Key Points
EPO is crucial for maintaining RBC balance, and its dysregulation can cause conditions such as anemia and polycythemia.
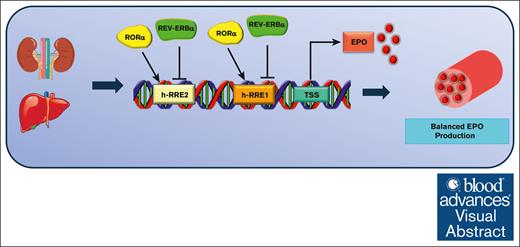
This study reveals a new discovery that 2 NRs, Rev-erbα and RORα, act oppositely, suggesting a potential therapy for EPO-related disorders.
Visual Abstract
The regulation of red blood cell (RBC) homeostasis by erythropoietin (EPO) is critical for O2 transport and maintaining the adequate number of RBCs in vertebrates. Therefore, dysregulation in EPO synthesis results in disease conditions such as polycythemia in the case of excessive EPO production and anemia, which occurs when EPO is inadequately produced. EPO plays a crucial role in treating anemic patients; however, its overproduction can increase blood viscosity, potentially leading to fatal heart failure. Consequently, the identification of druggable transcription factors and their associated ligands capable of regulating EPO offers a promising therapeutic approach to address EPO-related disorders. This study unveils a novel regulatory mechanism involving 2 pivotal nuclear receptors (NRs), Rev-ERBA (Rev-erbα, is a truncation of reverse c-erbAa) and RAR-related orphan receptor A (RORα), in the control of EPO gene expression. Rev-erbα acts as a cell-intrinsic negative regulator, playing a vital role in maintaining erythropoiesis at the correct level. It accomplishes this by directly binding to newly identified response elements within the human and mouse EPO gene promoter, thereby repressing EPO production. These findings are further supported by the discovery that a Rev-erbα agonist (SR9011) effectively suppresses hypoxia-induced EPO expression in mice. In contrast, RORα functions as a positive regulator of EPO gene expression, also binding to the same response elements in the promoter to induce EPO production. Finally, the results of this study revealed that the 2 NRs, Rev-erbα and RORα, influence EPO synthesis in a negative and positive manner, respectively, suggesting that the modulating activity of these 2 NRs could provide a method to target disorders linked with EPO dysregulation.
Introduction
The body maintains optimal oxygenation and blood viscosity by carefully regulating red blood cell (RBC) levels through erythropoiesis. This process, controlled by an oxygen-sensing system, adjusts RBC production to match oxygen demand.1,2 Central to this regulation is erythropoietin (EPO), a hormone crucial for RBC growth and survival.3 Balanced EPO levels are vital to prevent conditions such as erythrocytosis or anemia, ensuring RBC numbers stay within a healthy range. EPO is primarily produced in the adult kidney in response to hypoxia, with minimal production in other organs such as the liver and brain. During fetal development, the liver is the main site of EPO synthesis, shifting to the kidney in adulthood.4-6 The activation of EPO production is a systemic response to hypoxia, resulting in enhanced RBC production and increased blood O2 carrying capacity when pO2 is low.7-9
O2-dependent EPO induction is controlled by the hypoxia-inducible factor (HIF) complex, consisting of HIF1α, HIF2α, and HIF1β. In hypoxic condition, stabilized HIFα forms a complex with HIFβ, binding to EPO gene enhancers and inducing transcription with coactivators such as p300/CREB-binding protein (CBP).9-11 Prolyl hydroxylase domain protein 2 (PHD2) and HIF2α notably control renal and hepatic EPO production in hypoxia, highlighting the pivotal role of PHD2-HIF2α-Von Hippel-Lindau tumor suppressor (pVHL) axis in EPO regulation under low-oxygen conditions.12,13 However, apart from this, many other transcription factors (TFs) such as hepatocyte nuclear factor 4 alpha, Wilms tumor protein 1, and retinoid X receptor alpha are also shown to contribute in the EPO gene regulation.14-18 Clock proteins such as basic helix-loop-helix ARNT like 1 (BMAL1) and cryptochromes 1 and 2 regulate circadian EPO expression, and diurnal EPO variations are observed in humans.19,20 Moreover, the cross talk between the HIFα and BMAL1 is already well-known. The control of EPO gene expression under normoxic conditions, on the contrary, is far less understood.
EPO gene regulation primarily operates at the messenger RNA (mRNA) level, yet its molecular mechanisms are not fully grasped despite its vital role. This regulation is intricate, involving positive and negative regulatory regions in the promoter and 3’ enhancers that finely tune gene expression. Alongside hypoxia response elements (HREs), various DNA regulatory regions were initially discovered in the EPO gene using transgenic mice.21,22 These regulatory elements are believed to be involved in modulating EPO gene transcription, although the TFs or coregulatory proteins that bind in these regions are not well characterized. Mutations in these regions in transgenic mice cause severe polycythemia most likely due to excessive EPO production.23 All these studies suggest the existence of unexplored mechanisms and molecular players in erythropoiesis, which requires further investigation.
