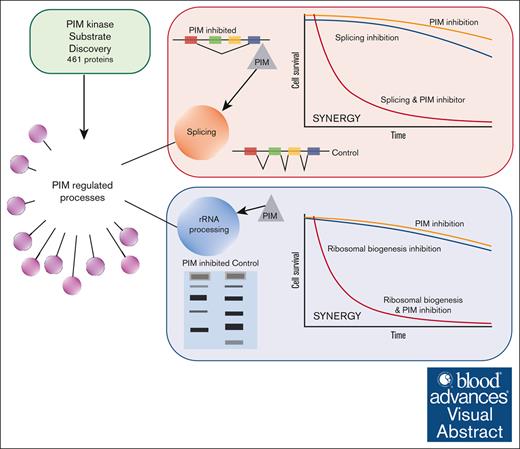
Proteomic profiling of PIM kinase substrates identifies RNA splicing and ribosome biogenesis as PIM-controlled cellular processes.
PIM inhibitors alter splicing and rRNA processing and synergize with splicing modulators or an RNA polymerase I inhibitor to kill AML cells.
Visual Abstract
Provirus integration site for Moloney murine leukemia virus (PIM) family serine/threonine kinases perform protumorigenic functions in hematologic malignancies and solid tumors by phosphorylating substrates involved in tumor metabolism, cell survival, metastasis, inflammation, and immune cell invasion. However, a comprehensive understanding of PIM kinase functions is currently lacking. Multiple small-molecule PIM kinase inhibitors are currently being evaluated as cotherapeutics in patients with cancer. To further illuminate PIM kinase functions in cancer, we deeply profiled PIM1 substrates using the reverse in-gel kinase assay to identify downstream cellular processes targetable with small molecules. Pathway analyses of putative PIM substrates nominated RNA splicing and ribosomal RNA (rRNA) processing as PIM-regulated cellular processes. PIM inhibition elicited reproducible splicing changes in PIM-inhibitor–responsive acute myeloid leukemia (AML) cell lines. PIM inhibitors synergized with splicing modulators targeting splicing factor 3b subunit 1 (SF3B1) and serine-arginine protein kinase 1 (SRPK1) to kill AML cells. PIM inhibition also altered rRNA processing, and PIM inhibitors synergized with an RNA polymerase I inhibitor to kill AML cells and block AML tumor growth. These data demonstrate that deep kinase substrate knowledge can illuminate unappreciated kinase functions, nominating synergistic cotherapeutic strategies. This approach may expand the cotherapeutic armamentarium to overcome kinase inhibitor–resistant disease that limits durable responses in malignant disease.
Introduction
Deregulation of kinase signaling networks is a common feature of nearly all malignancies.1 Targeting these networks using small-molecule drugs, principally kinase inhibitors (KIs), is revolutionizing cancer therapy.2 However, the promise of this therapeutic approach is tempered by the reality that durable responses are limited in most patients by the emergence of drug-resistant disease.3 KI resistance develops through multiple molecular mechanisms that reactivate the targeted signaling pathway, including kinase mutations that preclude drug binding, activation of downstream signaling, and upregulation of compensatory signaling pathways.4 These changes can emerge through de novo alterations in the bulk tumor cell population induced by therapeutic pressure or through the selection of preexisting subclones owing to cancer cell heterogeneity. KI polypharmacology approaches, wherein multiple signaling pathways are simultaneously inhibited, are widely anticipated to extend time to relapse by increasing the roadblocks that must be circumvented to permit cancer cell survival. However, choosing efficacious drug combinations remains a critical challenge.4 For well-characterized pathways, it may be feasible to target multiple components of a single pathway to prevent downstream signaling reactivation. A prominent exemplar of this strategy is the combination of B-raf inhibitors and mitogen-activated protein kinase (MEK) inhibitors in B-raf V600E-mutant melanomas.5 In cases where compensatory signaling that fuels resistance has been defined, increased therapeutic efficacy can also be achieved by simultaneously targeting the driver and compensating pathways. For example, in triple-negative breast cancer, MEK inhibition leads to upregulation of several receptor tyrosine kinases that activate extracellular signal-regulated protein kinase. Combining MEK inhibitors with sorafinib, which targets multiple receptor tyrosine kinases, led to increased tumor growth inhibition.6
Where such rational choices are less obvious, high-throughput combinatorial drug screens and genetic approaches that illuminate drug-phenotype relationships may be informative.7,8 Recently, patient–individualized system approaches that combine in silico prediction and ex vivo analyses of patient samples have identified synergistic combinations among 218 compounds, including KIs.9
From the examples cited above, it is axiomatic that knowledge of kinase signaling input and output is central to rationalizing potential drug combinations to inform multihit strategies on a single pathway, to combat compensatory signaling, and to train system-level algorithms. In terms of output, knowledge of direct physiological kinase substrates provides a clear route to elucidating signaling outcomes and rationalizing drug combinations, as in the case of B-raf and MEK inhibitors. Multiple biochemical and genetic approaches have been developed to profile protein kinase substrates.10 Despite substantial progress in our understanding of kinase-substrate interactions, comprehensive substrate repertoires are lacking even for the most well-studied kinases.11 This partial knowledge of a key dimension of kinase biology12 restricts our ability to rationalize cotherapeutics for many KIs.
Reasoning that deepened knowledge of direct kinase-substrate interactions would nominate new rational KI cotherapeutic strategies, we sought to comprehensively profile substrates of a well-studied oncogenic kinase family in a leukemia cell line. Provirus integration site for Moloney murine leukemia virus (PIM) kinases have long been known to be drivers of oncogenesis in acute myeloid leukemia (AML) and to augment oncogenic processes in other malignancies.13,14 PIM1, 2, and 3 are constitutively active serine threonine kinases, and triple loss-of-function affects growth but not viability or fertility in mice.15 This dispensability positions them as attractive therapeutic targets, and multiple small-molecule PIM KIs have been tested or are currently under evaluation in clinical trials for cancer treatment.16 PIM kinases are known to directly phosphorylate substrates involved in cell cycle progression, survival, translation, transcription, and drug resistance, and this knowledge has been leveraged to suggest rational, mechanism-based drug combinations that have shown considerable preclinical promise against multiple forms of cancer.17 PIM kinases have also been strongly implicated in the emergence of therapeutic resistance in cancer through multiple mechanisms, including direct phosphorylation of drug transporters.18-21
Here, we describe a high-throughput iteration of the reverse in-gel kinase assay22 (RIKA) and demonstrate, using PIM1 as a paradigm, that broadening understanding of kinase-substrate interactions identifies previously underappreciated cellular processes under kinase control and efficiently nominates rational drug combinations that synergize to block AML cell growth. Extending this approach to other oncogenic drivers and supportive kinases has the potential to considerably expand therapeutic options to delay time to relapse and extend overall survival in AML and other cancers treated with KIs.
