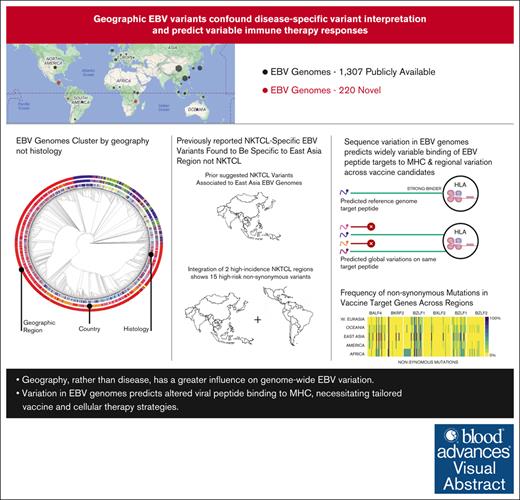
Key Points
Geography, rather than disease, has a greater influence on genome-wide EBV variation.
Variation in EBV genomes predicts altered viral peptide binding to major histocompatibility complex, necessitating tailored vaccine and cellular therapy strategies.
Visual Abstract
Epstein-Barr virus (EBV) is a potent carcinogen linked to hematologic and solid malignancies and causes significant global morbidity and mortality. Therapy using allogeneic EBV-specific lymphocytes shows promise in certain populations, but the impact of EBV genome variation on these strategies remains unexplored. To address this, we sequenced 217 EBV genomes, including hematologic malignancies from Guatemala, Peru, Malawi, and Taiwan, and analyzed them alongside 1307 publicly available EBV genomes from cancer, nonmalignant diseases, and healthy individuals across Africa, Asia, Europe, North America, and South America. These included, to our knowledge, the first natural killer (NK)/T-cell lymphoma (NKTCL) EBV genomes reported outside of East Asia. Our findings indicate that previously proposed EBV genome variants specific to certain cancer types are more closely tied to geographic origin than to cancer histology. This included variants previously reported to be specific to NKTCL but were prevalent in EBV genomes from other cancer types and healthy individuals in East Asia. After controlling for geographic region, we did identify multiple NKTCL-specific variants associated with a 7.8-fold to 21.9-fold increased risk. We also observed frequent variations in EBV genomes that affected peptide sequences previously reported to bind common major histocompatibility complex alleles. Finally, we found several nonsynonymous variants spanning the coding sequences of current vaccine targets BALF4, BKRF2, BLLF1, BXLF2, BZLF1, and BZLF2. These results highlight the need to consider geographic variation in EBV genomes when devising strategies for exploiting adaptive immune responses against EBV-related cancers, ensuring greater global effectiveness and equity in prevention and treatment.
Introduction
Epstein-Barr virus (EBV), which infects 95% of the world’s adult population, is classified as a group 1 carcinogen by the International Agency for Research on Cancer.1 However, only a small fraction of the population will develop EBV-associated cancer.
Furthermore, the incidence of specific EBV-associated cancers is strongly associated with geographic regions. Examples include Burkitt lymphoma (BL) in regions with holoendemic Plasmodium falciparum malaria in Sub-Saharan Africa, nasopharyngeal carcinoma (NPC) in East Asia, and natural killer (NK)/T-cell lymphoma (NKTCL) in East Asia, Central America, and Western South America. These patterns suggest that regional host or EBV variation influences the pathogenesis of these malignancies. Indeed, geographic variation in the EBV genome has been demonstrated in several studies.2 Single nucleotide polymorphisms (SNPs) in RPMS1, BALF2, and EBER2 were shown to be associated with a high risk for developing NPC in NPC-endemic areas.3-5 Several nonsynonymous mutations were also associated with NKTCL,6 however, this was limited to a single region and did not include genomes from other NKTCL global hot spots. A significant challenge to uncovering potential EBV variants that might influence the development or natural history of hematologic malignancies has been the difficulty in dissociating regional viral variants from disease-specific variants, especially for diseases that are prevalent in understudied populations such as those in Central and South America and Sub-Saharan Africa.
EBV has been considered a promising therapeutic target for nearly 30 years in hematologic and nonhematologic malignancies. Autologous or allogeneic T cells that target EBV-encoded peptides first showed efficacy in posttransplant lymphoproliferative disorder.7-12 Since then, cellular therapies targeting peptides derived from EBV proteins, including latent membrane protein 1 (LMP1), latent membrane protein 2 (LMP2), BARF1, Epstein-Barr nuclear antigen 1 (EBNA1), BMLF1, and BZLF1 for the treatment of NPC,13-17 NKTCL,18 and EBV–associated rituximab-refractory lymphoma,18 have demonstrated limited efficacy. Many of these approaches are major histocompatibility complex (MHC) restricted and depend on cellular selection using in vitro exposure to the full-length EBV reference genome B95-8 or peptides derived from EBV reference genomes. The design and development of prophylactic vaccine strategies similarly have been dependent on reference genomes using EBV antigens important to viral entry into the cell, including gp350 (encoded by BLLF1),19 and messenger RNA–based strategies that additionally targeting glycoprotein (gp)42 (encoded by BZLF2), glycoprotein (g)B (encoded by BALF4), glycoprotein (g)H (encoded by BXLF2), and glycoprotein (g)L (encoded by BKRF2) (ClinicalTrials.gov identifier NCT05164094). The efficacy of these approaches for a global population makes 2 assumptions: (1) variation in the EBV sequences that encode these antigenic targets will not affect the desired immune response and (2) binding across HLA types will be effective.
