Key Points
CD14+ monocytes from patients with CH stimulate inflammation through increased cytokine expression.
T cells from CH are deficient in GIMAP expression, suggesting CH may impair T-cell differentiation.
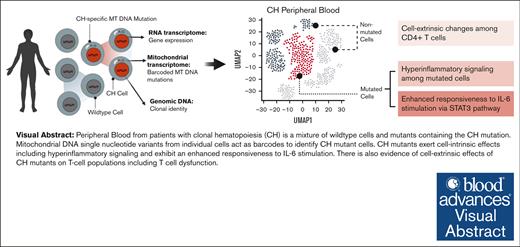
Visual Abstract
Clonal hematopoiesis (CH) is an age-associated phenomenon that increases the risk of hematologic malignancy and cardiovascular disease. CH is thought to enhance disease risk through inflammation in the peripheral blood.1 Here, we profile peripheral blood gene expression in 66 968 single cells from a cohort of 17 patients with CH and 7 controls. Using a novel mitochondrial DNA barcoding approach, we were able to identify and separately compare mutant Tet methylcytosine dioxygenase 2 (TET2) and DNA methyltransferase 3A (DNMT3A) cells with nonmutant counterparts. We discovered the vast majority of mutated cells were in the myeloid compartment. Additionally, patients harboring DNMT3A and TET2 CH mutations possessed a proinflammatory profile in CD14+ monocytes through previously unrecognized pathways such as galectin and macrophage inhibitory factor. We also found that T cells from patients with CH, although mostly unmutated, had decreased expression of GTPase of the immunity associated protein genes, which are critical to T-cell development, suggesting that CH impairs T-cell function.
Introduction
With age, hematopoietic stem cells acquire mutations in driver genes such as DNA methyltransferase 3A (DNMT3A) and Tet methylcytosine dioxygenase 2 (TET2), resulting in a selective advantage and clonal hematopoiesis (CH). CH is a risk factor not only for hematologic malignancy but also for multiple diseases of aging including cardiovascular disease, kidney disease, and osteoporosis.1-5 Although many epidemiological analyses consider CH as a single entity, the literature reveals that significant associations are gene specific. For example, TET2 is more strongly associated with a proinflammatory disease mechanism across multiple forms of cardiovascular disease,1,6 whereas DNMT3A CH is associated with heart failure7,8 and osteoporosis.5
Much attention has focused on how CH mutations lead to skewed hematopoiesis in hematopoietic stem and progenitor cells.9,10 However, little attention has focused on the peripheral compartment. Circulating immune cells with CH mutations are morphologically and immunophenotypically similar to their nonmutated counterparts, making direct comparisons difficult in primary human tissues. Whether primary cell-intrinsic transcriptional changes or secondary microenvironment effects, or both drive pathological phenotypes is unknown.
Although transcriptional profiling of single cells has become routine, it remains difficult to extract genotype and transcriptional data out of the same cell. Because DNMT3A and TET2 CH blood samples are a mixture of mutated and nonmutated cells, both genotyping and transcriptomic sequencing modalities are necessary to delineate these cells. Cell-intrinsic consequences may arise directly from the somatic mutation, whereas extrinsic, indirect consequences may arise from altered cell-cell interactions or secreted immune effectors. These phenomena could be distinguished by identifying mutant and nonmutant cells from the same sample. Several technologies have sought to close this gap by selectively amplifying the messenger RNA transcriptome and using this to genotype cells.11-14 This approach is effective in hematopoietic stem and progenitor cells that express DNMT3A and TET2; however, genotyping is less efficient in cells that do not express these genes, such as fully differentiated cells in the peripheral blood.12,13,15
To overcome this, we combined single-cell RNA sequencing (scRNA-seq) with cell-specific mitochondrial DNA (MT-DNA) barcoding to simultaneously resolve single-cell DNA mutation status for 100% of cells.15 Our analysis of 66 968 single cells from 17 individuals with TET2 CH or DNMT3A CH and 7 age-matched controls finds novel mechanisms of CH-driven inflammation and enables direct comparison between peripheral CH mutated and wildtype (WT) cells across individuals.
Methods
Primary patient samples
Fresh peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll separation. After low-speed centrifugation, pelleted cells were resuspended in freezing media (88% fetal bovine serum and 12% dimethyl sulfoxide) and placed in liquid nitrogen.
DNA extraction and CH variant calling
All enrolled patients underwent targeted sequencing to evaluate for the presence of CH mutations. DNA was extracted using Qiagen Mini kits (catalog no. 27104) according to manufacturer's recommendations. We sequenced samples using a custom capture panel designed to tile known CH genes, targeting 600× read depth coverage as previously described.16 Somatic mutations were called using publicly available methods in workflow description language in Mutect2 on the Terra platform (https://terra.bio/). A putative variant list was formulated and then cross-referenced with a list of known CH driver mutations.1 Variants were then filtered for read quality including sequencing depth and minimum alternate allele read depth.
scRNA-seq library preparation
Cryopreserved PBMCs from patients with CH and controls were thawed at 37°C and washed with complete Roswell Park Memorial Institute ([RPMI] RPMI + 10% fetal bovine serum + 1% physiologic saline) to remove freezing media. PBMCs from each sample (500 000 cells) were plated. Cells were pooled after staining with unique hashtag antibody oligonucleotide–conjugate (30 minutes) staining (BioLegend, TotalSeq-B). Pooled samples were immediately run on a 10× Chromium Controller after preparation with a 10× Chromium 3’ library preparation kit (10× Genomics) per manufacturer’s instructions. Briefly, 30 000 cells were loaded per well and captured in gel bead emulsions. Captured messenger RNA was reverse transcribed into complementary DNA (cDNA) and amplified to create whole-transcriptome analysis libraries. Further library construction was carried out after fragmentation, adapter ligation, and a sample index polymerase chain reaction (PCR) to create scRNA-seq libraries.
10× single-cell sequencing and data preparation
Next-generation 150-nt paired-end sequencing was conducted on an Illumina Novaseq6000 using the cDNA libraries produced by the 10× Chromium library (supplemental Table 6). CellRanger Count (10× Genomics) was used to filter low quality reads and align to the GRCh38 reference genome using STAR as described elsewhere.17 Resulting matrices from the CellRanger pipeline were then converted into Seurat18,19 objects. Demultiplexing was performed using the HTODemux function in the R package Seurat, applying a positive quantile value of 0.99. Cells containing ≥15% reads mapping to the mitochondrial genome were filtered. Similarly, we filtered cells with less than 250 genes and 500 unique molecular indices. Remaining doublets were removed with the R package DoubletFinder,20 using the first 10 principal components and a doublet formation rate of 7.5%.21 One lane had an abnormally high number of doublets, so a more stringent filter was applied for that lane using the DoubletFinder metric pANN of 0.28. Batch correction was performed with the R package Harmony (v0.1.1).