Nuclear receptors (NRs) are ligand-regulated TFs controlling gene expression, vital in processes such as cell differentiation and metabolism.24-31 Due to their modulation by small molecules, they are intrinsically druggable TFs and have long been recognized as drug candidates for new therapeutics.32-37 Rev-ERBA (Rev-erbα, is a truncation of reverse c-erbAa) is an adopted orphan receptor from the NR superfamily that plays an important role in molecular clock regulation, cellular differentiation, and metabolism. It is a transcriptional repressor and suppresses gene transcription by recruiting several corepressors.38-40 Heme, a structural component of hemoglobin (HGB), acts as an endogenous ligand for Rev-erbα, playing a vital role in erythropoiesis, regulating erythroid-specific genes and its own synthesis.41-43 However, Rev-erbα has been discovered to be a heme sensor that can also cross talk with other members of the NR superfamily to maintain cellular and metabolic homeostasis. But its role in erythropoiesis remains to be explored. Therefore, the goal of this study is to see whether the Rev-erbα regulates erythropoiesis and how it affects the process.
For the first time, to our knowledge, we describe Rev-erbα as a novel regulator of erythropoiesis and EPO gene transcription. We first analyzed the erythroid phenotypes in Rev-erbα knockout (KO) mice and showed that KO mice exhibit increased numbers of RBCs, HGB content, and hematocrit (HCT) values compared with that of the wild-type (WT) mice. Moreover, the expression level of EPO is also increased in KO mice compared with that of WT mice. The gain of function through ligand (GSK4112 and SR9011) treatment also confirms the transcriptional repression of the EPO gene by Rev-erbα in mice and human hepatic cell lines. Furthermore, chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (EMSA) results uncovered that Rev-erbα and RAR-related orphan receptor A (RORα) bind to the same response element in the EPO promoter and cross talk with each other to regulate EPO gene transcription. Therefore, these findings suggest that ligands (agonists) of Rev-erbα and RORα can regulate EPO production, and this approach could be used as a promising therapeutic strategy for treating pathological conditions, such as polycythemia and anemia, associated with EPO dysregulation.
Methods and materials
Cell culture
HepG2 cells were procured from the National Center for Cell Science, Pune, India and were maintained in Roswell Park Memorial Institute (RPMI) media (Gibco, Invitrogen), supplemented with 1% penicillin-streptomycin (Gibco, Invitrogen) and 10% fetal bovine serum (Gibco, Invitrogen).
Animal protocol and ethics statement
Rev-erbα KO mice (C57BL/6 background) were procured from the Jackson Laboratory. All animals including WT C57BL/6 mice were housed in a healthy and pathogen-free animal facility at the Institute of Microbial Technology (IMTech). For in vivo experiments, 6- to 8-week-old male C57BL/6 mice were used. Animals were intraperitoneally administered with SR9011 (100 mg/kg) for 24 hours between CT6 to CT8 and dimethyloxalyl glycine (DMOG; 300 mg/kg) for 6 hours before the completion of 24 hours for 3 alternate days to generate a polycythemia mice model. For chronobiology experiment of EPO, SR8278 was given subcutaneously at 25 mg/kg of mice body weight at around CT0/CT24, and samples were collected at indicated time points. The kidneys and liver were removed and kept in RNAlater (Sigma-Aldrich) solution until the mRNA was extracted.
Hematological parameters
For complete blood count (CBC) analysis, blood was collected from WT and Rev-erbα KO mice and examined by an auto hematology analyzer (BC-2800VET, Mindray).
ChIP
EMSA
EMSA was performed with 5′-end-labeled oligonucleotides, using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]-adenosine triphosphate (ATP). Recombinant mouse Rev-erbα and RORα proteins were obtained by in vitro transcription and translation of pCMV2-Rev-erbα and pcDNA3-RORα plasmids using the 1-Step Human Coupled IVT Kit (Thermo Scientific) according to the manufacturer’s instructions. DNA binding to recombinant proteins was determined as described previously.40,45,46
Statistical analysis
Unless otherwise stated, all data are presented as mean ± standard deviation. For statistical analysis, Sigma Plot and GraphPad were used. P values were calculated using 2-tailed Student t tests. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 were used to determine statistical significance.
The institutional animal ethics committee at IMTech approved all animal studies, which were carried out in accordance with the rules provided by the Committee for the Purpose of Supervision of Experiments on Animals (number 55/1999/CPCSEA), Ministry of Environment and Forest, Government of India.
Results
Increased EPO production and altered peripheral blood composition in Rev-erbα KO mice
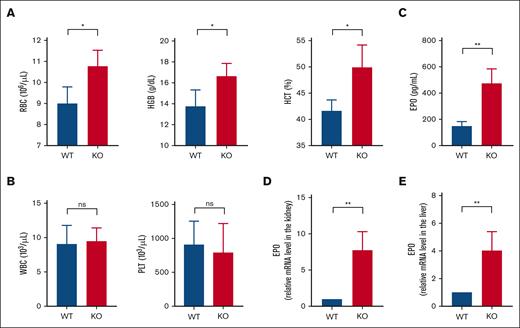
To investigate whether Rev-erbα plays a role in blood cell homeostasis, we first looked at the peripheral blood composition of Rev-erbα KO and WT mice by performing a CBC analysis to characterize the erythropoietic phenotypes. The CBC analysis demonstrated that major erythrocyte indices such as HCT, HGB, and RBC were significantly increased in Rev-erbα KO mice compared with the WT littermate controls, as shown in Figure 1A. However, we did not find any significant change in white blood cell counts or platelets between the WT and Rev-erbα KO littermates (Figure 1B). Moreover, the other blood parameters also remained unaffected (supplemental Figure 1A). As a result, the considerably higher red cell index, that is, increased HCT, HGB content, and RBC counts in Rev-erbα KO mice suggests increased erythropoiesis in mutant genotypes compared with the WT littermate controls. As previously mentioned, erythropoiesis is primarily regulated by the glycoprotein hormone EPO. Therefore, we evaluated the serum EPO level in WT and Rev-erbα KO mice by enzyme-linked immunosorbent assay (ELISA) and observed a significant increase in the EPO level in the sera of Rev-erbα KO mice compared with the WT mice (Figure 1C). We next used real-time quantitative reverse transcription PCR (qRT-PCR) to examine renal and hepatic EPO transcript levels, which revealed that EPO mRNA levels were significantly higher in Rev-erbα KO animals than in WT mice, both in the kidney (Figure 1D) and the liver (Figure 1E). Moreover, the relative mRNA expression level of Rev-erbα is also assessed in the kidney (supplemental Figure 1B) and liver (supplemental Figure 1C) of WT and KO mice. As a result, all these findings corroborate that Rev-erbα KO mice had significantly higher EPO levels than WT littermate controls, indicating that EPO may be a target gene for Rev-erbα.