Capture-based sequencing has rapidly expanded the number and diversity of EBV genomes available for study. However, many regions worldwide are still underrepresented in the publicly available genomes, and these regions have a high incidence of EBV-associated malignancies. Therefore, in the first phase of our study, we sought to improve global representation by generating, to our knowledge, the first EBV genomes from Malawi and the first from any country in Central or Western South America. We then incorporated these genomes into the previous analysis of disease-specific variants to determine if specific EBV variants can explain the incidence of specific EBV-associated hematologic malignancies. We next analyzed variation among EBV peptides across geographic regions and predicted binding to the most common HLA subtypes. Finally, we evaluated the global variability of EBV loci that encode the current vaccine targets.
Materials and methods
Selection of publicly available EBV genomes
The National Center for Biotechnology Information (NCBI) database was queried using the search terms “Human gammaherpesvirus 4” and filtered by sequence lengths of 120 000 to 200 000 base pairs. If genomes were not well annotated to include country of origin and phenotype or multiple genomes were sequenced from the same patient, preference was given to biopsy samples, and the remaining genomes were excluded (n = 96). After a literature review, we included 37 genomes6 that were not in the NCBI database but that were included in an alternative database (www.biosino.com) and 1 additional genome20 of 119 450 bp excluded in our original search, comprising a total of 1307 publicly available genomes.
Selection of novel EBV+ lymphomas for sequencing
Novel genomes were generated from cohorts previously identified and phenotyped.21,22 All novel cases were approved by institutional review boards of the Fred Hutchinson Cancer Center, The Ohio State University, Stanford University, the La Liga Nacional Contra el Cáncer y Instituto Nacional de Cancerología, the Malawian College of Medicine Research and Ethics Committee, the National Taiwan University Hospital, and the Instituto Nacional de Enfermedades Neoplásicas Peru. The research was conducted according to the Declaration of Helsinki.
DNA isolation, quantification of EBV, and case selection
Blocks from cases identified as EBV+ NKTCL, EBV+ classical Hodgkin lymphoma (cHL), or EBV+ diffuse large B-cell lymphoma (DLBCL) were identified, and 2 × 10 mm sections were cut. DNA was isolated using the AllPrep DNA/RNA FFPE (Qiagen). DNA quantification and the presence of EBV was confirmed by reverse transcriptase polymerase chain reaction or droplet digital polymerase chain reaction. Detailed methods and results are in the supplementary text.
Library preparation and sequencing
Standard next-generation sequencing (NGS) used fragmented DNA using a Covaris S Series sonicator. A unique barcode was added to each DNA sample using the KAPA HyperPrep Kit from Roche. Samples were pooled based on the EBV load and prepared according to the xGen hybridization capture of DNA libraries for the NGS sequencing enrichment protocol. Duplex sequencing was conducted using the Enzymatic Fragmentation Module and Duplex-sequencing Universal Kit (Twinstrand Biosciences). The capture probes were synthesized as custom hybridization capture panels designed for EBV type 1 and EBV type 2. EBV capture samples were sequenced using a NovaSeq 6000 (Illumina). Two samples were sequenced on standard NGS and duplex-sequencing systems (E0205 and E0981).
Genome assembly and sequencing quality assessment
Quality control of the raw Illumina paired-end targeted and whole-genome sequencing reads was performed using BBDuk.23 Standard Illumina adapters and low-quality sequences (Q ≤ 6) were trimmed across paired-end reads. Paired reads with a read length of <100 bases were removed. The filtered reads were mapped to the hg38 human reference genome with the EBV-associated contigs removed. Reads unmapped to the host genome were extracted from the BAM files and stored as paired-end FASTQ files.
Variant detection
Variant calling on whole-genome sequencing data sets was performed using the BAM files containing host-removed, EBV-aligned paired reads using the GATK HaplotypeCaller framework.27 Variants were annotated using SnpEff28 in which we first created a database for EBV variant annotation using the EBV type 1 reference genome from the NCBI (NC_007605). Annotated variant calls from SnpEff28 were used to generate a consensus assembly from the VCF using the FastaAlternateReferenceMaker utility from GATK.27
Phylogenetic analysis
Geographic regions were identified and classified as previously described.29 Complex repeats in the type I EBV genome were identified, and corresponding regions in the draft de novo assemblies and consensus assemblies were masked using RepeatMasker.30 Prokka26 was used to annotate the masked genomes. Draft assemblies with an LGA50 ≤2 and number of misassemblies ≤1, determined using Quast,31 were selected for phylogenetic analysis. A phylogenetic tree of the selected draft assemblies was performed using ParSnp.32
Identification of EBV variants in peptide targets
We identified a set of 30 peptide sequences from different regions of the EBV genome that are currently being studied for their efficacy in eliciting and/or serving as targets for CD8+ or CD4+ T-cell responses.13,33-35 We identified the variations within each of the 30 epitopes in the draft or consensus assemblies by creating a Basic Local Alignment Search Tool (BLAST) reference database of the assembly using makeblastdb36 and querying the 30 peptide sequences against this reference using tblastn.37 BLAST hits with an evalue ≤0.05 and percentage identity ≥75% were retained for further analysis.
