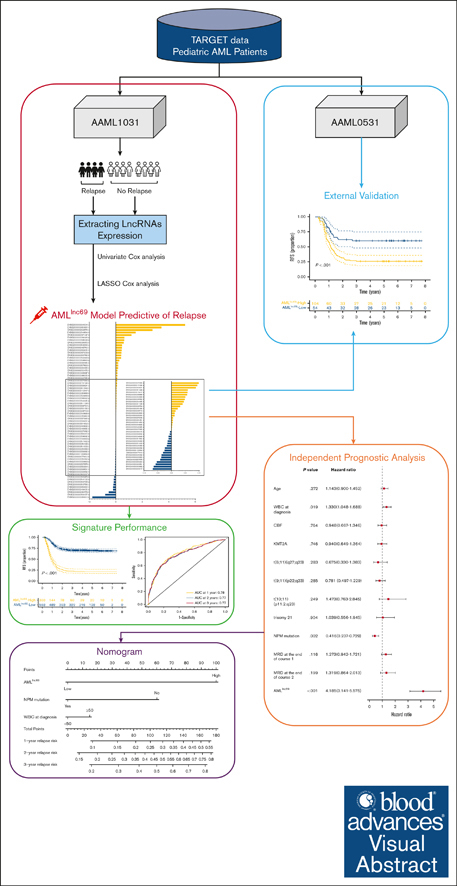
Key Points
A comprehensive prognostic model of 69 lncRNAs was generated to predict RFS in pedAML and further refine the current risk stratification.
A nomogram incorporating the 69 lncRNA signature, nucleophosmin mutation, and white blood cell count was developed to refine stratification.
Visual Abstract
Risk stratification using genetics and minimal residual disease has allowed for an increase in the cure rates of pediatric acute myeloid leukemia (pedAML) to up to 70% in contemporary protocols. Nevertheless, ∼30% of patients still experience relapse, indicating a need to optimize stratification strategies. Recently, long noncoding RNA (lncRNA) expression has been shown to hold prognostic power in multiple cancer types. Here, we aimed at refining relapse prediction in pedAML using lncRNA expression. We built a relapse–related lncRNA prognostic signature, named AMLlnc69, using 871 transcriptomes of patients with pedAML obtained from the Therapeutically Applicable Research to Generate Effective Treatments repository. We identified a 69 lncRNA signature AMLlnc69 that is highly predictive of relapse risk (c-index = 0.73), with area under the receiver operating characteristic curve (AUC) values for predicting the 1-, 2-, and 3-year relapse-free survival (RFS) of 0.78, 0.77, and 0.77, respectively. The internal validation using a bootstrap method (resampling times = 1000) resulted in a c-index of 0.72 and AUC values for predicting the 1-, 2-, and 3-year RFS of 0.77, 0.76, and 0.76, respectively. Through a Cox regression analysis, AMLlnc69, nucleophosmin mutation, and white blood cell at diagnosis were identified as independent predictors of RFS. Finally, a nomogram was build using these 2 parameters, showing a c-index of 0.80 and 0.71 after bootstrapping (n = 1000). In conclusion, the identified AMLlnc69 will, after prospective validation, add important information to guide the management of patients with pedAML. The nomogram is a promising tool for easy stratification of patients into a novel scheme of relapse-risk groups.
Introduction
The 5-year overall survival (OS) rate for pediatric acute myeloid leukemia (pedAML) is now up to 70% with the application of the most contemporary protocols.1 This improvement has been achieved through treatment intensification, the optimization of transplant procedures and supportive care, and the introduction of risk-adapted treatment strategies.2 Risk stratification for pedAML depends mainly on the presence or absence of cytogenetic and molecular abnormalities known to be associated with the achievement of complete remission, OS, and relapse-free survival (RFS).3-5 The achievement of minimal residual disease (MRD) during treatment has been shown to be another important risk stratification indicator enabling treatment adaptation.6-8 Nevertheless, pedAML remains a therapeutic challenge, with high relapse rates (∼30%) despite intensive therapy.9 Relapse is a major cause of pedAML treatment failure and an indicator of poor prognosis.10 Recent coding gene expression analyses revealed the ability of the leukemic stem cell 6 (LSC6) and LSC17 stemness signatures, reflecting the expression of 6 and 17 coding messenger RNAs, respectively, to predict the event-free survival (EFS) and OS of patients with pedAML.11,12 Similarly, the LSC47 signature was developed to predict EFS in the context of existing cytogenetic and molecular risk stratification.13 Recently, the lncScore, a long noncoding RNA (lncRNA)–based predictor of the EFS and OS of patients with pedAML, was developed.14 However, none of these signatures predicts RFS or has been implemented in a clinical setting. Thus, the further refinement of relapse–based risk stratification for pedAML remains an urgent need.