Methods
Cell culture
All cell lines were maintained at 5% CO2 and 37°C. Culture media were supplemented with antibiotics, antimycotics, and antimycoplasma agents.
Recombinant proteins
PIM1 and PIM2 genes (Open Biosystems) were cloned into pQE-80L and expressed in E. coli BL21. Recombinant PIM kinases were purified using nickel resin. Recombinant PIM substrates were generated by cloning complementary DNAs (cDNAs) reverse transcribed from RNA extracted from HeLa cells into pQE-80L and purified as described above for PIM kinases.
RIKA
RIKAs were performed essentially as described.22 For shotgun RIKAs, ∼1 mg hydrogen fluoride (HF)–treated K562 protein extract was analyzed in each of four 5-mL RIKA gels containing PIM1 (50 ug/mL). HF treatment was used to dephosphorylate and deglycosylate proteins as described.23 RIKA gels were cut into 8 sections, and sections of the same molecular weight from all 4 gels were combined and homogenized in 50 mM NaHCO3. Phosphopeptides were enriched using a titanium dioxide resin kit (Pierce).
NanoUPLC-MS/MS analysis of phosphopeptidome
Titanium dioxide–enriched tryptic phosphopeptides were separated on a nano-ACQUITY ultra performance liquid chromatography analytical column (BEH130 C18, 1.7 μm, 75 μm × 150 mm, Waters) over a 95-minute linear acetonitrile gradient (3%-35%) with 0.1 % formic acid on a Waters nano-ACQUITY UPLC system and analyzed on a coupled Thermo Scientific Orbitrap Fusion Tribrid mass spectrometer. Full scans were acquired at a resolution of 120 000, and precursors were selected for fragmentation by a real-time data-dependent decision tree logic based on the precursor charge state and mass-to-charge ratio using higher-energy collisional dissociation (normalized collision energy at 30%) or electron-transfer dissociation for a maximum 3-second cycle as described by Swaney et al.24 Tandem mass spectra were searched against UniProt Homo sapiens reference proteome using the Sequest HT algorithm described by Eng et al25 and MS Amanda algorithm developed by Dorfer et al26 with a maximum precursor mass error tolerance of 10 ppm. Phosphorylations of serine and threonine (monoisotopic mass change +79.9663 Da) were treated as dynamic modifications. The resulting hits were validated at a maximum false discovery rate of 0.01 using a semisupervised machine learning algorithm Percolator developed by Käll et al.27 The probabilities of phosphorylation sites were calculated by using phosphoRS algorithm developed by Taus et al.28
Inhibitor treatment
AZD1208 (pan-PIM inhibitor) (MedKoo Biosciences, catalog no. 205763), PIM447 (pan-PIM inhibitor) (MedChemExpress, catalog no. HY-19322B), LY2584702 (p70S6 kinase inhibitor) (Selleck chemicals, catalog no. S7704), pladienolide B (SF3B complex inhibitor) (Santacruz Biotechnology, catalog no. 445493-23-2) SPHINX31 (serine-arginine protein kinase 1 [SRPK1] inhibitor) (MedChemExpress, catalog no. HY-117661), and sudemycin D6 (SD6) were solubilized in dimethyl sulfoxide (DMSO). BMH-21 was provided as presolubilized in citrate buffer (pH 6). For vehicle treatment, a solvent volume equal to the volume of the inhibitor stock solution was used.
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays
Metabolic activity was quantified using MTT (GoldBio, catalog no. T-030) by measuring optical density at 570 nm on a Biotek Synergy 2 plate reader. Fraction affected (Fa) values were determined using CompuSyn to calculate the combination indices (CI) at ED50, ED75, ED90, and ED95 (where ED indicates the effective dose of combination).
Flow cytometry
EOL-1 cells were treated with DMSO, PIM inhibitor, SRPK1 inhibitor, or a combination of PIM and SRPK1 inhibitors for 48 hours. Cells were seeded 24 hours before treatment. Cells were harvested and resuspended in cold annexin-binding buffer (ThermoFisher, catalog no. V13241) to a cell density of ∼2e6 cells per mL. Cells were stained with annexin V-Alexa Fluor 647 (Thermo Fisher, catalog no. A23204) and propidium iodide on ice for 10 minutes. Flow cytometry was performed on BD FACSCanto II cytometer equipped with 488 nm and 633 nm lasers. A total of 50 000 singlet events from 2 biological replicates of each treatment condition were recorded in duplicate. Compensation was performed based on single-color control cells stained with annexin V or propidium iodide alone. Data analysis was performed using FCSExpress 7 (version 7.16, De Novo Software). Statistical analysis was performed using 1-way analysis of variance followed by multiple comparisons test with Tukey correction on GraphPad Prism (version 10.1.1).
Microarray analysis of RNA splicing
RNA samples extracted from AML cell lines with, and without, AZD1208 treatment were analyzed on Affymetrix GeneChip Human Transcriptome Analysis 2.0 microarrays. Data were analyzed using Transcriptome Analysis Console software using .CHP files from 3 biological replicates.
RT-PCR analysis
cDNA from DMSO control–treated and inhibitor-treated AML cell lines was analyzed by reverse transcription polymerase chain reaction (RT-PCR) to interrogate changes in glutathione-specific gamma-glutamylcyclotransferase 1 (CHAC1) splicing. Primers were designed by querying messenger RNA (mRNA) sequences using the National Center for Biotechnology (NCBI) nucleotide reference sequence database. NCBI/Primer-BLAST was used to identify primers that recognized sequences flanking alternatively spliced exons. Myeloid cell leukemia seqeunce 1 (MCL1) transcripts MCL-1L (assay name: Hs00172036_m1) and MCL-1S (assay name: Hs00766187_m1) were quantified using TaqMan assays (Thermo Fisher Scientific). About 30 ng of total cDNA was used for each reaction. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, assay name: Hs99999905_m1) was used as a loading control. A CFX96 Touch real-time PCR detection system was used for the reaction. Each sample was tested in triplicates. Data analysis was performed using the BioRad CFX Manager and CFX Maestro software. Ratios of MCL-1S to MCL-1L were quantified, and statistical analysis was performed using 1-way analysis of variance, with corrected standard deviations, followed by multiple comparisons test with Tukey correction on GraphPad Prism (version 10.1.1).