Prediction of EBV T-cell epitope binding affinity to MHC molecules
We calculated the binding affinity of a given MHC allele with all variations of the reference epitope sequence observed in the draft or consensus assemblies using NetMHCpan.38 We also assessed the binding affinity of each putative epitope sequence against different MHC class I and class II alleles to understand the effect of variation in the MHC alleles on MHC restriction.
Validation of EBV T-cell epitope binding affinity to MHC molecules
Reference genome encoded and variant peptides for 2 HLA-A∗02:01-restricted T-cell epitopes encoded by BMLF1 (GLCTLVAML,33 GLCTLMAML, and GLCTLVGML) and LMP2-2A (FLYALALLL13,34,35 and FLYKLALLL) were synthesized (GenScript, Piscataway, NJ) and reconstituted to 10 mg/mL in dimethyl sulfoxide (Invitrogen). Binding of reference genome encoded and variant peptides to HLA-A∗02 was assessed by quantifying the extent to which each peptide could stabilize the expression of HLA-A∗02 on the surface of T2 cells. Aliquots of 50 000 T2 cells were incubated in 200 mL LCL media supplemented with each peptide at serial 10-fold dilutions from 1 mM to 10 pM for 20 hours at 37°C. The cells were washed twice, stained with 1:50 dilution of anti-human HLA-A2-PE antibody (clone BB7.2, BD Biosciences, San Jose, CA), resuspended in DAPI (4′,6-diamidino-2-phenylindole) solution, and analyzed by flow cytometry (BD FACSymphony, BD Biosciences). The mean fluorescence intensity of the DAPI-negative live population was calculated.
NKTCL disease–specific variant detection
We identified and eliminated all variants that were present in >95% or <3% of the genomes. There were 2 geographic regions, namely Africa and Oceania, where there were no NKTCL cases, and these regions were excluded. A binomial generalized linear model was used in which cases were classified as either NKTCL or non-NKTCL and controlled for geographic region. A Bonferroni adjustment was applied to all P values to account for multiple comparisons.
Results
Frequency and characteristics of novel EBV+ cases
In the Guatemala cohort, a total of 636 cases of lymphoma were previously identified with interpretable Epstein-Barr encoded RNA (EBER) staining, including 169 (26.6%) that were EBV+. The EBV+ cohort included 24 of 252 DLBCL (9.5%), 65 of 89 (73%) cHL, 3 of 5 (60%) BL, and 55 of 55 (100%) NKTCL.22 The cohort identified from Malawi were all EBV+ , representing cHL (n = 1), BL (n = 80), lymphoma undefined (n = 6), and miscellaneous (leukemia, lymphoid leukemia, non-HL not otherwise defined, neuroblastoma, hepatocellular carcinoma, nongonadal germ cell tumor, and rhabdomyosarcoma). We identified additional EBV+ NKTCL lymphoma cases from Peru (n = 8), Taiwan (n = 3), and the United States (n = 16).
EBV genomes show a strong association with the geographic region rather than with phenotype
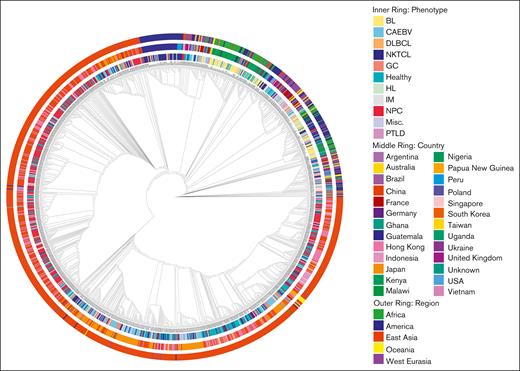
Novel sequences were combined with all publicly available EBV genomes for further analysis (supplemental Table 1). Among these EBV genomes, 1192 of 1307 (91%), and among novel sequences, 184 of 217 (85%) passed the predetermined quality metrics (Figure 1A). These represented 25 unique countries in the initial cohort and in those filtered for quality metrics (Figure 1B). Phylogenetic analysis demonstrated that EBV genomes primarily clustered based on geographic origin (Figure 2). The region with clades furthest from the type 1 reference genome were East Asian samples, representing the largest regional cohort in our analysis. Most cases were from Mainland China (n = 407) or Hong Kong (n = 245) and were distributed evenly throughout this clade. Most cases from Japan (61/79) all clustered into the same subclade, whereas the remaining 18 clustered together within East Asia. Novel cases in our cohort were from Malawi, Guatemala, Peru, Taiwan, and the United States. This led to an enrichment of cases from both America and Africa to the global EBV genome. We found that the largest cluster of samples from Guatemala clustered with each other and with the largest cluster of samples from Peru and also clustered more broadly with samples from East Asia. In contrast, most samples from Malawi clustered within the same major clade that included other samples from Africa, America, and most cases from West Eurasia (Figure 2).
Selection of novel and publicly available EBV genomes for analysis. (A) Schematic illustrating the source, histologic type, and number of EBV+ hematologic malignancies from Guatemala, Peru, Malawi, Taiwan, and the United States that underwent EBV-genome sequencing and the source of publicly available reference and cancer-associated EBV genomes that were utilized for this study. (B) Maps demonstrating the country of origin and number of EBV genomes after filtering based on sequence quality. Publicly available EBV genomes are indicated in black, and novel EBV genomes generated by this study are indicated in red. CAEBV, chronic active EBV disease; GC, gastric cancer; IM, infectious mononucleosis; misc, miscellaneous; PTLD, posttransplant lymphoproliferative disorder; QC, quality control.