After performing dimensionality reduction with the function RunUMAP from Seurat and calculating clusters with the function FindClusters (resolution = 0.75), cell type assignment was performed using ScType (v1.0). Low confidence cell types were annotated manually. Mitochondrial and ribosomal genes were removed.
Single-cell mitochondrial enrichment
Mitochondrial enrichment of 10× Genomics v3 3’ cDNA was performed using primer sequences as described in Miller et al.22 Briefly, mitochondrial enrichment was achieved through 2 additional PCR reactions using the 10× 3’ cDNA as a template. First, cDNA was amplified using custom primers encompassing the entire mitochondrial genome along with a barcoded i5 primer for sample indexing (supplemental Table 5). The samples were mixed and diluted to equal 20 ng DNA in 16 uL. The primer mix was added to KAPA HiFi Hotstart Readymix (Roche), and PCR was performed. The resultant PCR product was incubated with 1.0× AMPure XP Beads to remove the primers. The second PCR adds the Illumina indexes for sequencing. Both i5 and i7 indexes were added by combining the eluted DNA from PCR1 and KAPA HiFi HotStart ReadyMix. The DNA was purified with 0.8× AMPure XP beads and then eluted in TE Buffer.
Single-cell MT-DNA sequencing, read processing, and variant calling
MT-DNA processing and variant calling were carried out as previously published.15 Briefly, fastq files from MT-DNA enrichment were filtered for reads associated with low-frequency cell barcodes (CBs) and trimmed to remove the unique molecular index and CB. Reads were aligned using STAR to hg38. Next, we used maegatk to call variants across the mitochondrial genome.15,23 Maegatk calls MT-DNA variants using combined CB from both scRNA-seq and MAESTER enrichment for variants with at least 5 supporting reads.
scDNA-seq via Mission Bio Tapestri
Single-cell DNA sequencing (scDNA-seq) was performed using the Tapestri platform. Cells were stained with the BioLegend Total-Seq D Heme Oncology Panel and the Human TruStain FcX antibody. Targeted DNA amplification was carried out using custom designed probes from Mission Bio (supplemental Table 8). The amplified DNA was released from individual oil droplets using AMPure XP beads. The final product was quantified using a Qubit fluorometer from Thermo Fisher and assessed for quality on an Agilent Bioanalyzer. Samples were pooled before sequencing with a 25% spike-in of PhiX and run on a NovaSeq 6000 S4 flow cell from Illumina to generate 150 bp paired-end reads. Sequencing was performed at the Vanderbilt Technologies for Advanced Genomics sequencing core.
Pipeline processing and variant filtering for Tapestri scDNA-seq
Single-cell DNA samples were processed using the Tapestri Pipeline v1.8.4. Adapters were trimmed, and reads were aligned to the hg19 reference genome. Variants were called using GATK 3.7 and filtered based on quality scores, read depth, and genotype frequency. Informative variants were annotated, and cells were clustered based on their genotypes.
To annotate cell populations, unsupervised hierarchical clustering was performed on the antibody-oligo conjugate (AOC) data. Reads were normalized, and AOCs with low expression were removed. Principal component analysis was conducted on the normalized AOCs, and the first 10 principal components were used for uniform manifold approximation and projection (UMAP) coordinate calculation. The resulting cells were clustered using a k state of 100, and clusters with noisy AOC expression were eliminated. The remaining clusters were annotated based on expert knowledge of surface marker expression.
Identification and selection of informative single-cell MT-DNA variants
An allele frequency matrix was constructed using all possible variants in the mitochondrial genome for each patient. We anticipated to find the CH variant among a clone with a myeloid lineage bias based on understanding from prior publications.24-26 Because the same cDNA was used for MT-DNA enrichment and single-cell RNA expression, cell type annotations were assigned using the CBs from the corresponding RNA expression data set. MT-DNA variants enriched within monocytes and absent or very low-frequency in lymphoid cells were considered candidate markers of CH variants. Alignment of candidate MT-DNA variants with CH variants was confirmed via single-cell DNA sequencing (Tapestri) using cells from the same patient samples. MT variants that co-occurred with CH variants as expected were used for downstream classification of CH mutant status. Through this single-cell DNA sequencing process, both MT-DNA variants and CH variants can be simultaneously identified. This enabled the putative variants gathered from the MT-DNA enrichment in scRNA-seq samples to be verified.
Differential expression analysis and pathway analysis
Differential gene expression was calculated using a pseudobulk-like approach in which measurements from groups of similar cells were summed. Cells were separated by genotype, and cell type was then clustered using the Python module Metacell-2 (v0.8.0). We followed standard procedures from the Metacell-2 vignette. We excluded gene modules that had an average correlation with cell cycling genes of ≥0.75. Differential expression was calculated for genes that had at least 10 transcripts in at least 85% of metacells using the R package DESeq2. We performed Wald tests of significance with Benjamini-Hochberg multiple testing correction. Genes that were sex specific and red blood cell specific were removed. Pathway analysis was performed on differential expression results using the function gseGO with ont = “ALL,” minGSSize = 50, maxGSSize = 800, and nPermSimple = 10 000 from the R package clusterProfiler (v4.8.1).
Cell signaling interactions were predicted from scRNA-seq data with the R package CellChat (v1.6.1). Genes with extremely high or low expression were removed with the parameter trim = 0.1. Comparisons were restricted to cell types that had at least 25 cells with the parameter min.cells = 25.
To compare mutant and WT monocyte cell states, we performed pairwise differential gene expression (DGE) analysis by using the model-based analysis of single-cell transcriptomics method with false discovery rate (FDR) correction. To reduce transcriptional noise before DGE, we only included genes that were detected in at least 10 cells. We then applied the Hurdle model from the model-based analysis of single-cell transcriptomics R package (v.1.24.1) and adjusted for the cellular detection rate to determine significant differences in gene expression (threshold: absolute value of the log fold-change coefficients > 0.25; FDR > 0.05).
Phospho-specific flow cytometry
Cryovials of cryopreserved cells from healthy donors and patients with CH were thawed and washed with 10 mL of complete RPMI. The cells were stained for viability with AlexaFlour700 (Invitrogen; catalog no. P10163) and counted. An aliquot of 500 000 cells were plated in 200 μL of media in a 96-well plate and stimulated with 20 ng/mL of interleukin-6 (IL-6; Peprotech) for 15 minutes. Cells were then fixed with 1.6% PFA at room temperature and permeabilized with 150 μL of methanol at –80°C for at least 30 minutes. Cells were resuspended in 180 μL of phosphate buffered saline and fluorescence cell barcoding performed as previously described27 with serial dilutions of Pacific Blue (LifeTechnologies; catalog no. P10163) and Pacific Orange (Invitrogen; catalog no. P30253) dyes for 30 minutes in the dark at room temperature. Two concentrations of Pacific Blue were prepared (20 and 4 μg/mL), whereas 6 concentrations of Pacific Orange were prepared (7.00, 2.99, 1.27, 0.54, 0.23, and 0.10 μg/mL). Barcoding was quenched with 80 μL of cell staining media. The barcoded cells were then collected into a single tube and stained with a cocktail of antibodies: CD33 PECy7 (5 μL per 100 μL stain; Biolegend; catalog no. 303434; clone WM53) and pSTAT3 AlexFlour488 (2.5 μL per 100 μL stain; Biolegend; catalog no. 651006; clone 13A3-1) for 30 minutes before acquisition on a BD 5-laser Fortessa flow cytometer.