Analysis of rRNA precursors
MOLM-16, EOL-1, and OCI-M1 cells were treated with PIM inhibitor (AZD1208 or PIM447) or S6 kinase inhibitor (LY2584702). RNA (5 μg) was resolved on 0.9% Agarose gels in 1× NorthernMax Gly gel prep/running buffer (Ambion), transferred to Nytran membranes (GE Whatman 10416096), and crosslinked via UV irradiation. Northern blot analysis for ribosomal RNA (rRNA) precursors was performed essentially as described in Tafforeau et al.29
Western blotting
Cells were lysed in denaturing buffer containing urea (7 M urea, 2 M thiourea, 20 mM dithiothreitol, 1% C7BZ0, 20 mM Tris pH 7.5) and resolved proteins were transferred to polyvinylidene difluoride membranes. Phospho-rpS235/236 antibody (Cell Signaling Technology, catalog no. 4858) was used.
In vivo analysis of drug efficacy
5 × 106 EOL-1 cells in phosphate-buffered saline were injected subcutaneously in the flank of 8- to12-week-old Non-Obese Diabetic Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) male and female mice (Jackson Laboratory; strain 005557). Tumor volume was measured every other day until tumor volume reached 1500 mm3 (Institutional Animal Care and Use Committee (IACUC)–mandated end point for euthanasia).
The animal studies were performed under a protocol approved by the IACUC of the University of Maryland, Baltimore County.
Results
Shotgun RIKA identifies >450 new potential PIM kinase substrates
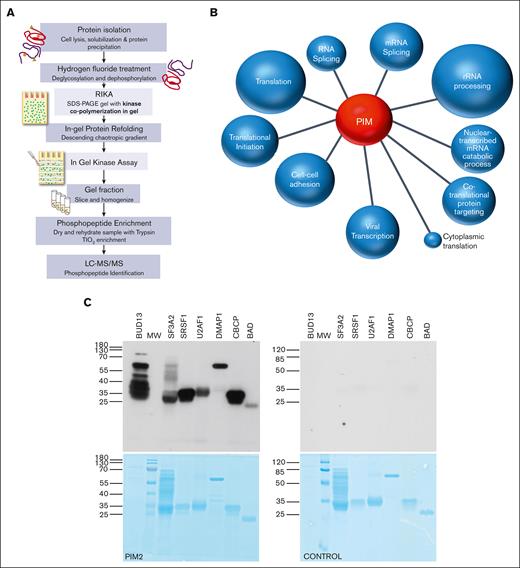
Protein kinase substrates can be identified, and phosphorylation stoichiometry can also be accurately quantified using the RIKA; however the assay throughput is low.22,30 To accelerate kinase substrate discovery, we developed a shotgun RIKA (Figure 1A). K562 chronic myeloid leukemia whole-cell protein extracts were first quantitatively dephosphorylated and deglycosylated using HF23 and resolved on a denaturing sodium dodecyl sulfate polyacrylamide gel with recombinant 33 kDa PIM1S (hereafter, PIM1) polymerized within and a parallel control gel without PIM1. After in-gel protein refolding, in-gel kinase assays were performed. Both gels were then divided into 8 regions based on apparent molecular weight, tryptic peptides were extracted, and phosphopeptides were enriched by TiO2 chromatography. Eluted phosphopeptides were fractionated by reverse-phase liquid chromatography and analyzed using high-resolution tandem mass spectrometry (MS/MS). A total of 1035 phosphopeptides present exclusively in the PIM1-containing gel were mapped to 778 putative PIM1 substrate proteins using Proteome Discoverer31 to query the Uniprot database (false discovery rate <0.05; supplemental Table 1). Of these, 461 were not previously reported as PIM substrates.32,33 Database for Annotation, Visualization, and Integrated Discovery (DAVID) pathway analysis34 demonstrated a strong link between PIM kinase substrates and RNA biology (Figure 1B). Ribosome, spliceosome, RNA transport, ribosome biogenesis, mRNA surveillance, and RNA degradation were among the top 10 DAVID hits (Figure 1B). To confirm specific RNA-linked and other pathway (ie, cell cycle control and DNA repair) components as PIM kinase substrates, full-length recombinant proteins produced in E. coli were analyzed in standard 32P-ATP RIKAs.22 Of 20, 19 were phosphorylated with high catalytic efficiency; 6 are shown in Figure 1C and the remainder in supplemental Figure 1. These data demonstrate that this high-throughput workflow accurately identifies, in most cases, proteins that are robust in vitro PIM substrates.
Shotgun RIKA nominates PIM-controlled pathways and potential physiological substrates. (A) RIKA workflow to survey the cellular proteome for protein substrates of PIM1. (B) Top 10 pathways nominated by DAVID analysis of 778 PIM substrates identified in K562 cell extracts. P values were calculated for all PIM involved pathways. The top 10, based on P value and the presence of >10 genes in each pathway, were plotted. Distance of each pathway from center reflects the average enrichment (further = more enriched), and the pathway circle area is proportional to the number of pathway proteins. Gene ontology analysis was performed with DAVID database, as described by Huang et al.35 Kinase substrates identified by mass spectrometry were used for gene overrepresentation analysis. An enrichment P value of < .05 was used in the enriched functional annotation to identify significant biological processes associated with identified kinase substrates. (C) Top, autoradiogram of standard RIKA analysis of recombinant putative PIM substrates. Left, PIM2-containing gel; right, control gel without PIM2; bottom, Coomassie blue-stained RIKA gels from which autoradiograms were obtained. Identical results were obtained with PIM1-containing gels. PIM2 has a lower degree of autophosphorylation compared with PIM1, resulting in higher signal-to-noise in a RIKA.
Shotgun RIKA nominates PIM-controlled pathways and potential physiological substrates. (A) RIKA workflow to survey the cellular proteome for protein substrates of PIM1. (B) Top 10 pathways nominated by DAVID analysis of 778 PIM substrates identified in K562 cell extracts. P values were calculated for all PIM involved pathways. The top 10, based on P value and the presence of >10 genes in each pathway, were plotted. Distance of each pathway from center reflects the average enrichment (further = more enriched), and the pathway circle area is proportional to the number of pathway proteins. Gene ontology analysis was performed with DAVID database, as described by Huang et al.35 Kinase substrates identified by mass spectrometry were used for gene overrepresentation analysis. An enrichment P value of < .05 was used in the enriched functional annotation to identify significant biological processes associated with identified kinase substrates. (C) Top, autoradiogram of standard RIKA analysis of recombinant putative PIM substrates. Left, PIM2-containing gel; right, control gel without PIM2; bottom, Coomassie blue-stained RIKA gels from which autoradiograms were obtained. Identical results were obtained with PIM1-containing gels. PIM2 has a lower degree of autophosphorylation compared with PIM1, resulting in higher signal-to-noise in a RIKA.