Selection of novel and publicly available EBV genomes for analysis. (A) Schematic illustrating the source, histologic type, and number of EBV+ hematologic malignancies from Guatemala, Peru, Malawi, Taiwan, and the United States that underwent EBV-genome sequencing and the source of publicly available reference and cancer-associated EBV genomes that were utilized for this study. (B) Maps demonstrating the country of origin and number of EBV genomes after filtering based on sequence quality. Publicly available EBV genomes are indicated in black, and novel EBV genomes generated by this study are indicated in red. CAEBV, chronic active EBV disease; GC, gastric cancer; IM, infectious mononucleosis; misc, miscellaneous; PTLD, posttransplant lymphoproliferative disorder; QC, quality control.
Phylogenetic analysis of reference, noncancer-associated, and cancer- associated EBV genomes. The inner circle represents the histology for cancer- associated genomes or other phenotype, the middle circle represents the country of origin, and the outer circle represents the region of origin. Genomes are aligned to the type I reference genome NC_007605 assembled from data for B95-8 (V01555) and Raji (M35547) genomes. CAEBV, chronic active EBV disease; GC, gastric cancer; IM, infectious mononucleosis; misc, miscellaneous; pLELC, primary lymphoepithelioma-like carcinoma; PTLD, posttransplant lymphoproliferative disorder.
Phylogenetic analysis of reference, noncancer-associated, and cancer- associated EBV genomes. The inner circle represents the histology for cancer- associated genomes or other phenotype, the middle circle represents the country of origin, and the outer circle represents the region of origin. Genomes are aligned to the type I reference genome NC_007605 assembled from data for B95-8 (V01555) and Raji (M35547) genomes. CAEBV, chronic active EBV disease; GC, gastric cancer; IM, infectious mononucleosis; misc, miscellaneous; pLELC, primary lymphoepithelioma-like carcinoma; PTLD, posttransplant lymphoproliferative disorder.
Consistent with visual clustering, the phylogenetic positioning of each genome was more strongly associated with geographic origin than with phenotype as quantified with a permutational analysis of variance pseudo-F statistic (F = 83.5 vs F = 26.6).39
Variants and disease phenotype
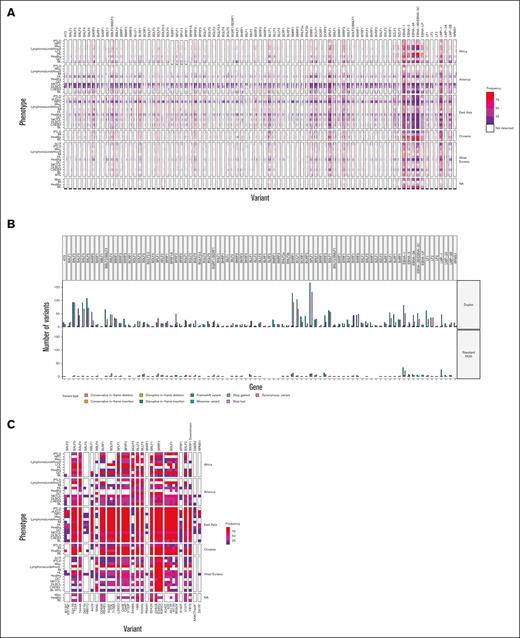
In agreement with previous work, we saw the highest frequency of variant calls in EBNA (highest in EBNA-3B/3C followed by EBNA-1), LMP-1, LMP-2, and BPLF1.40,41
Additional hot spots were at BARF0, BLLF1, BNRF1, BOLF1, and BRRF2 (Figure 3A). NKTCL and DLBCL samples from America had more variant calls, but these cohorts were mostly represented by duplex sequencing, which has greater sensitivity for detecting variants. We verified this by sequencing 1 of our novel samples using both NGS and duplex sequencing, which showed that duplex sequencing detected >10-fold more variants (Figure 3B). However, the relative hot spots for variant detection were unchanged. Variant allele frequency ranged from 50% to 100% in standard NGS and could be detected at a frequency of <10% in duplex samples. Despite the lack of clear disease clustering from phylogenetic analysis, we sought to determine if individual variants were associated with disease as previously suggested. Previous reports have demonstrated that SNPs in RPMS1 (G155391A, resulting D51N), BALF2 (T162476C, resulting I613V), BALF2 (C163364T, resulting V317M), and a 4 bp deletion in EBER2 (7188-7191) were associated with an increased risk for NPC.3-5 Our approach confirmed that these variants were present in nearly 100% of NPC cases from East Asia. We also found that nearly 100% of cases of pulmonary lymphoepithelioma-like carcinoma carried these same 3 variants. In agreement with previous studies, these variants were found in 50% or less of all other EBV genomes from East Asia. Africa was the only other region with an NPC–derived EBV genome, and these variants were not present in this single genome (Figure 3C).
Map of variant frequency across EBV genomes. (A) Identification and frequency by region and histology of all variants identified in 1376 EBV genomes meeting predetermined quality metrics. (B) Comparison of variant detection in standard NGS and duplex sequencing. Bar colors represent the nonsynonymous variant type detected. (C) Comparison of frequency of previously identified disease–associated nonsynonymous mutations in NKTCL and NPC. Variants previously associated with NPC are starred, all others were previously associated with NKTCL.