All patients in this study consented to all study procedures under Vanderbilt University Medical Center institutional review board approved research protocols (identifiers: 210022 and 201583) in accordance with the Treaty of Helsinki. Adult patients able to give consent were recruited from Vanderbilt University Medical Center clinics who had known CH mutations as a result of clinical evaluation or who were at risk of having a CH mutation. All patients were confirmed to be without active hematologic malignancy at the time of enrollment.
Results
We used scRNA-seq with cell-specific mitochondrial DNA sequencing to resolve single-cell genomic DNA mutation status and investigate pathological mechanisms of CH (Figure 1A; see “Methods”). PBMCs from 8 TET2, 9 DNMT3A, and 7 age-matched controls (age, 47-89 years) were selected from a prospective CH observational study that was designed to capture patients at high risk of CH through a robust referral network (Figure 1A-B; Table 1).
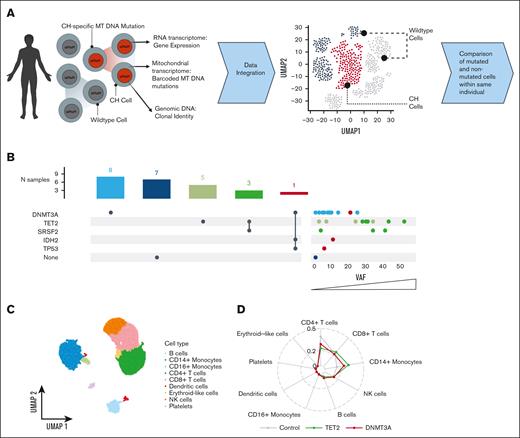
scRNA-seq reveals distinct cell type profiles in CH. (A) Cells from 17 patients with CH and 7 controls were processed via scRNA-seq. Mutational lineage tracing was performed to link genomic variants to scRNA-seq results. (B) UpSet plot displaying CH mutations for each patient with accompanying dot plot showing variant allele frequency for each mutation. Color corresponds to mutational group. Dots correspond to mutations, so patients with multiple CH mutations have separate dots for each mutation. (C) UMAP displaying cell type clusters, as defined by unsupervised clustering. Clusters were annotated using ScType. (D) Radar plot showing cell type proportions for controls, patients with TET2 mutations, and patients with DNMT3A mutations. NK cell, natural killer cell.
scRNA-seq reveals distinct cell type profiles in CH. (A) Cells from 17 patients with CH and 7 controls were processed via scRNA-seq. Mutational lineage tracing was performed to link genomic variants to scRNA-seq results. (B) UpSet plot displaying CH mutations for each patient with accompanying dot plot showing variant allele frequency for each mutation. Color corresponds to mutational group. Dots correspond to mutations, so patients with multiple CH mutations have separate dots for each mutation. (C) UMAP displaying cell type clusters, as defined by unsupervised clustering. Clusters were annotated using ScType. (D) Radar plot showing cell type proportions for controls, patients with TET2 mutations, and patients with DNMT3A mutations. NK cell, natural killer cell.
Demographic features of CH and control patients
| . | Control (n = 7) . | DNMT3A (n = 9) . | TET2 (n = 8) . |
|---|---|---|---|
| Sex | |||
| Female | 4 (57.1%) | 2 (22.2%) | 2 (25.0%) |
| Male | 3 (42.9%) | 7 (77.8%) | 6 (75.0%) |
| Race | |||
| White | 7 (100%) | 9 (100%) | 6 (75.0%) |
| Asian | 0 (0%) | 0 (0%) | 1 (12.5%) |
| Missing | 0 (0%) | 0 (0%) | 1 (12.5%) |
| Age | |||
| Mean (SD) | 75 (14) | 67 (15) | 75 (8.8) |
| Missing | 1 (14.3%) | 0 (0%) | 0 (0%) |
| Hx tobacco use | 3 (42.9%) | 5 (55.6%) | 3 (37.5%) |
| Hx CAD | 3 (42.9%) | 3 (33.3%) | 2 (25.0%) |
| Hx CHF | 1 (14.3%) | 1 (11.1%) | 1 (12.5%) |
| Systolic BP, mean (SD) | 130 (14) | 120 (16) | 130 (11) |
| Diastolic BP, mean (SD) | 73 (16) | 70 (9.1) | 74 (11) |
| Heart rate | |||
| Mean (SD) | 70 (11) | 78 (14) | 68 (9.4) |
| Missing | 0 (0%) | 1 (11.1%) | 0 (0%) |
| . | Control (n = 7) . | DNMT3A (n = 9) . | TET2 (n = 8) . |
|---|---|---|---|
| Sex | |||
| Female | 4 (57.1%) | 2 (22.2%) | 2 (25.0%) |
| Male | 3 (42.9%) | 7 (77.8%) | 6 (75.0%) |
| Race | |||
| White | 7 (100%) | 9 (100%) | 6 (75.0%) |
| Asian | 0 (0%) | 0 (0%) | 1 (12.5%) |
| Missing | 0 (0%) | 0 (0%) | 1 (12.5%) |
| Age | |||
| Mean (SD) | 75 (14) | 67 (15) | 75 (8.8) |
| Missing | 1 (14.3%) | 0 (0%) | 0 (0%) |
| Hx tobacco use | 3 (42.9%) | 5 (55.6%) | 3 (37.5%) |
| Hx CAD | 3 (42.9%) | 3 (33.3%) | 2 (25.0%) |
| Hx CHF | 1 (14.3%) | 1 (11.1%) | 1 (12.5%) |
| Systolic BP, mean (SD) | 130 (14) | 120 (16) | 130 (11) |
| Diastolic BP, mean (SD) | 73 (16) | 70 (9.1) | 74 (11) |
| Heart rate | |||
| Mean (SD) | 70 (11) | 78 (14) | 68 (9.4) |
| Missing | 0 (0%) | 1 (11.1%) | 0 (0%) |
Values are listed by counts and by percentages for categorical variables and by mean and SD for continuous variables. BP, blood pressure; CAD, coronary artery disease; CHF, congestive heart failure; Hx, history; SD, standard deviation.