PIM kinase inhibition profoundly and reproducibly alters RNA splicing
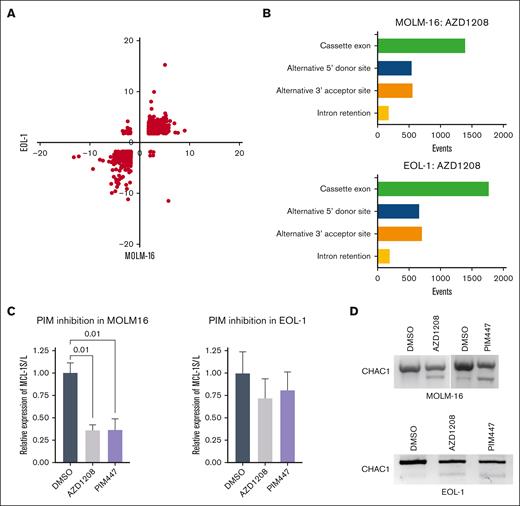
Among the 461 novel PIM substrates, 3 core mRNA splicing components (BUD13, SF3A1, and U2AF1) and 7 auxiliary splicing factors (SRSF1, SRSF2, SRSF5, SRSF6, SRSF7, SRASF9, and SRSF10) were observed, suggesting that PIM kinases may control mRNA splicing. To determine if modulating PIM activity alters mRNA splicing patterns, PIM-inhibitor–responsive MOLM-16 and EOL-1 acute myeloid leukemia cell lines were treated with the pan-PIM inhibitor (R,Z)-5-((2-(3-aminopiperidin-1-yl)-[1,1'-biphenyl]-3-yl)methylene)thiazolidine-2,4-dione (AZD1208) for 6 hours, and microarray analyses (Human Transcriptome Analysis 2.0; Affymetrix) were performed. A total of 2427 quantitative changes in alternative splicing events mapping to 1093 genes were observed in both cell lines (supplemental Table 2). Importantly, the direction of change in these splicing events was concordant in most cases, as indicated by quadrant plotting (Figure 2A). Among alternative splicing events, exon skipping/inclusion predominated, followed by alternative 5’ donor and 3’ acceptor usage (Figure 2B). Intron retention was the least frequently observed event (Figure 2B). To validate the PIM-inhibitor inducibility of specific splicing events predicted by the microarray data, RT-PCR analyses were performed. TaqMan quantitative RT-PCR analyses to quantify the long and short myeloid cell leukemia-1 (MCL-1) mRNA isoforms demonstrated a significant increase in the MCL-1L:MCL-1S ratio as observed in the microarray analysis in MOLM-16 cells (Figure 2C). Likewise, semiquantitative RT-PCR analysis of CHAC1 mRNA isoforms concordant with the microarray data were also observed (Figure 2D). To demonstrate that AZD1208-induced changes in MCL-1 and CHAC1 splicing are not due to the off-target effects of AZD1208, AML cells were treated with N-[4-[(1R,3S,5S)-3-amino-5-methylcyclohexyl]pyridin-3-yl]-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide (PIM447). Essentially identical changes in MCL-1 and CHAC1 mRNA splicing in MOLM-16 were observed by RT-PCR (Figure 2C-D). Similar changes in MCL-1 and CHAC1 splicing were also observed by RT-PCR in AZD1208- and PIM447-treated EOL1 cells (Figure 2C-D), despite not being detected as significant in the microarray analysis. These data demonstrate that PIM kinase blockade, using 2 chemically distinct small-molecule inhibitors, results in extensive alterations in specific mRNA isoform abundance in PIM-inhibitor–sensitive AML cells. In contrast, no consistent changes in MCL1 or CHAC1 splicing were observed in drug-treated AZD1208-resistant OCI-M1 cells36 (supplemental Figure 2A-B).
PIM inhibition alters RNA splicing in AML cell lines. (A) Quadrant plot of PIM-inhibitor–induced splicing index changes for 2427 events occurring in EOL-1 and MOLM-16 cells. Quadrant 1, up in MOLM-16, down in EOL-1; quadrant 2, up in MOLM-16 and EOL-1; quadrant 3, down in MOLM-16, down in EOL-1; quadrant 4, down in MOLM-16, up in EOL-1. (B) Incidence of alternative splicing event changes by type in AZD1208-treated MOLM-16 and EOL-1 cells compared to control (vehicle treated) cells. (C) TaqMan quantitative RT-PCR validation of microarray data for MCL-1L and MCL-1S isoforms in AZD1208-treated MOLM-16 and EOL-1 cells. Normalized expression (n = 3 biological replicates) with standard error of the mean is plotted. Data analysis was performed in BioRad CFX Manager 3.1., and significance values (P value) of normalized relative expression of MCL-1S to MCL-1L are displayed for 1-way analysis of variance comparisons with P <.05. (D) RT-PCR to detect CHAC1 alternative splicing in PIM-inhibitor–treated MOLM-16 or EOL-1 cells.
PIM inhibition alters RNA splicing in AML cell lines. (A) Quadrant plot of PIM-inhibitor–induced splicing index changes for 2427 events occurring in EOL-1 and MOLM-16 cells. Quadrant 1, up in MOLM-16, down in EOL-1; quadrant 2, up in MOLM-16 and EOL-1; quadrant 3, down in MOLM-16, down in EOL-1; quadrant 4, down in MOLM-16, up in EOL-1. (B) Incidence of alternative splicing event changes by type in AZD1208-treated MOLM-16 and EOL-1 cells compared to control (vehicle treated) cells. (C) TaqMan quantitative RT-PCR validation of microarray data for MCL-1L and MCL-1S isoforms in AZD1208-treated MOLM-16 and EOL-1 cells. Normalized expression (n = 3 biological replicates) with standard error of the mean is plotted. Data analysis was performed in BioRad CFX Manager 3.1., and significance values (P value) of normalized relative expression of MCL-1S to MCL-1L are displayed for 1-way analysis of variance comparisons with P <.05. (D) RT-PCR to detect CHAC1 alternative splicing in PIM-inhibitor–treated MOLM-16 or EOL-1 cells.