Map of variant frequency across EBV genomes. (A) Identification and frequency by region and histology of all variants identified in 1376 EBV genomes meeting predetermined quality metrics. (B) Comparison of variant detection in standard NGS and duplex sequencing. Bar colors represent the nonsynonymous variant type detected. (C) Comparison of frequency of previously identified disease–associated nonsynonymous mutations in NKTCL and NPC. Variants previously associated with NPC are starred, all others were previously associated with NKTCL.
Previous reports have suggested that specific variants are associated with NKTCL.6,40 Similar to NPC, these previous reports were exclusively from East Asian populations. Although these variants were present in NKTCL samples from East Asia, they were not specific to NKTCL. Instead, these variants were found in most cases from East Asia regardless of the disease phenotype and were also found in EBV sequences from healthy donors. Furthermore, several variants previously associated with NKTCL were present across phenotypes and geographic regions (Figure 3C), making it unlikely that these variants are specific to NKTCL.
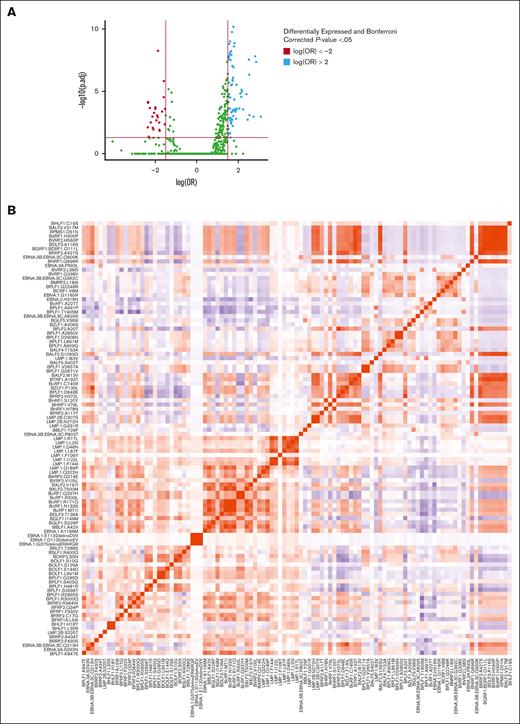
Alternatively, our novel sequences contained 74 NKTCL cases outside East Asia, and 2 additional cases were publicly available from West Eurasia. East Asia, America, and West Eurasia had 768, 83, and 89 cases of EBV genomes not associated with NKTCL, respectively. Among these genomes, there were 16 380 nonsynonymous mutations. After excluding those that occurred in <3% or >95% of cases, 1307 variants remained. We then used a bimodal general linearized regression model to compare the EBV genomes from cases of NKTCL and other EBV genomes while controlling for geographic region. After Bonferroni adjustment, 103 variants had P values <.05 and log(overall risk [OR]) >1.5 or < −1.5.
Fifteen variants demonstrated a log(OR) >2 (Figure 4A). Two of the top 5 high-risk variants for NKTCL were the BFRF1 mutations L59I and F332V with a log(OR) of 3.1 and 2.7, respectively, and they were weakly correlated with each other (Pearson correlation coefficient, 0.16; Figure 4B; supplemental Tables 2 and 3). Genes containing the highest frequency of high-risk variants, defined as those with a log(OR) >1.5, included BPLF1 (12), LMP-1 (11), and BcRF1 (6) (supplemental Figure 1). The BPLF1 variants S2696T and G2248R and the BcRF1 variant N133S had a log(OR) >2. An additional BPLF1 variant, H481, had been identified previously by Peng et al,40 but it was not reported when the Peng et al data set was combined with novel EBV genomes from NKTCL cases by Xiong et al.6 This was also found for the BRLF1 variant T288S and the EBNA3B/3C variant P803T.
Specific EBV genome variants are enriched in or depleted from NKTCL. (A) Volcano plot demonstrating all variants identified. Variants above the horizontal red line indicate variants with a Bonferroni corrected P value <.05. Significant variants with log(OR) less than −1.5 are shown in red (lower risk), and log(OR) >1.5 in blue (higher risk). (B) Heat map demonstrating a correlation between variants significantly enriched in NKTCL–derived EBV genomes with log(OR) >1.5.
Specific EBV genome variants are enriched in or depleted from NKTCL. (A) Volcano plot demonstrating all variants identified. Variants above the horizontal red line indicate variants with a Bonferroni corrected P value <.05. Significant variants with log(OR) less than −1.5 are shown in red (lower risk), and log(OR) >1.5 in blue (higher risk). (B) Heat map demonstrating a correlation between variants significantly enriched in NKTCL–derived EBV genomes with log(OR) >1.5.
EBV heterogeneity and HLA variations predict modified responses to therapeutic targets
We identified 30 peptide sequences encoded by BMLF1, BZLF1, EBNA1, EBNA3, LMP1, and LMP2 that have been used previously to induce a therapeutic immune response.13,33-35 Using our novel and publicly available sequences, we found variants at all peptide sequences with specific variant sequences occurring in 0.07% to 98.98% of the genomes.