Rev-erbα KO mice had altered RBC indices and increased EPO levels. A comparison of total blood counts in Rev-erbα KO and WT mice shows the values for RBCs, HGB, and HCT (A) and the number of white blood cells (WBCs) and platelets (PLTs) (B). (C) Serum EPO levels in Rev-erbα KO were compared with WT littermate controls. (D-E) qRT-PCR was used to determine the relative EPO mRNA levels in the kidney (D) and liver (E) of WT and KO mice. Values are mean ± standard deviation (SD; n = 4). ∗P < .05; ∗∗P < .01, as indicated.
Rev-erbα KO mice had altered RBC indices and increased EPO levels. A comparison of total blood counts in Rev-erbα KO and WT mice shows the values for RBCs, HGB, and HCT (A) and the number of white blood cells (WBCs) and platelets (PLTs) (B). (C) Serum EPO levels in Rev-erbα KO were compared with WT littermate controls. (D-E) qRT-PCR was used to determine the relative EPO mRNA levels in the kidney (D) and liver (E) of WT and KO mice. Values are mean ± standard deviation (SD; n = 4). ∗P < .05; ∗∗P < .01, as indicated.
Rev-erbα suppresses EPO production in HepG2 cells
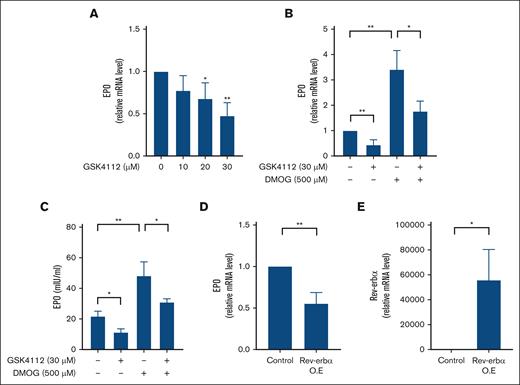
A reliable renal cell culture model capable of producing EPO in a hypoxia-dependent manner was lacking. Therefore, to assess the effect of Rev-erbα on EPO production, we treated the hepatic EPO-producing cell line HepG2 with an increasing concentration of GSK4112 (a specific agonist for Rev-erbα) for 24 hours, and the expression level of EPO was measured with qRT-PCR. As shown in Figure 2A, GSK4112 treatment significantly reduces the mRNA expression level of EPO in a dose-dependent manner. However, maximum inhibition was found at a concentration of 30 μM. Hypoxia is the principal inducer of EPO production, and the human hepatic cells HepG2 have been shown to express hypoxia–induced EPO expression. Therefore, to investigate the effect of Rev-erbα on EPO production under hypoxic conditions in HepG2 cells, we used DMOG, a PHD inhibitor, and a well-known hypoxia mimetic agent. As a result, treatment of DMOG in HepG2 cells leads to the induction of EPO expression in comparison with the solvent control (Figure 2B). GSK4112 treatment, on the contrary, suppressed DMOG-induced mRNA expression of EPO in HepG2 cells, as shown in Figure 2B. The inhibitory effect of Rev-erbα on EPO production was also monitored at the protein level by performing an ELISA with HepG2 cell culture supernatant. As expected, treatment with GSK4112 led to a significant reduction in EPO protein levels in cell culture media in comparison with the solvent control (Figure 2C). Furthermore, as shown in Figure 2C, concomitant treatment with GSK4112 inhibits DMOG-induced EPO production in HepG2 cells. Furthermore, another widely used Rev-erbα ligand, SR10067, demonstrated a similar inhibitory effect on EPO expression (supplemental Figure 2A-B). Finally, the overexpression of Rev-erbα in HepG2 cells also reduces the mRNA expression level of the EPO gene (Figure 2D). Overexpression of Rev-erbα in HepG2 cells was confirmed by qRT-PCR analysis (Figure 2E). In conclusion, all of these results show that Rev-erbα suppresses EPO gene expression under both normoxic and hypoxic conditions.
Rev-erbα inhibits EPO production in vitro. (A) HepG2 cells were treated with an increasing concentration of GSK4112 for 24 hours and expression of EPO is measured with qRT-PCR. (B-C) HepG2 cells were treated with DMOG or GSK4112 for 24 hours, and EPO expression was evaluated by qRT-PCR at the mRNA level (B) and ELISA at the protein level (C) in the cell culture supernatant. (D) qRT-PCR analysis of EPO in HepG2 cells ectopically expressed with Rev-erbα. (E) Overexpression (O.E) of Rev-erbα in HepG2 cells determined with the qRT-PCR analysis. Data are representative and mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01, compared with control or as indicated.