PIM kinase inhibition synergizes with splicing disruptors to kill AML cells
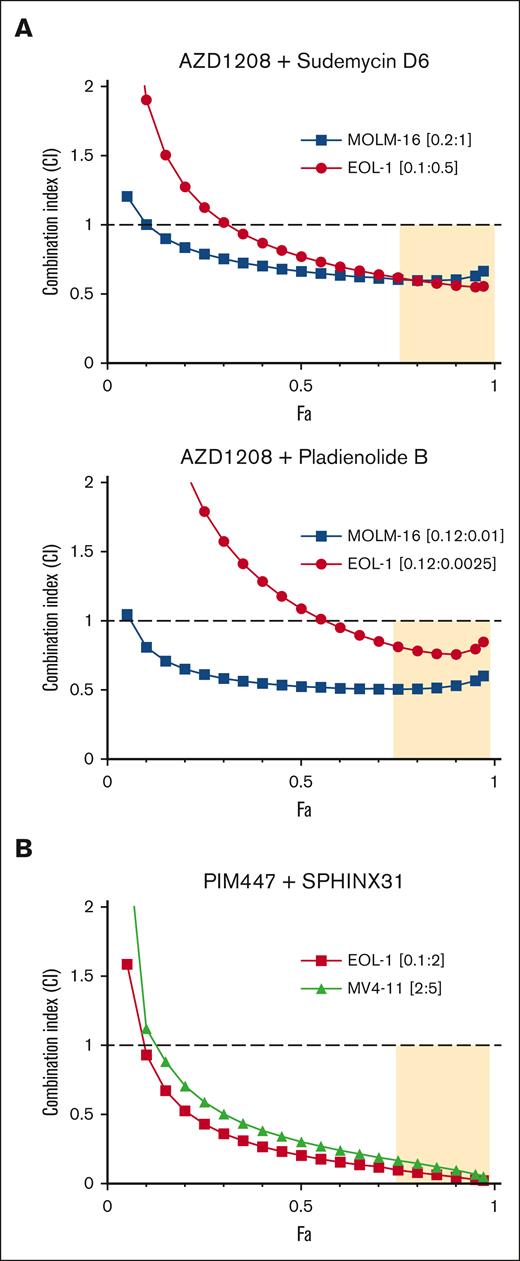
Alternative splicing has recently been identified as a therapeutic vulnerability in AML and other cancers. Recurrent hot spot mutations have been identified in 3 spliceosome components (SF3B1, SRSF2, and U2AF1) in multiple hematologic malignancies, including AML.37 These mutations lead to altered splice site recognition. Interestingly, in AML, spliceosome mutations are almost always heterozygous and mutually exclusive. This suggests that while some degree of spliceosome component perturbation may be pro-oncogenic, further alteration may be incompatible with viability.38 We therefore reasoned that, PIM inhibition might induce chemical synthetic lethality in SF3B1 wild-type cells by providing a first hit to alternative splicing, rendering them more susceptible to SF3B1-targeting agents. To test this hypothesis, we determined, using MTT assays, the effects of combining PIM inhibition with the SF3B1 inhibitors pladienolide B39 or SD6.40 CI values were determined using the Chao-Talalay median-effect equation41 for combinations of AZD1208 plus the SF3B1 inhibitors pladienolide B39 or SD6.40 Synergy in the AZD1208 plus pladienolide B and the AZD1208 plus SD6 combinations was observed in both MOLM-16 and EOL-1 cell lines (Figure 3A).
PIM inhibitors synergize with splicing disruptors to kill AML cell lines. (A) MOLM16 and EOL-1 cells were treated with AZD1208 and SD6 or pladienolide B or each drug alone, at 5 concentrations (see supplemental Methods) in the ratios shown. (B) MLL-rearranged EOL-1 and MV4-11 cells were treated with PIM447 + SPHINX31 or each drug alone at 5 concentrations (see supplemental Methods) in the ratios shown. CI values were calculated using CompuSyn. Fa-CI plots were generated based on constant dose ratio drug combination data. CI < 1 indicates synergy; CI > 1 indicates antagonism; CI = 1 indicates additivity. The tan shaded area indicates synergy and strong effect (fraction affected, Fa > 0.75).
PIM inhibitors synergize with splicing disruptors to kill AML cell lines. (A) MOLM16 and EOL-1 cells were treated with AZD1208 and SD6 or pladienolide B or each drug alone, at 5 concentrations (see supplemental Methods) in the ratios shown. (B) MLL-rearranged EOL-1 and MV4-11 cells were treated with PIM447 + SPHINX31 or each drug alone at 5 concentrations (see supplemental Methods) in the ratios shown. CI values were calculated using CompuSyn. Fa-CI plots were generated based on constant dose ratio drug combination data. CI < 1 indicates synergy; CI > 1 indicates antagonism; CI = 1 indicates additivity. The tan shaded area indicates synergy and strong effect (fraction affected, Fa > 0.75).
In addition to SF3B1-targeting agents, inhibitors of splicing kinases are also being developed as splicing-targeting therapeutics. SRPK1-3 phosphorylate serine-arginine–rich (SR) proteins in the cytoplasm to facilitate transport to the nucleus where they regulate splice site selection.42 SRPK1 also facilitates SR protein release from CDC2-like kinase 1–SR protein complexes in the nucleus.43 Genetic and pharmacologic SRPK1 inhibition has been shown to alter splicing and block growth in MLL-rearranged AML.44,45 To determine if PIM kinase inhibition would synergize with the SRPK1 inhibitor SPHINX31, MLL-rearranged AML EOL-1, MV4-11, and MLL-wild type MOLM-16 cells were treated with the combination.45 Very strong synergy in cell killing was observed in EOL-1 and MV4-11 (Figure 3B). To further explore the cellular basis of the observed synergy, flow cytometry and trypan blue exclusion analyses were performed in EOL1 cells (supplemental Figure 3). As expected, PIM kinase and SRPK1 inhibition alone increased the population of apoptotic and dead cells. The combination significantly increased that population (supplemental Figure 3).