All published peptide sequences were accompanied by MHC restriction annotated at 2- or 4-digit resolution based on in vitro T-cell activation assays.13,33-35 We sought to determine if the variants identified in our global EBV genome cohort would affect binding within and outside the previously published MHC-restricted subtypes using a previously validated MHC binding platform, NetMHCpan-4.0.42 We first verified if previously demonstrated HLA type and peptide pairs showed high-affinity binding. For example, the HLA-A∗02:01-binding peptide BSLF2/BMLF1 GLCTLVAML showed weak predicted binding affinity across all HLA-A∗02 subtypes with decreasing affinity for other HLA types. Similarly, the HLA-B∗35-binding peptide EBNA-1 HPVGEADYFEY showed high predicted affinity for all HLA-B∗35 subtypes, but poor predicted binding to HLA- A∗02 (Figure 5A). Indeed, the predicted binding of peptides strongly depended on the MHC allele with very few peptides showing strong binding outside their designated MHC allele and most having no predicted binding. There were 12 peptides with 15 unique variants in >10% of all cases in each geographic region (supplemental Table 4).
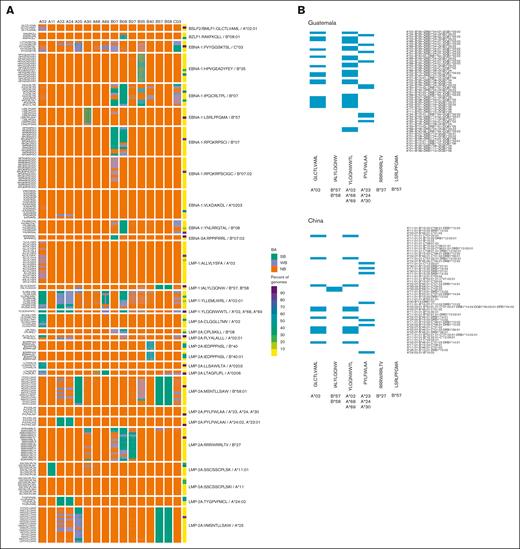
EBV genome variation in intervals encoding prototypic T-cell epitopes is predicted to generate variant peptides with altered binding to class I MHC molecules. (A) Heat map of predicted binding affinity (BA) of 30 prototypic EBV- encoded CD8+ T-cell epitopes (rows) to 16 class I MHC alleles (columns) and of the corresponding peptides encoded by EBV genome variants. The far-right column contains a heat map of the frequency of the EBV genome variant that encodes each peptide in the 1376 EBV genomes meeting predetermined quality metrics in this study. (B) Heat map indicating the presence (blue) or absence (white) of the class I MHC alleles known to present 6 prototypic EBV-encoded peptides to CD8+ T cells in the 50 most frequent class I MHC haplotypes in Guatemala (left) and China (right). The associated class I MHC allele is shown in bold beneath each peptide. NB, no binding; SB, strong binder; WB, weak binder.
EBV genome variation in intervals encoding prototypic T-cell epitopes is predicted to generate variant peptides with altered binding to class I MHC molecules. (A) Heat map of predicted binding affinity (BA) of 30 prototypic EBV- encoded CD8+ T-cell epitopes (rows) to 16 class I MHC alleles (columns) and of the corresponding peptides encoded by EBV genome variants. The far-right column contains a heat map of the frequency of the EBV genome variant that encodes each peptide in the 1376 EBV genomes meeting predetermined quality metrics in this study. (B) Heat map indicating the presence (blue) or absence (white) of the class I MHC alleles known to present 6 prototypic EBV-encoded peptides to CD8+ T cells in the 50 most frequent class I MHC haplotypes in Guatemala (left) and China (right). The associated class I MHC allele is shown in bold beneath each peptide. NB, no binding; SB, strong binder; WB, weak binder.
Variants in EBNA-1 (RPQKRPSCI > RPKKRPSCI, IPQCRLTPL > VPQCRITPL, and YNLRRGTAL > YNLRRGIAL), LMP-1 (YLLEMLWRL > YLLEILWRL), and LMP-2A (SSCSSCPLSK > SSCSSCPLTK) had strong predicted binding. The LMP-2A variant VMSNTLLSAW > MMTNTLLSAW had higher predicted binding. However, several peptides and their variants, including the BSLF2/BMLF1 variant GLCTLVAML, EBNA-1 variants HPVGEADYFEY and RPQKRPSCIGC, LMP-1 variant ALLVLYSFA, and LMP-2A variant IEDPPFNSL, were predicted to have no binding, weak binding, or strong binding that was dependent on both the sequence of the variant peptide and the specific MHC allele (Figure 5A).
To validate our HLA binding predictions for a subset of peptides, we measured the extent to which 2 reference (NC_007605) EBV peptides (BMLF1: GLCTLVAML, LMP-2A: FLYALALLL), both of which are known to bind HLA-A∗02, and their variants (GLCTLVGML, GLCTLMAML, and FFYKLALLL) could stabilize the expression of HLA-A∗02 on T2 cells (supplemental Figure 2). The peptide predicted by our analysis as a strong binder to HLA-A∗02 (FLYALALLL) was 100-fold more potent at stabilizing HLA-A∗02 expression on the surface of T2 cells than the 2 predicted nonbinding peptides (FFYKLALLL and GLCTLVGML). The 2 predicted weak binders to HLA-A∗02 (GLCTLVAML and GLCTLMAML) were sixfold more potent at stabilizing HLA-A∗02 expression on T2 cells than the predicted nonbinding peptides.