LncRNAs are transcripts longer than 200 nucleotides that are not translated into proteins and have highly tissue-specific expression.15 They have been shown to play important roles in normal development and the development of diseases,16,17 including pedAML.18 The aberrant expression of key lncRNAs involved in hematopoietic stem cell maintenance and differentiation has been shown to result in the development of hematological malignancies.19,20 LncRNA expression also has prognostic power for malignancies such as adult AML,21 breast cancer,22 and neuroblastoma.23 Hence, the inclusion of lncRNA expression in pedAML risk stratification could add value for RFS prediction.
In this study, we aimed to identify an lncRNA signature that predicts pedAML RFS using publicly available RNA sequencing (RNA-seq) data from patients with pedAML from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) repository, including cases from Children’s Oncology Group studies AAML053124 and AAML1031.25,26
Methods
TARGET data acquisition
RNA-seq data and corresponding clinical information on patients with pedAML were retrieved from the TARGET repository (https://ocg.cancer.gov/programs/target) and downloaded from the National Cancer Institute Genomic Data Commons (https://portal.gdc.cancer.gov). We included pedAML cases from Children’s Oncology Group studies AAML053124 and AAML1031.25,26 During enrollment in those clinical trials, which were conducted in accordance with the Declaration of Helsinki, participants provided written informed consent to the use of their data for correlative biological studies. We excluded cases evaluated by low-depth sequencing and sequencing data obtained at nondiagnostic time points, such as after treatment or relapse. Because we focused on relapse, only patients whose first event was relapse and those who were censored were included. Patients from the AAML1031 study were allocated to the discovery cohort (n = 871), and those from the AAML0531 study were allocated to the validation cohort (n = 158; supplemental Figure 1). Detailed information on each of the included patients is provided in supplemental Table 1, and the value labels used in supplemental Table 1 are provided in supplemental Table 2. Gene annotation was performed using Homo_sapiens.GRCh38.110 from the Ensembl project (http://www.ensembl.org).27
LncRNA prognostic signature construction
Univariate Cox regression analysis was used to identify lncRNAs associated significantly with RFS (P < .05). Then, least absolute shrinkage and selection operator Cox regression was used to further filter for lncRNAs associated strongly with RFS and to estimate their coefficients for linear predictor establishment. During this process, the optimal parameter “λ” was obtained through crossvalidation to maintain equilibrium between model deviation and variance. The caret, tidyverse, tibble, data.table, survival, survminer, glmnet, pbapply, and magrittr R packages were used for these analyses. Next, an outcome-oriented method was applied using the surv_cutpoint function of the survminer R package to determine an optimal cutoff value that maximized survival differences between low- and high-risk lncRNA signature groups. Finally, the risk score distribution was plotted and RFS status mapping was performed to further analyze the influence of the selected signature on RFS.
Outcome analysis and validation
Using the survival and survminer R packages and the Kaplan-Meier (KM) method, RFS and OS probabilities were estimated. Standard errors were calculated using the Greenwood formula, and the data were compared using the log-rank test. Receiver operating characteristic (ROC) curves were drawn, and the area under the receiver operating characteristic curve (AUCs) were calculated using the survminer and timeROC R package to assess model performance at 1, 2, and 3 years. Concordance indices (c-indices) were calculated for signature assessment using the concordance function.28 The bootstrap method, based on 1000-fold resampling, was used for internal validation.29,30 Univariate and multivariate Cox regression analyses were performed to obtain independent prognostic values for the lncRNA signature, and c-indices of these values were calculated. Finally, a nomogram was generated using the regplot, survival, rms, ggDCA, and timeROC R packages.31 The coefficients of lncRNAs in the model generated from discovery cohort data were used for external validation.
Comparison of clinical characteristics
Fisher exact test and the χ2 test were used to assess distributional differences in categorical data between the low- and high-risk groups. A heat map of these differences was generated using the ComplexHeatmap R package. The normality of data distributions was assessed using the D'Agostino-Pearson, Anderson-Darling, Shapiro-Wilk, and Kolmogorov-Smirnov tests. Nonnormally distributed quantitative data were evaluated using the Mann-Whitney U test. Box plots of quantitative results were generated using GraphPad Prism 9.
Functional and biological pathway enrichment analyses
Gene set enrichment analysis (GSEA) was performed using the limma, org.Hs.eg.db, clusterProfiler, and enrichplot R packages (P < .05).32 The hallmark, gene ontology, and Kyoto Encyclopedia of Genes and Genomes gene sets were downloaded from Molecular Signatures Database (http://www.broadinstitute.org/msigdb)33 and used for this purpose.