PIM kinase inhibition alters rRNA processing and synergizes with ribosome biogenesis disruptors to kill AML cells
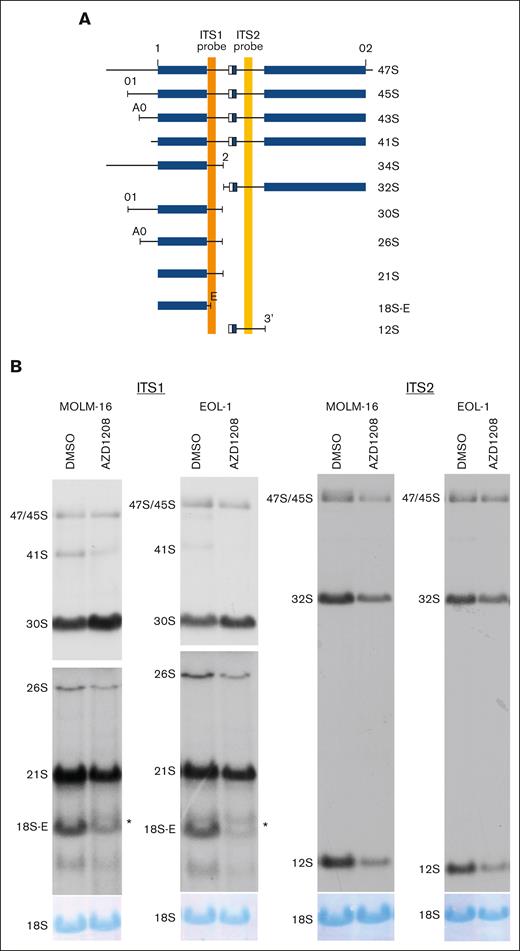
In addition to mRNA splicing, pathway analysis also nominated ribosome biology as a major target of PIM activity (Figure 1B). PIM inhibition is known to alter protein translation but has not been reported to affect ribosome biogenesis.46 Systematic comparison of the RIKA–generated PIM substrate profile to a database of 286 validated rRNA processing factors (exclusive of ribosomal proteins) revealed 50 PIM substrates known to be validated rRNA processing factors.29 To determine whether PIM inhibition alters rRNA processing in AML, pre-rRNA abundance was quantified by northern blot analyses of rRNA from AZD1208-treated MOLM-16 and EOL-1 cells. rRNA processing was reproducibly perturbed at multiple steps, resulting in the accumulation of 30S pre-rRNA, the reduction of 26S, 21S, and 18S-E pre-rRNAs, and the appearance of an aberrant rRNA intermediate larger than the 18S-E pre-rRNA (Figure 4). In contrast, PIM inhibition in AZD1208-resistant OCI-M1 cells failed to induce detectable rRNA processing changes (supplemental Figure 3C). To demonstrate that AZD1208-induced changes in rRNA processing are not due to off-target effects of AZD1208, MOLM-16 cells were treated with PIM447, and identical changes in pre-rRNA processing were observed (supplemental Figure 4A). These data demonstrate that PIM inhibition alters specific steps in rRNA processing. In contrast, inhibition of S6 kinase, a well-known regulator of ribosome biogenesis, with LY2584702 in MOLM-16 cells did not induce rRNA processing changes, despite a clear decrease in pRPS6-S235-236 upon LY2584702 treatment (supplemental Figure 4B-C). These data demonstrate that PIM inhibition profoundly and specifically alters rRNA processing at multiple steps, strongly suggesting that PIM kinases regulate a key step in ribosome biogenesis.
PIM inhibition results in pre-rRNA processing defects. (A) Schematic showing pre-rRNA isoforms and positions of internal transcribed spacer (ITS) 1 (orange vertical bar) and 2 (yellow vertical bar) probes. Mature rRNA sequences within precursors are depicted as blue rectangular boxes, and spacers are depicted as black lines. Adapted from Tafforeau et al29 with permission. (B) Northern blot analysis for AZD1208-treated MOLM-16 and EOL-1 cells using ITS1 (left) and ITS2 (right) probes. Autoradiograms showing ITS1 probe hybridization are presented at 2 different exposures (top and bottom panels) to permit optimal visualization of all rRNA intermediates. ∗aberrant 18S-E intermediate. The 18S rRNA abundance was analyzed by methylene blue staining of northern blot membranes.
PIM inhibition results in pre-rRNA processing defects. (A) Schematic showing pre-rRNA isoforms and positions of internal transcribed spacer (ITS) 1 (orange vertical bar) and 2 (yellow vertical bar) probes. Mature rRNA sequences within precursors are depicted as blue rectangular boxes, and spacers are depicted as black lines. Adapted from Tafforeau et al29 with permission. (B) Northern blot analysis for AZD1208-treated MOLM-16 and EOL-1 cells using ITS1 (left) and ITS2 (right) probes. Autoradiograms showing ITS1 probe hybridization are presented at 2 different exposures (top and bottom panels) to permit optimal visualization of all rRNA intermediates. ∗aberrant 18S-E intermediate. The 18S rRNA abundance was analyzed by methylene blue staining of northern blot membranes.
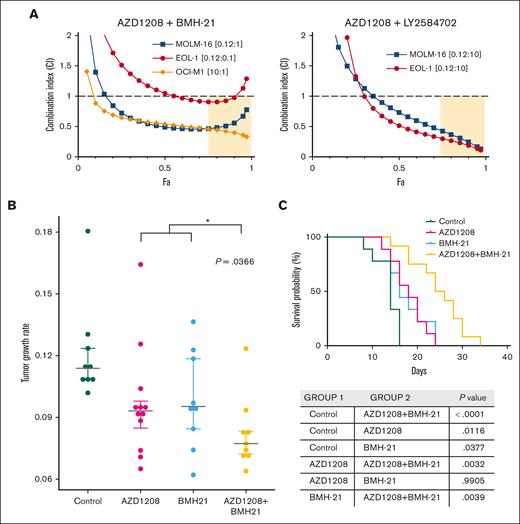
The identification of ribosome biogenesis as a PIM-inhibitor–responsive pathway suggested that combining PIM inhibition with disruptors of this pathway could potentially synergize. To test this hypothesis, we determined the effects of co-targeting PIM kinases and RNA polymerase I (Pol I) or S6 kinase. CI values determined using the Chao-Talalay median-effect equation for combinations of PIM inhibitors plus the RNA pol I inhibitor BMH-21,47 or the S6 kinase inhibitor48 showed synergistic cell killing effects against PIM-inhibitor–sensitive MOLM-16 and EOL-1 cells (Figure 5A). In OCI-M1 cells, synergy was also observed between AZD1208 and BMH-21, although a relatively high AZD1208 concentration was required due to the intrinsic resistance of OCI-M1 cell to PIM kinase inhibition (Figure 5A). To determine whether the combination of AZD1208 and BMH-21 would have a significantly greater therapeutic effect in vivo, flank EOL-1 tumors were established in NSG mice. Both AZD1208 and BMH-21 treatment alone significantly reduced the rate of tumor growth compared with vehicle treatment (Figure 5B). Cotreatment with AZD1208 plus BMH-21 was significantly (P = .0366) more potent than either agent alone in both tumor growth inhibitions. These data demonstrate potentiation between AZD1208 and BMH-21 in reducing the rate of EOL-1 tumor growth in vivo. In addition, the combination also significantly increased overall survival (Figure 5C).