We next sought to leverage the data to select the most globally conserved peptide candidates in each region. We set a threshold for peptides preserved in at least 75% of cases in each region. There were no peptides that met these criteria for the African region. Six peptides were present in at least 75% of samples from the remaining regions: BSLF2/BMLF1 (GLCTLVAML), LMP-1 (IALYLQQNW, YLQQNWWTL), LMP-2A (PYLFWLAA/PYLFWLAAI, RRRWRRLTV), and EBNA-1 (LSRLPFGMA). These were specific to HLA-A∗02:01, HLA-B∗57/58, HLA-A∗02/68/69, HLA-A∗24:02/23:01, HLA-B∗27, and HLA-B∗57, respectively. We then accessed an HLA frequency database (allelefrequencies.net) to generate the top 50 haplotypes in 2 distinct country populations, Guatemala and China, that were well represented in our data set.43,44 The majority of haplotypes would only be reactive to 2 of 5 peptides. HLA-B∗27 and HLA-B∗57 were not present in the top 50 haplotypes in either country, indicating likely futility in activating a response with the conserved peptides RRRWRRLTV and LSRLPFGMA in LMP-2A and EBNA-1, respectively (Figure 5B).
Heterogeneity at EBV loci encoding proposed vaccine targets
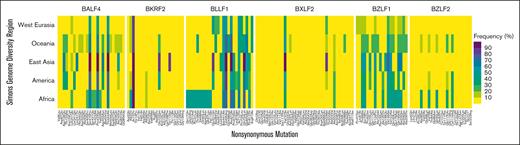
Several EBV-encoded proteins have been proposed as potential vaccines, including BXLF2 (gpH), BALF4 (gpB), BKRF2 (gp85), BLLF1 (gp350/250), BZLF1 (bzlf1), and BZLF2 (bzlf2). These include peptide-based strategies and, more recently, microRNA-based vaccines, which can deliver the entire coding sequence of 1 or more targeted antigenic proteins, thereby allowing for endogenous presentation of peptides. To determine if there is heterogeneity at the EBV loci encoding proposed vaccine targets, we evaluated these genes across geographic regions. BXLF2 had the most variants, but most of these occurred in <10% of cases across each geographic region. In contrast, BZLF1 had the fewest variants, but these variants occurred at higher frequencies (Figure 6).
Frequency of nonsynonymous single nucleotide variants in 6 canonical protein-coding genes in EBV genomes from 6 global regions. All 6 genes have been the focus of EBV vaccine development efforts. The frequency (%) is calculated as the (number of EBV genomes from each region carrying the indicated sequence variant)/(total number of EBV genomes analyzed from that region) × 100.
Frequency of nonsynonymous single nucleotide variants in 6 canonical protein-coding genes in EBV genomes from 6 global regions. All 6 genes have been the focus of EBV vaccine development efforts. The frequency (%) is calculated as the (number of EBV genomes from each region carrying the indicated sequence variant)/(total number of EBV genomes analyzed from that region) × 100.
Discussion
EBV is a critical driver in both hematologic and nonhematologic malignancies. Given the persistent expression of at least some EBV antigens across malignancies, these antigens would seem like ideal therapeutic targets. Despite great interest in preventive and disease-focused therapeutic strategies, success has been elusive. However, the decreasing cost of sequencing and advances in sequencing technology have dramatically increased the number and diversity of available EBV genomes for analysis. We used novel sequencing technology to add nearly 200 high-quality EBV genomes from poorly represented populations and disease phenotypes. We then combined our data with all publicly available EBV genomes to perform, to our knowledge, the largest global analysis of EBV phylogeny and genetic variation. Our analysis revealed that (1) EBV genomes are more closely associated with geographic region than with disease phenotype; (2) previous evaluations of disease-specific variants were limited and biased by the available genomes from specific regions, leading to confounding of associations between EBV genome variants and disease phenotypes; and (3) variants in the EBV genome are present in previously targeted EBV peptides with a variable impact on MHC binding.
We found that despite 11 broadly categorized EBV-associated phenotypes, the geographic origin of the EBV genome, rather than the phenotype, predicted where the genome would cluster in our phylogenetic tree. Our work, and others, is limited by where one draws distinctions for global regions. Instead of relying on arbitrary, politically influenced geographic borders, we based our analysis on the previous work that analyzed the phylogeny of diverse global populations.29 We found a striking parallel between EBV genomes and human genomic ancestry. Although many populations from America were admixed populations from Africa and West Eurasia, most of our novel sequences from Central and Western South America demonstrated a close phylogenetic relationship to samples from East Asia. Indeed, most of these samples came from populations within or partially within the indigenous communities with a common ancestor closely related to Asian regions.45 This parallel persisted despite the increasingly mobile global populations and EBV being an infection from childhood through young adulthood.
However, inferring associations based on self-reported and sociologically driven classifications can further complicate analysis and lead to unintended generalizations.46
Several groups have hypothesized that disease-specific variants may explain the pathogenesis and epidemiologic patterns. To our knowledge, our data are the first to include EBV genome sequencing from NKTCL cases outside of East Asia. We demonstrated that the previously suggested variants were not disease-specific but rather common to EBV in East Asia.
The difference between our data and those of Xiong et al6 is that, in the latter data set, EBV genomes from other disease phenotypes were not of East Asian origin, leaving geographic variation unaccounted for. Indeed, geography rather than disease was shown to be the strongest predictor of genome clustering within the global phylogeny of EBV. This does not eliminate the potential for disease variants to be identified but suggests that even greater representation in geographic diversity and disease phenotypes is needed to identify them and control for region-specific variants. The latter scenario is supported by our confirmation of previous studies on NPC in which there was an increase in variant frequency for several genes in NPC when compared with other phenotypes in East Asia included in the previous studies. Notably, pulmonary lymphoepithelioma-like carcinoma, not included in previous work, also demonstrated a high frequency of these variants, suggesting that variants could drive specific disease groups. It should be noted that none of these variants were present in our single NPC case from outside East Asia.