Rev-erbα inhibits EPO production in vitro. (A) HepG2 cells were treated with an increasing concentration of GSK4112 for 24 hours and expression of EPO is measured with qRT-PCR. (B-C) HepG2 cells were treated with DMOG or GSK4112 for 24 hours, and EPO expression was evaluated by qRT-PCR at the mRNA level (B) and ELISA at the protein level (C) in the cell culture supernatant. (D) qRT-PCR analysis of EPO in HepG2 cells ectopically expressed with Rev-erbα. (E) Overexpression (O.E) of Rev-erbα in HepG2 cells determined with the qRT-PCR analysis. Data are representative and mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01, compared with control or as indicated.
Rev-erbα represses EPO production without affecting HIF activity
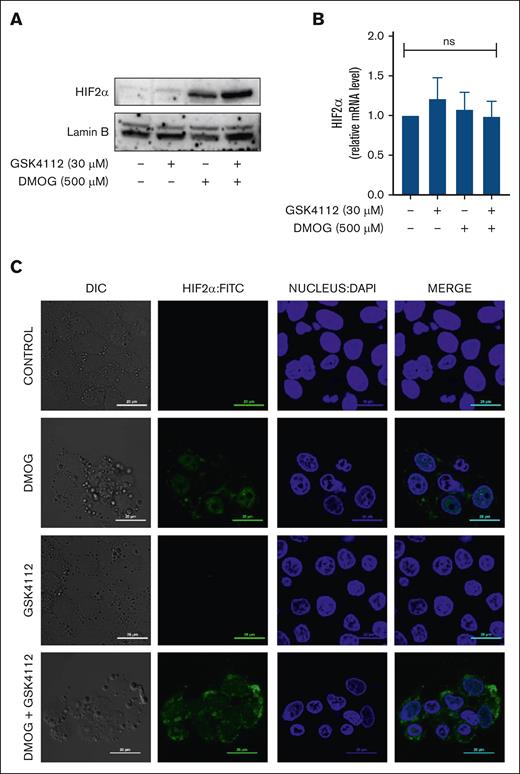
The HIF pathway, which is an O2-sensing mechanism, controls EPO production, and HIF2α has been shown to be a transcriptional regulator of the EPO gene. Under hypoxic conditions, HIF2α translocate into the nucleus to regulate the expression of various hypoxia-responsive genes. Therefore, to see whether Rev-erbα inhibited EPO production by affecting HIF2α activity, we used western blot analysis to detect HIF2α nuclear accumulation. Although Rev-erbα significantly reduced hypoxia–induced EPO mRNA expression in HepG2 (Figure 3), it had no effect on nuclear HIF2α accumulation under normoxic conditions or during the DMOG-induced hypoxic response (Figure 3A). Furthermore, Rev-erbα also had no influence on HIF2α mRNA expression level under either hypoxic or normoxic conditions (Figure 3B). We have checked the HIF2α translocation by confocal microscopy in the presence of DMOG, GSK4112, or combination. Image analysis depicts the HIF2α translocation to nucleus in the presence of DMOG, however, GSK4112 does not elicit any impact on the nuclear accumulation of HIF2α, under normoxic or DMOG-induced hypoxic condition (Figure 3C). As a result, these findings suggest that Rev-erbα specifically impedes EPO expression, whereas the expression or localization of HIF2α remains unaffected.
Effect of Rev-erbα on HIF2α translocation. (A) Immunoblot analysis of HIF2α in the nuclear extract in HepG2 cells. (B) Expression of HIF2α in HepG2 cells as measured by qRT-PCR. (C) Image analysis of HIF2α protein translocation by confocal microscopy as observed in HepG2 cells treated with DMOG or GSK4112 or combination for 24 hours. Data are representative and mean ± SD from 3 independent experiments.
Effect of Rev-erbα on HIF2α translocation. (A) Immunoblot analysis of HIF2α in the nuclear extract in HepG2 cells. (B) Expression of HIF2α in HepG2 cells as measured by qRT-PCR. (C) Image analysis of HIF2α protein translocation by confocal microscopy as observed in HepG2 cells treated with DMOG or GSK4112 or combination for 24 hours. Data are representative and mean ± SD from 3 independent experiments.