PIM inhibitors synergize with disruptors of ribosome biogenesis to kill AML cell lines in vitro and potentiate growth rate inhibition in vivo. (A) AML cell lines were treated with AZD1208 and BMH-21 (left) or AZD1208 and S6 kinase inhibitor LY2584702 or each drug alone at 5 concentrations (see supplemental Methods) in the ratios shown. Fa, fraction affected. (B) Tumor growth rates were calculated for EOL-1 tumors in NSG mice treated with vehicle (Control), 5 mg/kg AZD1208, 5 mg/kg BMH21, or 5 mg/kg AZD1208 + 5 mg/kg BMH21 . Mean tumor growth rates (mm3 per day) were as follows: control (vehicle treatment) = 0.1212; AZD1208 = 0.0967; BMH-21 = 0.0983; AZD1208 + BMH-21 = 0.0822. Asterisk (∗) indicates a statistically significant difference between the average of the mean tumor growth rates of AZD1208- and BMH-21-treated mice vs AZD1208 + BMH-21 (t test, P = .0366). For statistical analysis of tumor growth rates, we assumed that tumor volume follows an exponential growth model as described49:
where is the tumor volume, a and b are parameters of interest, and is an error term, which has a normal distribution with equal variance for each observation. For 4 different groups, including the control group, we estimated a and b from Eq 1, then computed the mean of growth rates. We denote , , , and to be the mean growth rate for each of 3 treatment groups (A = AZD1208, B = BMH-21, AB = AZD1208 + BMH-21) and one for control group (C = DMSO), respectively. For instance, , where is the number of mice in group A, is the estimate of b for i-th mouse, and is an indicator function. , , , and , which amounts to samples in total. We interpret as a growth rate of i-th observation, in other words, one expects that it may vary according to the group which i-th mouse belongs to. We then carried out a 2-sample t test to identify whether a significant difference exists among each of treatment groups and control group. We tested
where j = A, B, AB. We concluded that the growth rate of the control group was significantly higher than the growth rate of any of the treatment groups on average with 95% confidence level (P value <.05). The mean growth rates were as follows: , , , and . and did not appear to be statistically significantly different from each other. We provide the mean of predicted tumor volumes according to time, in Figure 5 which shows the similarity in predicted tumor volumes for groups A and B. The t test confirmed that there is no growth rate difference between A and B: the P value for testing was >.05. To determine if the therapeutic effect of AZD1208 + BMH-21 was statistically significantly greater than AZD1208 or BMH-21 alone, we compared the tumor growth rate in groups AB, A, and B. We tested this by conducting a t test to determine whether the growth rate of AB is statistically significantly lower than those of groups A and B. Because it was shown that there is no difference between groups A and B, we performed the following t test
where is the mean growth rate of both of A and B groups, that is, . This test yielded a P value of <.05, hence, we concluded that AZD1208 + BMH-21 reduced the tumor growth rate significantly, compared with the effectiveness of AZD1208 or BMH-21 alone. (C) Kaplan-Meier graph for survival. AZD1208 + BMH-21 extends survival significantly compared with AZD1208 alone (P = .0032) or BMH-21 alone (P = .0039). Log-rank test (Mantel-Cox) P values are tabulated.
PIM inhibitors synergize with disruptors of ribosome biogenesis to kill AML cell lines in vitro and potentiate growth rate inhibition in vivo. (A) AML cell lines were treated with AZD1208 and BMH-21 (left) or AZD1208 and S6 kinase inhibitor LY2584702 or each drug alone at 5 concentrations (see supplemental Methods) in the ratios shown. Fa, fraction affected. (B) Tumor growth rates were calculated for EOL-1 tumors in NSG mice treated with vehicle (Control), 5 mg/kg AZD1208, 5 mg/kg BMH21, or 5 mg/kg AZD1208 + 5 mg/kg BMH21 . Mean tumor growth rates (mm3 per day) were as follows: control (vehicle treatment) = 0.1212; AZD1208 = 0.0967; BMH-21 = 0.0983; AZD1208 + BMH-21 = 0.0822. Asterisk (∗) indicates a statistically significant difference between the average of the mean tumor growth rates of AZD1208- and BMH-21-treated mice vs AZD1208 + BMH-21 (t test, P = .0366). For statistical analysis of tumor growth rates, we assumed that tumor volume follows an exponential growth model as described49:
where is the tumor volume, a and b are parameters of interest, and is an error term, which has a normal distribution with equal variance for each observation. For 4 different groups, including the control group, we estimated a and b from Eq 1, then computed the mean of growth rates. We denote , , , and to be the mean growth rate for each of 3 treatment groups (A = AZD1208, B = BMH-21, AB = AZD1208 + BMH-21) and one for control group (C = DMSO), respectively. For instance, , where is the number of mice in group A, is the estimate of b for i-th mouse, and is an indicator function. , , , and , which amounts to samples in total. We interpret as a growth rate of i-th observation, in other words, one expects that it may vary according to the group which i-th mouse belongs to. We then carried out a 2-sample t test to identify whether a significant difference exists among each of treatment groups and control group. We tested
where j = A, B, AB. We concluded that the growth rate of the control group was significantly higher than the growth rate of any of the treatment groups on average with 95% confidence level (P value <.05). The mean growth rates were as follows: , , , and . and did not appear to be statistically significantly different from each other. We provide the mean of predicted tumor volumes according to time, in Figure 5 which shows the similarity in predicted tumor volumes for groups A and B. The t test confirmed that there is no growth rate difference between A and B: the P value for testing was >.05. To determine if the therapeutic effect of AZD1208 + BMH-21 was statistically significantly greater than AZD1208 or BMH-21 alone, we compared the tumor growth rate in groups AB, A, and B. We tested this by conducting a t test to determine whether the growth rate of AB is statistically significantly lower than those of groups A and B. Because it was shown that there is no difference between groups A and B, we performed the following t test
where is the mean growth rate of both of A and B groups, that is, . This test yielded a P value of <.05, hence, we concluded that AZD1208 + BMH-21 reduced the tumor growth rate significantly, compared with the effectiveness of AZD1208 or BMH-21 alone. (C) Kaplan-Meier graph for survival. AZD1208 + BMH-21 extends survival significantly compared with AZD1208 alone (P = .0032) or BMH-21 alone (P = .0039). Log-rank test (Mantel-Cox) P values are tabulated.