To trace the effects of CH mutations on peripheral blood cell type proportions, we derived cell type annotations based on known marker genes (Figure 1C-D; supplemental Figure 1). There were no significant differences in cell type proportions on routine clinical laboratories (supplemental Table 1). Notably, 4 patients had multiple CH mutations with concomitant cytopenias without bone marrow dysplasia, meeting the diagnostic criteria for clonal cytopenias of undetermined significance.28
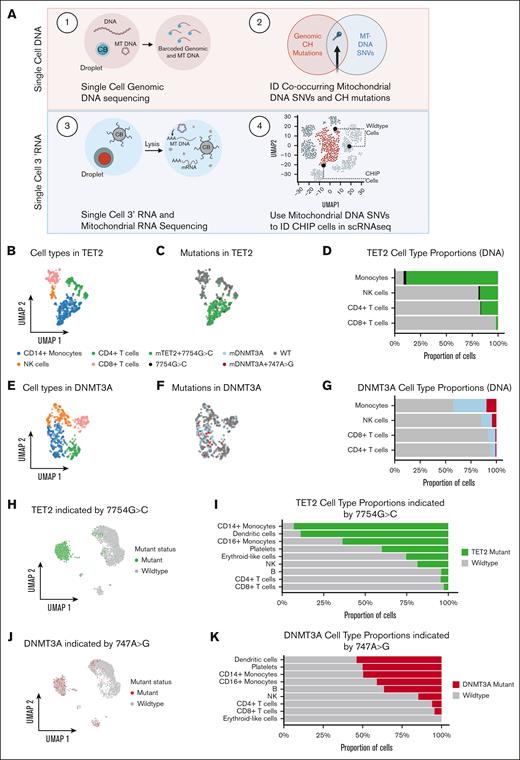
To annotate mutant and nonmutant cells from the same sample, we combined single-cell targeted amplicon sequencing (scDNA-seq) and 3' RNA mitochondrial lineage tracing in our patients with TET2 and DNMT3A (Figure 2A). PBMCs from both patients with TET2 and DNMT3A were processed through the scDNA-seq pipeline (Mission Bio) that captures known genomic CHIP (clonal hematopoiesis of indeterminate potential) mutations and co-occurring mitochondrial variants. We also profiled the immunophenotype of the samples by combining scDNA-seq with oligo-conjugated antibodies to annotate cell populations (Figure 2B,E; supplemental Figure 2B). In 1 patient with a known TET2 mutation at chr4:106157967 with 51% variant allele frequency (VAF) (supplemental Figure 2; supplemental Table 2), our scDNA-seq analysis revealed a single mitochondrial variant (MT 7754G>C) that was concordant with cells harboring the known TET2 mutation, suggesting common lineage. We found 492 cells that carried both the TET2 mutation and the MT 7754G>C variant and 492 cells that carried neither variant. We excluded a marginal number (n = 15) of cells in which only the 7754G>C variant was detected. The mature myeloid cell compartment was heavily enriched for both the TET2 mutation and the mitochondrial variant (Figure 2C-D; extended data Figure 3A). We repeated the analysis for a sample with a known DNMT3A mutation with 24% VAF (Figure 2F-G; supplemental Figure 2). The concomitant genomic and mitochondrial variants (chr2:25470560 and MT 747A>G) were detected in most myeloid cells and a moderate proportion of lymphocytes, consistent with previous knowledge regarding DNMT3A mutations in hematopoiesis24 (Figure 2F-G; extended data Figure 3B). There were 111 DNMT3A cells in which the 747A>G variant was not detected, indicating that 747A>G marks a cell population that is subclonal to DNMT3A. We validated the cooccurrence of the CH variant and 747A>G using primary template amplification of genomic DNA from single-cell colonies (extended data Figure 3D, see “Methods”). Subsequently, we were able to use the MT-DNA single nucleotide variant as an identifying “barcode” for the mutant cell annotation, allowing for the partition of mutant and WT populations within our scRNA-seq data (Figure 2H-K). In the corresponding single-cell RNA data set evaluating for the TET2 sample, we found a significant myeloid bias among cells identified with MT 7754G>C (log2[fold change] = 4.146; FDR = 0.001; Figure 2H-I). In DNMT3A scRNA-seq sample, we identified cells with 747A>G, finding a less severe monocytic skew accounting for the lower relative VAF than that of the TET2 sample (Figure 2J-K).
Single-cell DNA and RNA sequencing define cells carrying CH mutations. (A) Schematic showing experimental design. (B) UMAP showing cell types for cells from patient with TET2 CH-21-014. (C) UMAP showing mutational status for cells from patient with TET2 CH-21-014. (D) Stacked bar plot quantifying proportion of mutant cells per cell type for cells from patient with TET2 CH-21-014. (E) UMAP showing cell types for cells from patient with DNMT3A CH-20-046. (F) UMAP showing mutational status for cells from patient with DNMT3A CH-20-046. (G) Stacked bar plot quantifying proportion of mutant cells per cell type for cells from patient with DNMT3A CH-20-046. (H) UMAP showing predicted mutational status for cells from patient with TET2 CH-21-014, based on presence of MT mutation 7754G>C. (I) Stacked bar plot quantifying proportion of mutant cells per cell type for cells from patient with TET2 CH-21-014. (J) UMAP showing predicted mutational status for cells from patient with DNMT3A CH-20-046, based on presence of MT mutation 747A>G. (K) Stacked bar plot quantifying proportion of mutant cells per cell types for cells from patient with DNMT3A CH-20-046. NK, natural killer; SNVs, single nucleotide variant.
Single-cell DNA and RNA sequencing define cells carrying CH mutations. (A) Schematic showing experimental design. (B) UMAP showing cell types for cells from patient with TET2 CH-21-014. (C) UMAP showing mutational status for cells from patient with TET2 CH-21-014. (D) Stacked bar plot quantifying proportion of mutant cells per cell type for cells from patient with TET2 CH-21-014. (E) UMAP showing cell types for cells from patient with DNMT3A CH-20-046. (F) UMAP showing mutational status for cells from patient with DNMT3A CH-20-046. (G) Stacked bar plot quantifying proportion of mutant cells per cell type for cells from patient with DNMT3A CH-20-046. (H) UMAP showing predicted mutational status for cells from patient with TET2 CH-21-014, based on presence of MT mutation 7754G>C. (I) Stacked bar plot quantifying proportion of mutant cells per cell type for cells from patient with TET2 CH-21-014. (J) UMAP showing predicted mutational status for cells from patient with DNMT3A CH-20-046, based on presence of MT mutation 747A>G. (K) Stacked bar plot quantifying proportion of mutant cells per cell types for cells from patient with DNMT3A CH-20-046. NK, natural killer; SNVs, single nucleotide variant.