Results
Prognostic lncRNA signature establishment
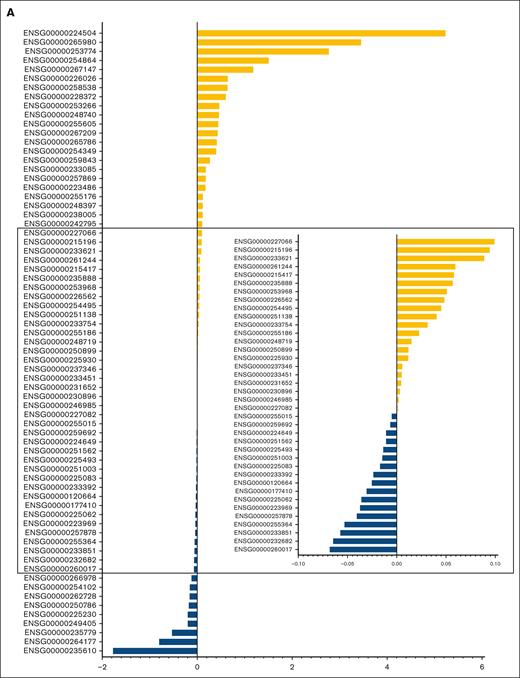
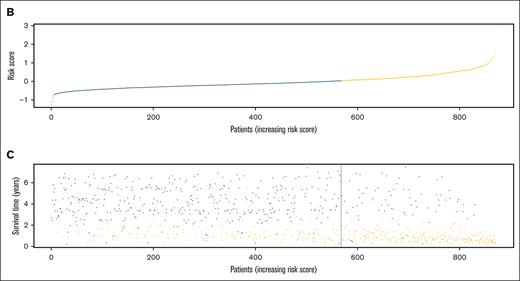
The characteristics of patients in the discovery and validation cohorts are summarized in supplemental Table 3, and RFS in the 2 cohorts is illustrated in supplemental Figure 2. Univariate Cox regression analysis of discovery cohort data yielded 1751 relapse-related lncRNAs (supplemental Table 4). To improve the prediction accuracy and avoid overfitting, a least absolute shrinkage and selection operator Cox regression analysis was performed to construct a linear prognostic model (AMLlnc69), in which 69 lncRNAs were retained (Figure 1A; supplemental Figure 3; supplemental Table 5). AMLlnc69 risk group information, survival data, and expression levels of the lncRNAs used in model construction are provided for all patients in the discovery cohort in supplemental Table 6. An optimal cutoff value was calculated using the surv_cutpoint function in the survminer R package and an outcome-oriented method that enabled patient assignment to low- and high-risk AMLlnc69 groups. The risk score distribution is shown in Figure 1B. The RFS status map for all patients with pedAML shows that the proportion of patients with relapse increases with the AMLlnc69 score (Figure 1C).
Identification of the 69 relapse–related lncRNAs prognostic signature for patients with pedAML. (A) Coefficients of the 69 lncRNAs originated from the LASSO Cox regression. (B) Risk score distribution and (C) RFS status map of patients with pedAML. LASSO, least absolute shrinkage and selection operator.
Identification of the 69 relapse–related lncRNAs prognostic signature for patients with pedAML. (A) Coefficients of the 69 lncRNAs originated from the LASSO Cox regression. (B) Risk score distribution and (C) RFS status map of patients with pedAML. LASSO, least absolute shrinkage and selection operator.