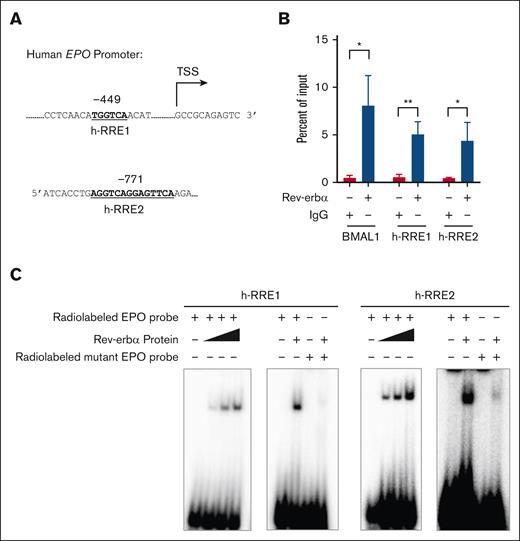
EPO is a direct target gene for Rev-erbα
Rev-erbα is a transcriptional repressor that functions by binding to the conserved motifs known as “Rev-erbα response element” (RRE; (A/G)GGTCA sequence preceded by a 5′ rich A/T). It binds as a monomer to the half-site motif and as a homodimer to the DR2, a direct repeat of RRE separated by 2 base pair (bp).38,47 To determine whether Rev-erbα modulates EPO expression at the genomic level, the promoter sequence of the human EPO gene is analyzed for the Rev-erbα binding sites by using bioinformatics tools, such as nuclear receptor binding site scanner (NUBIScan) and nuclear hormone receptor scanner (NHRscan), or manually. We discovered putative Rev-erbα binding sites in the EPO gene promoter region at 449 bp and 771 bp upstream of the transcription start site (TSS) and named them h-RRE1 and h-RRE2 (Figure 4A). The binding of Rev-erbα on these putative sites was confirmed by ChIP analysis in HepG2 cells. qRT-PCR analysis with primers showed that Rev-erbα interacted with the binding site. BMAL1 (circadian clock regulator), a well-known target gene of Rev-erbα served as a positive control (Figure 4B). Additionally, EMSA was carried out by using radiolabeled oligonucleotide sequences containing the Rev-erbα binding motif (h-RRE1 and h-RRE2) in the EPO promoter region (Figure 4C). To further verify the specific binding of Rev-erbα to binding sites, we generated mutant probes containing mutations in the core motif sequences (RRE1 and RRE2) and found that the binding was completely obliterated in the case of mutant probes (Figure 4C). We have also identified putative Rev-erbα binding sites on mice EPO gene promoter (supplemental Figure 3A). The Rev-erbα binding was observed with the WT probes, whereas it was abolished in case of the mutant probes (supplemental Figure 3B). This suggests that Rev-erbα binding is conserved across the species.
EPO is a direct target gene of Rev-erbα. (A) Pictorial representation of a RREs on the human EPO promoter. (B) ChIP analysis of Rev-erbα binding to the human EPO promoter region using chromatin extracted from HepG2 cells treated with GSK4112 for 24 hours. (C) EMSA with Rev-erbα protein by using WT (containing h-RRE1 and h-RRE2) and mutant radiolabeled probes. Data are representative and mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01, as indicated.
EPO is a direct target gene of Rev-erbα. (A) Pictorial representation of a RREs on the human EPO promoter. (B) ChIP analysis of Rev-erbα binding to the human EPO promoter region using chromatin extracted from HepG2 cells treated with GSK4112 for 24 hours. (C) EMSA with Rev-erbα protein by using WT (containing h-RRE1 and h-RRE2) and mutant radiolabeled probes. Data are representative and mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01, as indicated.
Rev-erbα regulates EPO production in vivo
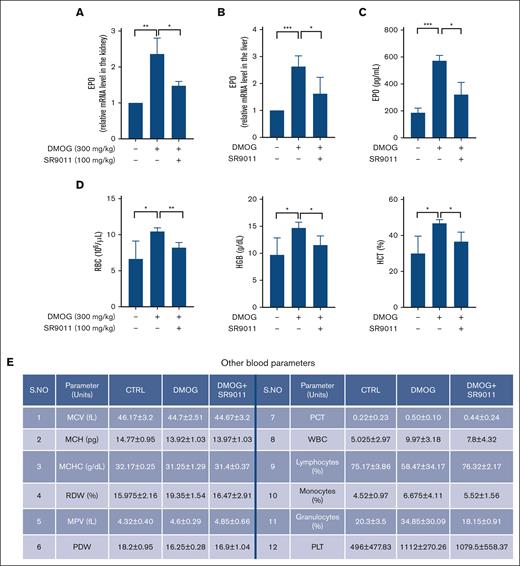
In vitro, GSK4112 has been extensively used to investigate Rev-erbα functions. However, due to its low pharmacokinetics profile and oral bioavailability, it is not suitable for studying Rev-erbα activities in vivo. Therefore, we used another potent Rev-erbα agonist, SR9011, which was synthesized based on the chemical structure of GSK4112 and proved to have better pharmacokinetic properties (suitable for in vivo studies). EPO is majorly produced in kidneys and liver under hypoxic conditions. Therefore, to study the effect of Rev-erbα activation on EPO production under hypoxic and in vivo conditions, mice were treated with DMOG alone or in combination with SR9011 for 3 days, and the expression level of EPO was measured with qRT-PCR analysis by isolating total RNA from the kidney and liver. As shown in Figure 5A-B, the expression of EPO was induced upon DMOG treatment in the kidney as well as in the liver. However, DMOG-induced EPO mRNA levels were significantly reduced upon treatment with SR9011, both in the kidney (Figure 5A) and liver (Figure 5B). Our findings also demonstrated a significant decrease in EPO expression in both the kidney and liver after SR9011 treatment (supplemental Figure 4A-B). We evaluated the serum EPO level by ELISA and observed a significant increase in the EPO level in the sera of DMOG treated mice compared with control. The increased EPO levels were significantly reduced upon SR9011 treatment (Figure 5C).
Rev-erbα regulates EPO production in vivo. C57BL/6 mice treated with DMOG (6 hours) alone or in combination with Rev-erbα agonist SR9011 (24 hours) for 3 alternate days were investigated for relative mRNA expression (A-B) of EPO in the kidney (A) and liver (B), and serum EPO level (C) and a comparison of hematological parameters (n = 4) (D-E). ∗P < .05; ∗∗P < .01; ∗∗∗P < .0001, as indicated. MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume; MPV, mean platelet volume; PCT, plateletcrit; PDW, platelet distribution width; RDW, red blood cell distribution width.