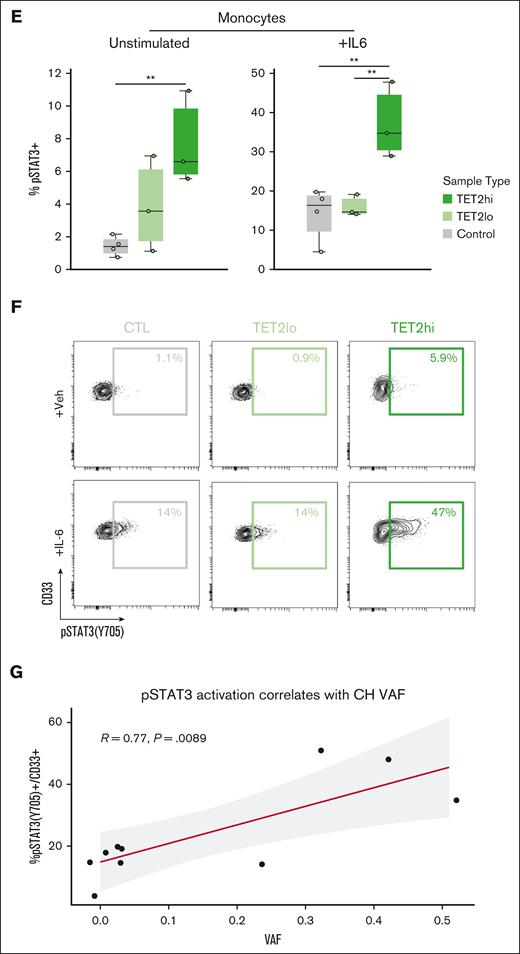
Mutant CD14+ monocytes from 4 patients with TET2 and 2 patients with DNMT3A highlight inflammatory cell-intrinsic effects of CH mutations. (A) Violin plots displaying expression of cytokines in control CD14+ monocytes and TET2 mutant CD14+ monocytes. (B) Violin plots displaying expression of cytokines in control CD14+ monocytes and DNMT3A mutant monocytes. (C) GSEA based on results from differential expression analysis comparing TET2 mutant CD14+ monocytes with control CD14+ monocytes. (D) GSEA based on results from differential expression analysis comparing DNMT3A mutant CD14+ monocytes with control CD14+ monocytes. (E) Quantification of phospho-flow cytometry displaying pSTAT3(Y705)+ in the CD33+ gate of large VAF TET2 CH samples (>25%; n = 3) compared with both low VAF (<25%; n = 3) and control samples (n = 4) (∗∗P < .01 by analysis of variance with Tukey's Honestly Significant Difference). (F) With IL-6 stimulation condition (similar to E). (G) Pearson correlation of %pSTAT3(Y705)+ in the CD33+ gate and VAF after IL-6 stimulation. For samples with multiple mutations, the highest VAF value was selected for the analysis. GSEA, gene set enrichment analysis.
Mutant CD14+ monocytes from 4 patients with TET2 and 2 patients with DNMT3A highlight inflammatory cell-intrinsic effects of CH mutations. (A) Violin plots displaying expression of cytokines in control CD14+ monocytes and TET2 mutant CD14+ monocytes. (B) Violin plots displaying expression of cytokines in control CD14+ monocytes and DNMT3A mutant monocytes. (C) GSEA based on results from differential expression analysis comparing TET2 mutant CD14+ monocytes with control CD14+ monocytes. (D) GSEA based on results from differential expression analysis comparing DNMT3A mutant CD14+ monocytes with control CD14+ monocytes. (E) Quantification of phospho-flow cytometry displaying pSTAT3(Y705)+ in the CD33+ gate of large VAF TET2 CH samples (>25%; n = 3) compared with both low VAF (<25%; n = 3) and control samples (n = 4) (∗∗P < .01 by analysis of variance with Tukey's Honestly Significant Difference). (F) With IL-6 stimulation condition (similar to E). (G) Pearson correlation of %pSTAT3(Y705)+ in the CD33+ gate and VAF after IL-6 stimulation. For samples with multiple mutations, the highest VAF value was selected for the analysis. GSEA, gene set enrichment analysis.
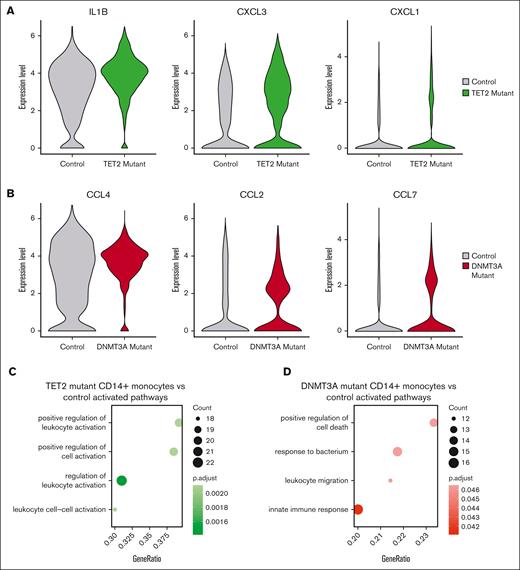
We applied our mitochondrial lineage tracing method to a total of 4 patients with TET2 and 2 with DNMT3A to identify CH clones (supplemental Figure 3). We first performed DGE testing on CD14+ monocytes comparing CH mutant cells with their WT counterparts. This resulted in 70 differentially expressed genes (DEGs) in TET2 mutants, whereas there were 0 DEGs in DNMT3A mutants compared with WT counterparts. We then evaluated mutant TET2 and DNMT3A CD14+ monocytes against unaffected control CD14+ monocytes. We identified 202 and 122 DEGs in TET2 and DNMT3A CD14+ monocytes, respectively (supplemental Table 7). There were 12 overlapping DEGs when comparing mutant TET2 CD14+ monocytes with WT cells and with controls (supplemental Figure 3). Notable among these were inflammatory mediators CXCL1, CXCL3, and IL1B (FDR < 1 × 10–20 for all comparisons; Figure 3A; supplemental Table 7). Top DEGs in mutant DNMT3A monocytes compared with controls included C-C motif chemokine ligand 4 (CCL4; FDR = 1.26 × 10–4), CCL2 (FDR = 2.78 × 10–12), and CCL7 (FDR = 1.34 × 10–7) (Figure 3B; supplemental Table 7). Pathway analysis showed upregulation of leukocyte activation and cell adhesion in mutant TET2 monocytes, whereas mutant DNMT3A monocytes had enrichment in regulation of cellular death pathways and leukocyte migration (Figure 3C-D).