Therefore, future studies outside East Asia are needed to determine the global importance of these variants. We identified several variants that were high risk for NKTCL, but the impact of these variants on EBV infection and malignant transformation is unknown.
Previous work has established that the response to specific EBV peptides is variable based on the presence of specific MHC polymorphisms. Genetic variants in the class I locus have been associated with both variability in the response to infection and the risk for EBV-associated malignancies.47-54 Indeed, when we evaluated the predicted binding affinity of previously proposed peptides to generate a therapeutic immune response, we found dramatic differences within and across HLA types. Reassuringly, the most high-frequency variants across EBV genomes in these peptides did not affect class I binding when compared with the reference sequence. However, this specificity of binding to distinct MHC alleles reaffirms that peptide approaches must account for both the EBV variants and the most common HLA alleles in a given region. Unknown at this time is whether the use of messenger RNA–based strategies, using sequences based on reference genomes, will generate responses to peptides presented on infected cells that may vary based on an individual’s EBV variant. A potential application of this work and the continued sequencing of EBV genomes from diverse populations is to equip vaccines with multiple variants of the vaccine target. Alternatively, rational design of population-specific therapeutics may be warranted.
Our study has several important limitations. Most public genomes do not have raw sequencing data available, thereby preventing an evaluation of the quality of the assemblies and variants. We sought to overcome this by setting thresholds for alignment to the reference genome, but this led to slightly biasing acceptable genomes to type 1 vs type 2 EBV. In addition, because of the limitations in short-read sequencing and the proximity of some genes to repeat regions of the genome, parts of the EBV genome could not be evaluated. Short-read sequencing also limits our ability to determine if allele frequency differences are caused by multiple strains or SNPs occurring secondary to rapid viral replication within the tumor. This could be resolved with targeted and/or long-read sequencing of these genes, but these efforts will require original sample material across the previously and still to be sequenced global populations. Finally, most EBV genomes, including several of our novel EBV genomes, were generated with standard NGS rather than with duplex sequencing. Using the latter technology, we were able to detect a greater number of variants in the same sample. The increased sensitivity of duplex sequencing for detecting rare variants means that studies using this method may identify additional mutations not detected by standard NGS. This difference complicates direct comparisons, because it is not always clear whether detected variants were truly absent in previous studies or simply missed because of lower sensitivity.
To our knowledge, our study, in addition to contributing nearly 200 novel EBV genomes from previously poorly represented populations and disease phenotypes, represents the largest global analysis of EBV phylogeny and gene variants. Our analysis emphasized that geographic region had a greater influence on EBV genomes than disease phenotypes, raising caution about drawing conclusions from variants identified in limited data sets. This extensive genomic diversity analysis has important implications for future therapeutic strategies, including cellular therapy and vaccine development. Continued efforts in sequencing EBV genomes from diverse populations will be pivotal in understanding the global impact of genetic variation in EBV and advancing appropriate and equitable therapeutic approaches.
Acknowledgments
The authors thank Vicki De Falla and the board of La Liga Nacional Contra el Cáncer de Guatemala y El Instituto de Cancerología y Hospital, Bernardo del Valle, and the patients and families whose tumor biopsies contributed to this study.
This study was supported by the Cush-It to The Limit Foundation, the Cancer Therapeutics Endowment, the National Cancer Institute (NCI) of the National Institutes of Health (NIH) of the US Department of Health and Human Services (HHS; grants R01CA217138, R01CA239287, P30 CA015704, and R01AI141531), the Office of Research Infrastructure Programs of NIH/HHS (grant S10OD028685), and NIH National Heart, Lung, and Blood Institute (grant 2T32HL007093-46).
Authorship
Contribution: E.L.B., S.R., and E.H.A. designed and performed experiments, analyzed the data, and drafted the manuscript; C.C.C.A., J.C.B.M., E.S.-O., P.S., D.E.-V., C.B., S.-C.Y., N.M., G.S.-R., and R.N. analyzed the data; O.S., M.M.S.T., Y.N., H.L.K., A.N., C.W., and Y.X. performed experiments, analyzed data, and edited the manuscript; C.O., F.V., and D.M.W. designed experiments and edited the manuscript; A.S. and R.A.B. edited the manuscript; and R.A.B. and E.H.W. designed experiments, analyzed data, and drafted the manuscript; A.W.W. analyzed the data and R.R. edited the manuscript.
Conflict-of-interest disclosure: D.M.W. is an employee of Merck and Co and owns equity in Merck & Co, Bantam, Ajax, and Travera. A.S. is an employee of Cellectar Biosciences Inc. R.A.B. is on the scientific advisory boards of Atara Biotherapeutics and Viracta Therapeutics, and is a consultant and has stock options in Viracta Therapeutics. The remaining authors declare no competing financial interests.
The current affiliation for D.M.W. is Merck & Co, Rahway, NJ.
Correspondence: Edus H. Warren, Hutchinson Cancer Center 1100, Fairview Ave N, S3-204, Seattle, WA; email: ehwarren@fredhutch.org.
References
Author notes
The project IDs for all of our sequences are now publicly available. The data reported in this article have been deposited in the BioProject (accession number PRJNA1063319; available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1063319).
The full-text version of this article contains a data supplement.