Rev-erbα regulates EPO production in vivo. C57BL/6 mice treated with DMOG (6 hours) alone or in combination with Rev-erbα agonist SR9011 (24 hours) for 3 alternate days were investigated for relative mRNA expression (A-B) of EPO in the kidney (A) and liver (B), and serum EPO level (C) and a comparison of hematological parameters (n = 4) (D-E). ∗P < .05; ∗∗P < .01; ∗∗∗P < .0001, as indicated. MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume; MPV, mean platelet volume; PCT, plateletcrit; PDW, platelet distribution width; RDW, red blood cell distribution width.
To determine the effect of SR9011 in blood cell homeostasis, we compared the hematological parameters of mice treated with DMOG alone or in combination with SR9011 by performing a CBC analysis. The CBC analysis demonstrated that major erythrocyte indices such as HCT, HGB, and RBC were significantly increased in DMOG treated mice compared with the littermate controls. The increased levels of these indices were significantly decreased upon SR9011 treatment (Figure 5D). However, the other blood parameters remained unaffected (Figure 5E). Hence, these findings revealed that Rev-erbα also regulates EPO expression in the polycythemia mice model. Given recent observations regarding EPO's circadian rhythm and Rev-erbα's involvement in circadian biology, we explored whether inhibiting Rev-erbα could disrupt EPO's circadian pattern. To accomplish this, we administered a Rev-erbα antagonist to mice and subsequently analyzed EPO levels in the kidney. Our results revealed the antagonist of Rev-erbα resulted in a significant increase in EPO mRNA levels across the circadian cycle (supplemental Figure 4C).
RORα also regulates EPO gene induction
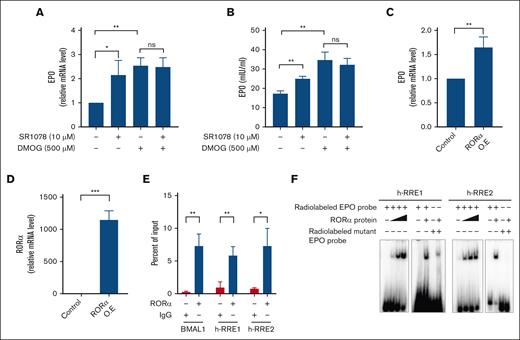
Similar to Rev-erbα, RORα belongs to the NR superfamily and plays a vital role in circadian rhythm and metabolic regulation. It has been reported that RORα and Rev-erbα share the same response element RRE/retinoic orphan receptor response element, even though they regulate gene transcription with opposite effects, which indicates the cross talk between these 2 NR signaling pathways.48,49RORα has been shown to be a target gene for HIF1α and play important role in hypoxia biology.50 Furthermore, RORα has recently been shown to play a role in cell differentiation by inducing EPO gene expression.51 Because both Rev-erbα and RORα recognize the same response elements, we hypothesized that RORα could directly regulate EPO production and induce its transcription. Therefore, to check the effect of RORα on EPO expression level, HepG2 cells were treated with ligand SR1078 (RORα agonist), and the level of EPO was measured at both mRNA and protein levels. As shown in Figure 6A-B, treatment with ligand led to a significant increase in EPO expression level compared with the solvent control. Furthermore, to investigate the impact of RORα on EPO expression under hypoxic conditions, HepG2 cells were treated with DMOG alone or in combination with SR1078. DMOG treatment leads to the significant induction of EPO expression in comparison with the solvent control. Interestingly, standalone SR1078 treatment led to a substantial increase in EPO expression; however, it did not significantly impact the response induced by hypoxia, as observed at both mRNA and protein levels (Figure 6A-B). Moreover, SR1078 did not affect HIF2α nuclear accumulation under normoxic and hypoxic conditions (supplemental Figure 5A). Further immunofluorescence analysis reveals the translocation of HIF2α to the nucleus in case of DMOG treatment, whereas no discernible effect is observed in the presence of the RORα ligand, SR1078, either alone or in combination with DMOG (supplemental Figure 5B).
RORα binds to the EPO promoter to regulate its gene expression. HepG2 cells were treated with DMOG or SR1078 or combination for 24 hours and EPO expression was evaluated for (A) relative mRNA expression by qRT-PCR and (B) the protein levels in the cell culture supernatant by ELISA. (C) qRT-PCR analysis of EPO in HepG2 cells ectopically expressed with RORα. (D) O.E of RORα in HepG2 cells determined with the qRT-PCR analysis. (E) ChIP analysis of RORα binding to the human EPO promoter region using chromatin extracted from HepG2 cells treated with SR1078 for 24 hours. (F) EMSA with RORα protein by using WT and mutant radiolabeled probes. Data are representative and mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, as indicated.
RORα binds to the EPO promoter to regulate its gene expression. HepG2 cells were treated with DMOG or SR1078 or combination for 24 hours and EPO expression was evaluated for (A) relative mRNA expression by qRT-PCR and (B) the protein levels in the cell culture supernatant by ELISA. (C) qRT-PCR analysis of EPO in HepG2 cells ectopically expressed with RORα. (D) O.E of RORα in HepG2 cells determined with the qRT-PCR analysis. (E) ChIP analysis of RORα binding to the human EPO promoter region using chromatin extracted from HepG2 cells treated with SR1078 for 24 hours. (F) EMSA with RORα protein by using WT and mutant radiolabeled probes. Data are representative and mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, as indicated.