Noting the increased expression of IL-1B, a prominent downstream mediator of the IL-6 pathway among mutant TET2 monocytes, we sought to further evaluate whether signaling along this axis was a cell-intrinsic or cell-extrinsic phenomenon. To do this, we used phospho-specific flow cytometry to measure response to IL-6 in high VAF TET2 mutant (TET2hi), low VAF TET2 mutant (TET2lo), and controls. The basal pSTAT3+ monocyte percentage was significantly higher in TET2hi monocytes than controls. All samples showed some response to IL-6, whereas TET2hi monocytes had the highest proportion of pSTAT3+ cells, significantly higher than both control and TET2lo samples, in response to IL-6 stimulation (Figure 3E-F). There was a linear increase in the proportion of pSTAT3+ cells after IL-6 stimulation in accordance with increasing VAF (R = 0.77; P = .008), suggesting cell-intrinsic–altered signaling among the mutant fraction (Figure 3G).
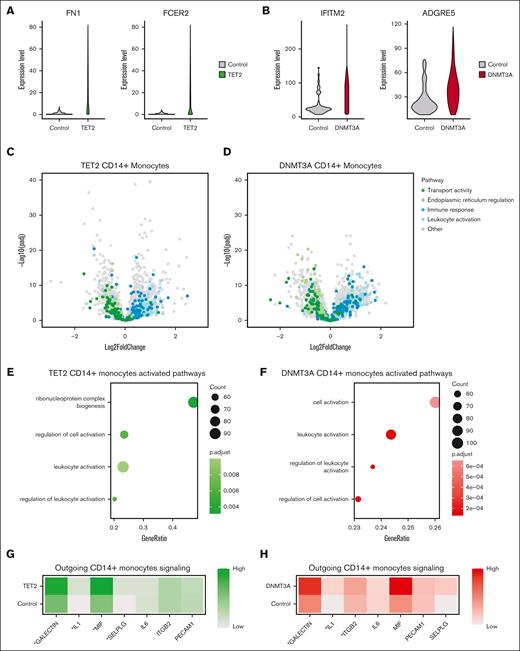
We then queried whether there were also cell-extrinsic effects of CH mutations in our cohort as has been recently reported.29 To determine this, we compared grouped RNA expression profiles from CD14+ monocytes between patients with DNMT3A or TET2 and controls, because this would include both mutated and nonmutated cells. To reduce potential for false discovery from high dropout rates, we partitioned our data set into metacells30 before performing DGE analysis. The top DEGs among patients with TET2 included fibronectin 1 (FN1; adjusted P = 2.39 × 10–26) and Fc epsilon receptor II (FCER2; adjusted P = 2.4 × 10–14), which encodes CD23. Both of these are important components of monocyte adhesion31,32 (Figure 4A). This contrasts with the top DEGs from DNMT3A CD14+ monocytes that included interferon-induced transmembrane protein 2 (IFITM2, adjusted P = 2.37 × 10-18) and adhesion G protein–coupled receptor E5 (ADGRE5; adjusted P = 4.4 × 10–14), genes involved in monocyte adhesion33 and differentiation34 (Figure 4B). Although the specific genes affected were different between TET2 and DNMT3A comparisons, the pathways they converged on were similar. In general, genes related to immune responses and leukocyte activation were upregulated, whereas genes related to transport activity and endoplasmic reticulum regulation were downregulated (Figure 4C-D). Similarly, gene set enrichment analysis highlighted convergent pathways between TET2 and DNMT3A CD14+ monocytes including leukocyte activation, regulation of leukocyte activation, and regulation of cell activation (Figure 4E-F). Using CellChat35 to infer intercellular interactions in our scRNA-seq data, we found CD14+ monocytes from TET2 samples exhibited enhanced signaling across IL-1, macrophage migration inhibitory factor (MIF), and galectin, all parts of the inflammatory signaling axis (Figure 4G). CD14+ monocytes from DNMT3A samples also exhibited enhanced IL-1 and galectin signaling and uniquely had elevated integrin beta 2 (ITGB2) signaling (Figure 4H; supplemental Table 7).
Genotype–grouped CD14+ monocyte vs control comparisons highlight inflammatory cell-extrinsic effects of CH mutations. (A) Violin plots displaying expression of FN1 and FCER2 in CD14+ monocytes from controls and CD14+ monocytes from patients with TET2 mutations. (B) Violin plots displaying expression of IFITM2 and ADGRE5 in CD14+ monocytes from controls and CD14+ monocytes from patients with DNMT3A mutations. (C) Volcano plot showing results of differential expression analysis comparing CD14+ monocytes from patients with TET2 mutations with CD14+ monocytes from controls, colored by biological pathway. (D) Volcano plot showing results of differential expression analysis comparing CD14+ monocytes from patients with DNMT3A mutations with CD14+ monocytes from controls, colored by biological pathway. (E) GSEA based on results from differential expression analysis comparing CD14+ monocytes from patients with TET2 mutations with CD14+ monocytes from controls. (F) GSEA based on results from differential expression analysis comparing CD14+ monocytes from patients with DNMT3A mutations with CD14+ monocytes from controls. (G) Heat map showing predicted outgoing signaling from CD14+ monocytes from patients with TET2 mutations and from controls for pathways involved in inflammatory response and immune cell migration, as determined by CellChat (∗P < .05/7). (H) Heat map showing predicted outgoing signaling from CD14+ monocytes from patients with DNMT3A mutations and from controls for pathways involved in inflammatory response and immune cell migration, as determined by CellChat (∗P < .05/7). GSEA, gene set enrichment analysis.
Genotype–grouped CD14+ monocyte vs control comparisons highlight inflammatory cell-extrinsic effects of CH mutations. (A) Violin plots displaying expression of FN1 and FCER2 in CD14+ monocytes from controls and CD14+ monocytes from patients with TET2 mutations. (B) Violin plots displaying expression of IFITM2 and ADGRE5 in CD14+ monocytes from controls and CD14+ monocytes from patients with DNMT3A mutations. (C) Volcano plot showing results of differential expression analysis comparing CD14+ monocytes from patients with TET2 mutations with CD14+ monocytes from controls, colored by biological pathway. (D) Volcano plot showing results of differential expression analysis comparing CD14+ monocytes from patients with DNMT3A mutations with CD14+ monocytes from controls, colored by biological pathway. (E) GSEA based on results from differential expression analysis comparing CD14+ monocytes from patients with TET2 mutations with CD14+ monocytes from controls. (F) GSEA based on results from differential expression analysis comparing CD14+ monocytes from patients with DNMT3A mutations with CD14+ monocytes from controls. (G) Heat map showing predicted outgoing signaling from CD14+ monocytes from patients with TET2 mutations and from controls for pathways involved in inflammatory response and immune cell migration, as determined by CellChat (∗P < .05/7). (H) Heat map showing predicted outgoing signaling from CD14+ monocytes from patients with DNMT3A mutations and from controls for pathways involved in inflammatory response and immune cell migration, as determined by CellChat (∗P < .05/7). GSEA, gene set enrichment analysis.