Discussion
Despite the clinical success of kinase inhibitor therapies, achieving durable responses remains a formidable challenge.4 Although combinatorial treatments with targeted agents represent a proven avenue to attack this problem through polypharmacology, we submit that a major bottleneck in choosing rational co-therapies is our incomplete knowledge of the substrate repertoire of known oncogenic driver kinases and our complete ignorance of physiological substrates for more than 150 less well-studied kinases.11 In an effort to expand our knowledge of kinase-substrate relationships, we adapted the RIKA to permit rapid, proteome-wide discovery of potential physiological kinase substrates. A major source of confounding background phosphorylation in protein extract-based kinase substrate discovery technologies is endogenous kinase activity, and multiple mitigation strategies have been used.50-57 Our electrophoretic approach virtually eliminates background phosphorylation from endogenous kinases, given that they can only access comigrating proteins, whereas the kinase of interest can access all proteins. In addition, nonenzymatic, quantitative removal of existing phosphorylation and glycosylation by HF treatment ensures the openness of phosphoacceptor sites to maximize assay sensitivity.23 To our knowledge, no other kinase-substrate profiling method incorporates a deglycosylation step, despite the well-documented tension between phosphorylation and glycosylation in vivo.58
Application of the RIKA approach to PIM1 essentially doubled the number of putative substrates. However, as in all in vitro proteomic kinase substrate discovery assays that eliminate cellular compartmentalization, establishing the bona fides of these putative relationships demands further experimental validation.59 PIM kinases are known to be both nuclear and cytoplasmic, with the potential to access a broad swath of the proteome, and well-studied substrates are found in both compartments.60 To satisfy a key (albeit in vitro) criterion for establishing the validity of putative kinase-substrate relationships, we demonstrated that 19 of 20 recombinant nuclear and cytoplasmic candidate PIM substrates were robustly phosphorylated by PIM kinases in standard kinase assays.59 These data also augur favorably for the accuracy of our MS/MS identification workflow for establishing protein identities. Moreover, the assignment of substrates into distinct pathways argues strongly for the physiological relevance of a substantial proportion of these putative kinase-substrate relationships. Most importantly, the demonstration that perturbing PIM kinase activity reproducibly alters the output of cellular processes predicted by the pathway analysis to be directly associated with PIM kinase activity provides functional data in living cells that further substantiates physiological relevance. It is important to note that we cannot, without extensive further time- and resource-intensive investigation, determine precisely what fraction of the substrates reported here may not, under any physiological condition, in any cell population, be directly phosphorylated by PIM kinases in vivo. Given that our identification process requires in-gel protein refolding, it is likely that some phosphoacceptor sites on partially refolded proteins could be falsely phosphorylated in the assay. Despite this limitation, it is unequivocal that the analysis of our substrate data set generated rational, actionable, pathway-based leads for combinatorial therapies that synergized to kill AML cells, nominating new treatments for further preclinical development, and providing a paradigm to discover cotherapeutics for other KIs using this functional validation approach.
The apparent convergence of PIM kinase activity on proteins associated with RNA biology is striking. Of the 461 novel PIM substrates we identified, >250 are classified as RNA-binding proteins. Of these, 65 have been associated with nearly all aspects of mRNA splicing. This highly regulated process initiates during RNA polymerase II transcription and, through alternative splicing, ultimately encodes a proteome with a complexity of species that far exceeds the number of annotated protein-coding genes. Several kinase families have long been known to regulate splicing, including SR-protein kinases (SRPKs), Cyclin-dependent–like kinases, as well as pre-mRNA processing factor 4 kinase, NIMA (Neven in Mitosis Gene A) related kinase, and AKT.61 The activity of these kinases comprises a complex splicing-regulatory network that is often substantially altered in cancers, with some, for example, SRPK1, having both oncogenic and tumor suppressive functions (reviewed in Czubaty and Piekielko-Witkowska61). To our knowledge, PIM kinases have not been reported to play a major splicing-regulatory role; however, the breadth of putative substrates reported here that are involved in this process and the extensive changes in splicing observed upon inhibition in AML cell lines strongly implicate PIM family kinases as splicing modulators. Despite the fact that PIM kinase activity is not required for viability in mice,62 the strong synergy in cell killing we observed between the SRPK inhibitor SPHINX31 and PIM inhibitors demonstrates that, in at least some cases, AML cells depend on the combined activity of SRPK and PIM kinases for survival. Although based on a limited number of cell lines, these data suggest that the combination of PIM inhibitors and splicing disrupters warrants further investigation as a potential treatment option for some patients with AML. Future experiments using patient samples will be critical to establishing the clinical viability of this approach. In addition, given that specific and quantifiable changes in alternative splicing were restricted to PIM-inhibitor–responsive cell lines, a panel of splicing changes could potentially serve as a pharmacodynamic biomarker of PIM-inhibitor responsiveness.
The PIM kinase substrate repertoire reported here includes 50 proteins implicated in ribosome biogenesis and blocking PIM activity in AML cells resulted in highly reproducible changes in rRNA processing. The diminution of the 26S species is particularly noteworthy. 26S pre-rRNA is generated by endonucleolytic cleavage of the 30S pre-rRNA at a site termed A0.63 Recent evidence suggests that UTP23, a putative PIM substrate reported here, is responsible.64 We speculate that PIM phosphorylation may be required for full UTP23 function in AML cells. Collectively, alterations in rRNA processing resulting from PIM kinase substrate hypophosphorylation can potentially account for the strong synergy in AML cell killing we observed between PIM and RNA polymerase I inhibitors in vitro, and the potentiation of tumor growth inhibition in vivo. The synergy observed in AML is also consistent with recent studies demonstrating that PIM inhibition cooperates with Pol I inhibition to block growth of MYC-driven prostate cancers.65
The approach reported here highlights the merits of illuminating kinase functions by expanding knowledge of their potential physiological substrates to inform new therapeutic approaches and nominate pharmacodynamic biomarkers. Given the strong linkage between aberrant kinase activity and malignant disease, extending this strategy across the kinome can yield mechanistic insights that identify new drug combinations to improve patient outcomes.
Acknowledgments
The authors thank Flow Cytometry Shared Service of the University of Maryland Marlene and Stewart Greenebaum Comprehensive Cancer Center for assistance with flow cytometric analyses.
This work was supported by a 2018 University of Maryland, Baltimore County Technology Catalyst Fund award (C.J.B.). This publication was supported by funds through the Maryland Department of Health's Cigarette Restitution Fund Program (grant CH-649-CRF) and the National Cancer Institute Cancer Center Support Grant (grant P30CA134274).
Authorship
Contribution: T.J., A.C., X.L., and C.J.B. conceptualized the study; T.J., A.C., X.L., A. Raja, W.H., and A. Rege conducted the investigation; T.J., A.C., W.H., X.L., A.V.-K., M.R., B.S., X.F., and C.J.B. conducted formal analysis; T.R.W., M.L., and M.K. gathered resources; T.J., X.L., and C.J.B. wrote the original draft of the manuscript; X.L. and C.J.B. acquired funding; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charles J. Bieberich, Department of Biological Sciences, University of Maryland, Baltimore County, 1000 Hilltop Cir, Baltimore, MD 21250; email: bieberic@umbc.edu; and Xiang Li, Department of Biological Sciences, University of Maryland, Baltimore County, 1000 Hilltop Cir, Baltimore, MD 21250; email: lxiang@umbc.edu.
References
Author notes
Microarray data are available on https://umbc.box.com/s/8lrvo3m1j0qy9fik2n7xvgd7lx0e4yhx. All other data and reagents are available on request from the corresponding authors, Charles J. Bieberich (bieberic@umbc.edu) and Xiang Li (lxiang@umbc.edu).
The full-text version of this article contains a data supplement.