In addition, ectopically overexpressing RORα in HepG2 cells also leads to a small but significant increase in EPO gene expression compared with that of the control (Figure 6C). The overexpression of RORα is confirmed with qRT-PCR analysis (Figure 6D). The recruitment of RORα in the promoter region of EPO was confirmed by performing a ChIP experiment with an anti-RORα antibody in HepG2 cells (Figure 6E). BMAL1 is also a target for RORα, therefore used as a positive control for ChIP experiments. To further check the binding, an in vitro DNA binding assay (EMSA) was performed with radiolabeled WT (containing h-RRE1 and h-RRE2) and mutant probes. As shown in Figure 6F, RORα also binds to both RREs in the WT radiolabeled probe, whereas the binding was lost after mutating the RRE half-sites. To check whether the binding is conserved across species, we checked the binding of RORα on mice EPO gene promoter (supplemental Figure 3A). The binding was observed with the WT probes, whereas it was abolished in case of mutant probes on the EPO gene promoter (supplemental Figure 6).
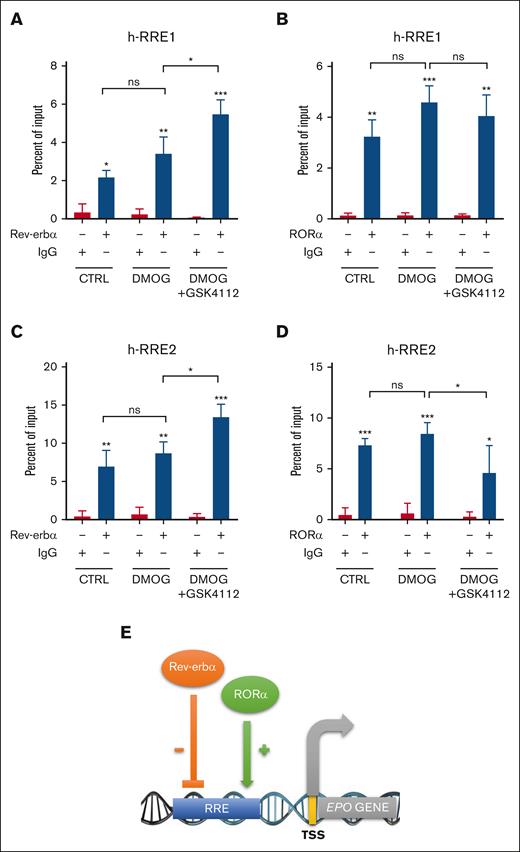
We investigated the cross talk of Rev-erbα and RORα on EPO promoter by ChIP analysis from chromatin extracted from HepG2 cells treated with DMOG alone or in combination with GSK4112. Immunoprecipitation was performed using RORα, Rev-erbα, and immunoglobulin G (IgG) antibody. The qRT-PCR was performed to check the fold enrichment of RORα and Rev-erbα at h-RRE1 and h-RRE2 sites in EPO gene promoter. We found that RORα and Rev-erbα interacts with the EPO gene promoter, compared with IgG (Figure 7A-D). Specifically, at the h-RRE1 site, Rev-erbα protein enrichment on the EPO promoter significantly increased with DMOG and GSK4112 treatment, whereas RORα enrichment did not notably change (Figure 7A-B). Conversely, at the h-RRE2, Rev-erbα binding increased with ligand treatment, whereas RORα binding decreased significantly in the presence of DMOG and GSK4112 (Figure 7C-D). The isotype IgG, Rev-erbα, and RORα antibodies did not pull down any nonspecific DNA region, which further confirm the specificity of the assay (supplemental Figure 7). These findings suggest the competitive binding of Rev-erbα to the promoter region of EPO in the presence of its ligand keeping the EPO production in the balanced state.
Rev-erbα cross talk with RORα to regulates EPO gene expression. ChIP analysis was performed using chromatin isolated from HepG2 cells treated with DMOG (500 μM) and GSK4112 (30 μM) for 24 hours. Immuno-precipitation was performed with Rev-erbα and RORα antibodies at h-RRE1 (A-B) and h-RRE2 (C-D). (E) Schematic representation of the proposed mechanism underlying the modulation of EPO gene expression by Rev-erbα and RORα. Data are representative or mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, compared with control or as indicated. CTRL, control.
Rev-erbα cross talk with RORα to regulates EPO gene expression. ChIP analysis was performed using chromatin isolated from HepG2 cells treated with DMOG (500 μM) and GSK4112 (30 μM) for 24 hours. Immuno-precipitation was performed with Rev-erbα and RORα antibodies at h-RRE1 (A-B) and h-RRE2 (C-D). (E) Schematic representation of the proposed mechanism underlying the modulation of EPO gene expression by Rev-erbα and RORα. Data are representative or mean ± SD from 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, compared with control or as indicated. CTRL, control.
Taken together, all these results show that EPO production is also regulated by RORα at the transcriptional level and that Rev-erbα cross talk with RORα by binding to the same response element in the EPO gene promoter (Figure 7E).
Discussion
The regulation of RBC homeostasis by EPO is critical for O2 transport in vertebrates, and dysregulation in EPO synthesis results in anemia or polycythemia.1,2 The PHD-HIF-pVHL pathway induces EPO production by activating HIF complex to bind to the EPO gene's HREs in response to anemia or hypoxia.9-11,52 Additionally, EPO gene regulation involves other TFs such as Wilms tumor protein 1, GATA binding protein (GATA), and hepatocyte nuclear factor 4 alpha.14-18,53,54 Moreover, the EPO gene promoter and 3′ enhancer region contains multiple uncharacterized DNA regulatory regions, and 2 additional HREs have recently been hypothesized to be present in the EPO 5′ promoter. The discovery of different positive and negative regulatory regions in the EPO gene revealed that its regulation is mediated through a complex interplay between HREs and other DNA regulatory elements found in the EPO gene promoter and 3' enhancer regions.21,22 This study reveals a novel mechanism for EPO gene regulation by Rev-erbα and RORα, as evidenced by dysregulated erythropoiesis and altered RBC indices in Rev-erbα KO mice, whereas white blood cell and platelet counts remain unaffected (Figure 1).