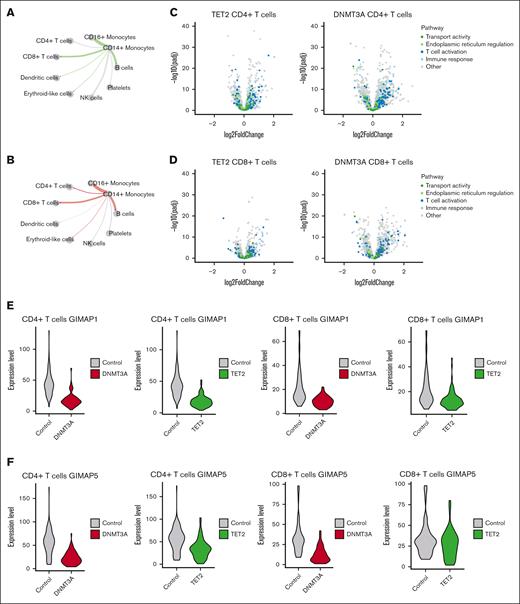
When evaluating signaling patterns between cell types, we noted increased signaling from both CH CD14+ monocytes to T cells, leading us to investigate the impact of CH on T cells (Figure 5A-B). We found that genes involved in T-cell activation and immune response were highly expressed in both TET2 and DNMT3A samples compared with controls (Figure 5C-D; supplemental Table 7). Each member of the GTPase of the immune associated nucleotide binding protein (GIMAP) family, which plays a critical role in proper T- and B-cell differentiation,36,37 was downregulated in CD4+ T cells and CD8+ T cells in both TET2 and DNMT3A samples (Figure 5E-F; supplemental Fig 4). GIMAP1 and GIMAP5, which both result in T-cell deficiency when knocked out in mice,36 were significantly downregulated (adjusted P < .05; log2[fold change] < –.5) in each comparison, except for GIMAP5 in TET2 CD8+ T cells, which had a P value of .235.
Genotype–grouped CD4+ and CD8+ T cell vs control comparisons highlight alterations to T-cell activation and differentiation in CH. (A) Circle plot displaying predicted differential interaction strength between CD14+ monocytes and relevant hematopoietic cells from TET2 samples compared with controls, as determined by CellChat. Line thickness corresponds to differential interaction strength. Green color indicates increased signaling in TET2 samples compared to controls. Gray indicates decreased signaling. (B) Circle plot displaying predicted differential interaction strength between CD14+ monocytes and relevant hematopoietic cells from DNMT3A samples compared with controls, as determined by CellChat. Line thickness corresponds to differential interaction strength. Red color indicates increased signaling in DNMT3A samples compared with controls. Gray indicates decreased signaling. (C) Volcano plot showing results of differential expression analysis comparing CD4+ T cells from patients with TET2 (left) or DNMT3A (right) mutations with CD4+ T cells from controls, colored by biological pathway. (D) Volcano plot showing results of differential expression analysis comparing CD8+ T cells from patients with TET2 (left) or DNMT3A (right) mutations with CD8+ T cells from controls, colored by biological pathway. (E) Violin plots displaying expression of GIMAP1 in CD4+ T cells and CD8+ T cells from controls and from patients with CH mutations. (F) Violin plots displaying expression of GIMAP5 in CD4+ T cells and CD8+ T cells from controls and from patients with CH mutations.
Genotype–grouped CD4+ and CD8+ T cell vs control comparisons highlight alterations to T-cell activation and differentiation in CH. (A) Circle plot displaying predicted differential interaction strength between CD14+ monocytes and relevant hematopoietic cells from TET2 samples compared with controls, as determined by CellChat. Line thickness corresponds to differential interaction strength. Green color indicates increased signaling in TET2 samples compared to controls. Gray indicates decreased signaling. (B) Circle plot displaying predicted differential interaction strength between CD14+ monocytes and relevant hematopoietic cells from DNMT3A samples compared with controls, as determined by CellChat. Line thickness corresponds to differential interaction strength. Red color indicates increased signaling in DNMT3A samples compared with controls. Gray indicates decreased signaling. (C) Volcano plot showing results of differential expression analysis comparing CD4+ T cells from patients with TET2 (left) or DNMT3A (right) mutations with CD4+ T cells from controls, colored by biological pathway. (D) Volcano plot showing results of differential expression analysis comparing CD8+ T cells from patients with TET2 (left) or DNMT3A (right) mutations with CD8+ T cells from controls, colored by biological pathway. (E) Violin plots displaying expression of GIMAP1 in CD4+ T cells and CD8+ T cells from controls and from patients with CH mutations. (F) Violin plots displaying expression of GIMAP5 in CD4+ T cells and CD8+ T cells from controls and from patients with CH mutations.
Discussion
Here, we present transcriptional profiling and characterization of DNMT3A and TET2 CH in human peripheral blood. By using a novel approach that integrates multimodal scRNA-seq with scDNA-seq to link mitochondrial mutations to somatic nuclear mutations, we simultaneously resolve DNA mutational status and cell state. Our study revealed CH mutation–specific aberrations in cellular state, allowing for several conclusions.
First, we identified CD14+ monocytes as drivers of CH-associated inflammation in the peripheral blood in both TET2 and DNMT3A CH. Specifically, we found TET2 CH mutant CD14+ monocytes harbored important differences, suggesting cell-intrinsic mechanisms are important to TET2 phenotypes. In relation to nonmutant WT monocytes from patients with TET2, mutant CD14+ monocytes exhibited significant differences across important inflammatory genes including IL1B, CXCL3, and CXCL1, a phenomenon that was not seen in DNMT3A. Furthermore, intracellular monocyte signaling via STAT3 in response to IL-6 exhibited a VAF-dependent increase, further supporting the notion that mutant TET2 monocytes exhibit cell-intrinsic signaling patterns. These experiments suggest that a precision medicine approach is possible in TET2 CH. A recent analysis of the canakinumab anti-inflammatory thrombosis outcomes trial found that IL-1B antagonist, canakinumab,38 reduced cardiovascular risk in TET2 but not patients with DNMT3A CH.2,39 Our data provide a mechanistic rationale for a genotype-specific approach to treat CH, a finding only possible with the ability to partition mutant and WT cells from the same sample.