The in vivo relevance of the Rev-erbα modulating EPO gene was confirmed by the increased production of EPO in the serum and enhanced mRNA level in the kidney and liver of Rev-erb KO mice compared with the WT littermates. Studies with HepG2 cells revealed Rev-erbα's function as a transcriptional repressor, inhibiting EPO gene expression under normoxic and hypoxic conditions (Figure 2). Importantly Rev-erbα did not affect nuclear translocation or mRNA level of HIF2α in HepG2 cells, which strongly indicated that Rev-erbα is associated with EPO gene suppression in a HIF-independent manner and directly regulates the EPO gene transcription (Figure 3). The 2 DNA response elements (RREs) located at 449 bp and 771 bp upstream of TSS were found in the EPO gene promoter. Furthermore, ChIP and EMSA results discovered that Rev-erbα binds to these newly identified response elements in the promoter region to regulate EPO gene transcription (Figure 4). These findings further validated the presence of DNA regulatory elements in the human EPO promoter regions that had been previously suggested.21,22,51 In addition to this, these in vitro results were supported by in vivo observations that Rev-erbα agonist (SR9011) treatment inhibited DMOG-induced renal and hepatic EPO mRNA expression levels in polycythemia mice model (Figure 5). These results indicated that the Rev-erbα is a cell-intrinsic negative regulator of erythropoiesis, and its activity is important for maintaining EPO production. The antagonist of Rev-erbα resulted in a significant increase in EPO mRNA levels across the circadian cycle, thus reaffirming the inhibitory role of Rev-erbα in regulating the EPO promoter (supplemental Figure 4C). These findings underscore the intricate interplay between Rev-erbα, circadian rhythms, and EPO expression, offering valuable insights into the underlying mechanisms of blood cell homeostasis. The cross talk between the Rev-erbα and RORα is well documented in the control of metabolic processes and circadian rhythm,48,49 although it has not been investigated in erythropoiesis and EPO gene regulation.
RORα agonist (SR1078) and overexpression stimulated EPO production in HepG2 cells. This is consistent with the recently reported role of RORα in EPO regulation.51 Further ChIP and EMSA analyses demonstrated the binding of RORα to the same response element on the EPO promoter (Figure 6). In contrast to Rev-erbα, which acts as a transcriptional repressor of the EPO gene, RORα functions as an activator. Collectively, these findings indicated that EPO gene expression is under negative and positive regulation by Rev-erbα and RORα, respectively, and thus, the balanced expression and activation of these 2 NRs is important for the dynamic regulation of EPO production (Figure 7).
The study of EPO regulation is necessary not just from a physiological standpoint but also because of the clinical consequences. Increased levels of EPO cause polycythemia or erythrocytosis, whereas less EPO production is linked with anemia or anemia associated with kidney complications such as anemia of chronic kidney disease.7,8 Currently, the medication used to treat patients with chronic kidney disease depends on the use of recombinant human erythropoietin, which is, however, costly and needs lifelong injections.55,56 As a result, PHD inhibitors are being tested in clinical trials as a possible alternative to recombinant human erythropoietin because they are less expensive and may be taken orally.57,58 Further research into EPO's negative effects and mechanisms is crucial, balancing its life-saving potential with risks such as increased blood viscosity and heart failure.7,8 In this scenario, identifying TFs in EPO regulation can inspire new, cost-effective treatments with NRs as promising targets. Therefore, modulating Rev-erbα and RORα could offer innovative ways to manage EPO levels in conditions such as anemia or polycythemia.
Acknowledgments
The authors thank Institute of Microbial Technology (IMTech; a constituent laboratory of the Council of Scientific and Industrial Research) for its facilities and financial support.
This work was supported by the Department of Biotechnology-India project GAP0162, the Council of Scientific and Industrial Research (CSIR), RC project OLP704, and Major Lab Project MLP039 (P.G.).
The funders had no role in study design, data collection, interpretation, or the decision to submit the work for publication.
Authorship
Contribution: S. Kumar and P.G. designed the experiments, analyzed the data, and wrote the manuscript; S. Kumar, R.A., S.G., N.A., E.B., R.N., R.K., A.K.K., S. Kumawat, V.K., and M.S. performed the experiments; and P.G. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S. Kumar is Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH.
The current affiliation for N.A. is Laboratory of Molecular Neurodegeneration and Gene Therapy, University of Oxford, Oxford, United Kingdom.
The current affiliation for E.B. is Champions Oncology Inc, Rockville, MD.
The current affiliation for R.N. is Center for Cancer Research, National Cancer Institute, Bethesda, MD.
The current affiliation for R.K. is Division of Hematology and Oncology, Heersink School of Medicine, The University of Alabama at Birmingham, Birmingham, AL.
Correspondence: Pawan Gupta, Department of Molecular Biology, CSIR-Institute of Microbial Technology, Sector 39A, Chandigarh 160036, India; email: pawan@imtech.res.in.
References
Author notes
The data that support the findings of this study will be made available to researchers. The data are also submitted to Council of Scientific and Industrial Research (CSIR)- Institute of Microbial Technology (IMTech) institutional repository (CSIR-IMTech communication number, 039/2022).
Materials are available on request from the corresponding author, Pawan Gupta (pawan@imtech.res.in).
The full-text version of this article contains a data supplement.