Second, collective differences between CD14+ monocytes from both TET2 and DNMT3A and controls identify novel gene targets and signaling pathways. Computationally inferred outgoing signaling in monocytes from patients with TET2 and DNMT3A indicated a notable increase in MIF signaling. MIF resides as a preformed peptide in a variety of cell types and binds with its receptors CXCR2 as well as CXCR4 to promote the recruitment of monocytes and T cells to sites of tissue injury.40 Recruitment of hyperinflammatory monocytes has been identified as the initiating event in the development of atherosclerotic plaques.41 It is notable that among the patients with TET2, 3 had co-occurring serine/arginine-rich splicing factor 2 (SRSF2) mutations. SRSF2 is also a CH mutation and associated with poor survival in myelodysplastic syndromes and more recently found responsible for monocytosis in the presence of TET2.25,42SRSF2 mutations are not readily detected with scDNA-seq due to high GC content in the region, therefore, we were unable to assess the effect of this mutation independently at the single-cell level.43 Comparison of CD14+ monocytes from patients with both SRSF2 and TET2 against those with only TET2 mutations yielded several DEGs, and further gene set enrichment analysis showed enhanced nucleic acid and RNA metabolic processes. The inflammatory profile noted between the TET2 and control CD14+ monocytes was not recapitulated in this, and the expression of MIF was not significantly different between TET2 only and TET2/SRSF2 samples (log2[fold change] = –0.118; adjusted P = .099). Importantly, the effect of VAF also confounds these analyses because patients with co-occurring SRSF2 mutations had higher VAFs than patients with only TET2 mutations. Additional work detailing the interdependent roles of TET2 and SRSF2 in hematopoiesis and inflammation is needed. ADGRE5, which encodes CD97, had significantly higher expression among monocytes from patients with DNMT3A. The protein product of this gene promotes adhesion and migration to sites of inflammation44 and has been associated with rheumatoid arthritis.45 Therefore, MIF and ADGRE5 may represent novel targets in treating inflammation associated with TET2 and DNMT3A CH.
Third, our study clarified the cell-extrinsic effects of CH mutations in peripheral blood. Comparison between T cells from CH samples and controls highlighted significant effects of CH on both T-cell differentiation and T-cell activation. We observed consistent downregulation of the GIMAP protein family in CD4+ and CD8+ T cells in both TET2 and DNMT3A samples. Work in mice has established that knockout of GIMAP proteins impairs the development of T and B cells, resulting in a relative T/B-cell deficiency and a myeloid skew,36 similar to what is observed in CH. GIMAP proteins are regulated together under the direction of the transcription factors RUNX1, GATA3, and TAL1.46,47DNMT3A directly binds to RUNX1 and GATA3,46 and TAL1 expression has been shown to be disrupted by knockout of both TET2 and DNMT3A.48 Further work investigating the effect of TET2 and DNMT3A mutations on GIMAP expression and subsequent differentiation is warranted.
Our study has several limitations. First, although TET2 and DNMT3A mutations make up approximately two-thirds of all CH mutations, CH represents a diverse set of mutations in >70 genes. These CH mutations are likely to have divergent effects from those we describe here. We also binned samples with co-occurring mutations additional to TET2 or DNMT3A mutations to increase our sample set size, although this may introduce a source of variability. Second, we cannot exclude that CH with small clones below our limit of detection are present in our control samples. However, we would expect minimal pathological effect given the marginal size of these clones. Third, a shortcoming of our work is the absence of neutrophils in PBMC samples. Given the myeloid bias of CH mutations, it is likely that neutrophils also harbor mutations, and so their functional consequences within the periphery require investigation.
Overall, our study provides mechanistic support for a genotype-specific precision medicine approach for future CH therapeutics.
Acknowledgments
The authors thank Angela Jones for her assistance with sequencing efforts associated with this study.
A.G.B. is supported by a Burroughs Wellcome Fund Career Award for Medical Scientists and the National Institutes of Health (NIH) Director’s Early Independence Award (DP5-OD029586). P.B.F. is supported by the NIH (K23HL138291) and a Mark Foundation Endeavor Award. P.v.G. is supported by the Ludwig Center at Harvard, the NIH (R00CA218832), Gilead Sciences, the Bertarelli Rare Cancers Fund, the Starr Cancer Consortium, the William Guy Forbeck Research Foundation, and is an awardee of the Glenn Foundation for Medical Research and American Federation for Aging Research Grant for Junior Faculty. M.R.S. is supported by the NIH (1R01CA262287 and 1U01OH012271), a Leukemia and Lymphoma Society Clinical Scholar Award, the Biff Ruttenburg Foundation, the Adventure Alle Fund, the Beverly and George Rawlings Research Directorship, and the Edward P. Evans Foundation.
Authorship
Contribution: J.B.H. and P.B. designed the study, facilitated data collection, conducted formal analysis and interpretation of results, generated figures, prepared the original draft of the manuscript, and edited the manuscript; A.C.P., C.V., and M.T.J. collected data, conducted formal analysis and interpretation of results, generated figures, prepared the original draft of the manuscript, and edited the manuscript; J.U., C.R.P., S.O., and N.H. facilitated sample curation and data collection; B.S. and A.A. provided analysis software; P.v.G. provided resources and analysis software and edited the manuscript; A.J.S. and M.R.S. facilitated sample curation, provided resources and project administration, and edited the manuscript; J.C.V.A. and D.B. facilictated data collection and edited the manuscript; and A.G.B. and P.B.F. conceived and supervised the study, provided funding for the study, provided resources and project administration, conducted formal analysis and interpretation of results, generated figures, prepared the original draft of the manuscript, and edited the manuscript.
Conflict-of-interest disclosure: M.R.S. reports personal fees from AbbVie, Bristol Myers Squibb, CTI Biopharma, Sierra Oncology, and Novartis; grants from Astex and Incyte; personal fees and other support from Karyopharm and Ryvu; personal fees from Sierra Oncology; and grants and personal fees from Takeda and TG Therapeutics, outside the submitted work. P.B.F. reports research funding from Novartis. A.G.B. is a scientific cofounder and has equity in TenSixteen Bio. The remaining authors declare no competing financial interests.
Correspondence: Alexander G. Bick, Vanderbilt University Medical Center, 2200 Pierce Ave, Nashville, TN 37232; email: alexander.bick@vumc.org; and P. Brent Ferrell, Division of Hematology and Oncology, Department of Medicine, Vanderbilt University Medical Center, 2220 Pierce Ave, 777 Preston Research Building, Nashville, TN 37232; email: brent.ferrell@vumc.org.
References
Author notes
J.B.H., P.B., and A.C.P. contributed equally to this study.
All filtered count matrices and differential gene expression tables are available on Open Science Framework at https://osf.io/rac5w/. Seurat objects will be made available through the Chan Zuckerberg Initiative database. All data analysis was completed using R (v4.1.2) on the Terra.bio cloud platform. All R files used to generate the figures and tables are publicly available on GitHub https://github.com/bicklab/Single_Cell_CHIP_Multiomics.
Data are available on request from the corresponding authors, Alexander G. Bick (alexander.bick@vumc.org) and P. Brent Ferrell (brent.ferrell@vumc.org).
The full-text version of this article contains a data supplement.