Signature validation
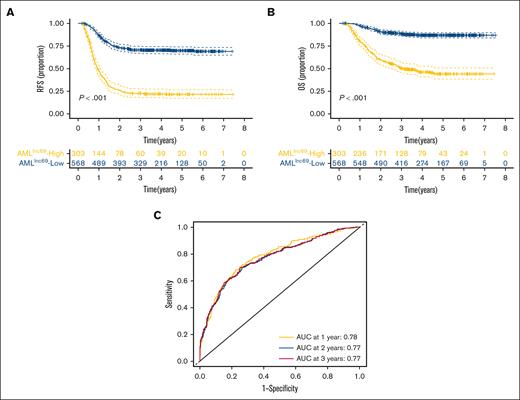
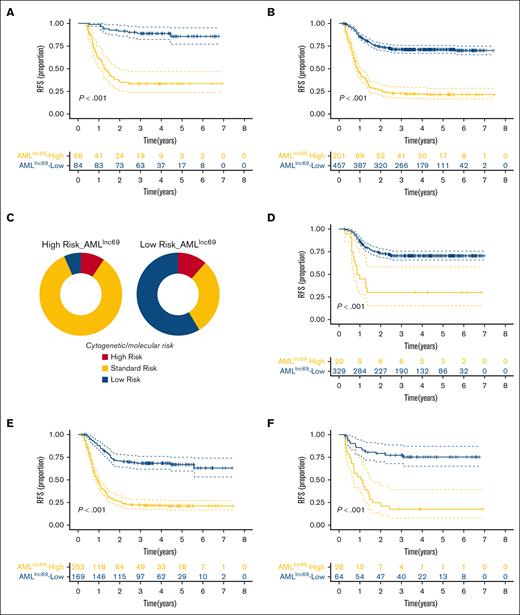
KM survival curves further illustrated that the RFS rate was significantly lower for patients with high-risk AMLlnc69 than those with low-risk AMLlnc69 in the discovery cohort (Figure 2A). In addition, the AMLlnc69 signature was significantly predictive of OS (Figure 2B). To further substantiate the predictive value of the model, we performed ROC analyses with the calculation of AUCs for the prediction of 1-, 2-, and 3-year RFS; these values were 0.78, 0.77, and 0.77, respectively, with a corresponding c-index of 0.73 (Figure 2C). Internal validation yielded a c-index of 0.72 and AUCs for the prediction of 1-, 2-, and 3-year RFS of 0.77, 0.76, and 0.76, respectively (supplemental Table 7). As some patients underwent hematopoietic stem cell transplantation (HSCT) during their first complete remission periods, which might confound the survival analysis, separate KM RFS curves were generated for patients who did and did not undergo this procedure. These curves illustrated that the model remains valid irrespective of HSCT performance (Figure 3A-B).
Validation of AMLlnc69. (A-B) KM survival curve of RFS (A) and OS (B) of patients with pedAML in the low- and high-risk groups. (C) ROC curves and AUCs for 1-, 2-, and 3-year RFS.
Validation of AMLlnc69. (A-B) KM survival curve of RFS (A) and OS (B) of patients with pedAML in the low- and high-risk groups. (C) ROC curves and AUCs for 1-, 2-, and 3-year RFS.
The performance of AMLlnc69 in terms of undergoing HSCT in first CR and cytogenetic/molecular risk. (A-B) KM survival curves of RFS for the of patients with pedAML receiving HSCT (A) and without receiving HSCT in the first CR (B). (C) The distribution of cytogenetic/molecular risk between low- and high-risk groups based on AMLlnc69. (D-F) KM survival curve of RFS of patients with pedAML based on AMLlnc69 in low-, standard-, and high-risk groups categorized by cytogenetic/molecular risk stratification. CR, complete remission.
The performance of AMLlnc69 in terms of undergoing HSCT in first CR and cytogenetic/molecular risk. (A-B) KM survival curves of RFS for the of patients with pedAML receiving HSCT (A) and without receiving HSCT in the first CR (B). (C) The distribution of cytogenetic/molecular risk between low- and high-risk groups based on AMLlnc69. (D-F) KM survival curve of RFS of patients with pedAML based on AMLlnc69 in low-, standard-, and high-risk groups categorized by cytogenetic/molecular risk stratification. CR, complete remission.
The potential of the LSC6,11 LSC17,12 and LSC4713 messenger RNA signatures and the lncScore14 for pedAML risk stratification has been demonstrated. Because these signatures were generated for the prediction of OS and EFS, we tested their predictive value for RFS in the discovery cohort. Our analyses confirmed this value. KM plots showed that all 3 models successfully stratified patients based on RFS. However, acceptable AUCs (∼0.65) for 1-, 2-, and 3-year RFS prediction were obtained only for the LSC47 signature and lncScore, and these values were significantly lower than those obtained for the AMLlnc69 (supplemental Figure 4).
Next, AMLlnc69 was validated with an independent validation cohort (n = 158 patients with pedAML; supplemental Figure 1). Patients in the validation cohort were categorized into low- and high-risk groups based on the AMLlnc69, yielding AUCs of 0.66, 0.68, and 0.70 for 1-, 2-, and 3-year RFS prediction, respectively (supplemental Figure 5A-B). Because the benefit of adding gemtuzumab ozogamicin was a randomized question in this study cohort, we evaluated whether AMLlnc69 was predictive for RFS on each of the randomization arms. Interestingly, KM survival analysis illustrated that AMLlnc69 remained predictive, irrespective of gemtuzumab ozogamicin randomization arm (supplemental Figure 5C-D).
Subsequently, we plotted the distribution of cytogenetic/molecular risk for the AMLlnc69 low- and high-risk groups in the discovery cohort. Importantly, a significant proportion of conventionally stratified standard-risk patients could be reclassified as high risk according to the AMLlnc69 (Figure 3C). Furthermore, KM plots showed that the AMLlnc69 could be used to reclassify patients in all (low, standard, and high) traditional risk groups based on cytogenetic/molecular characteristics (Figure 3D-F; supplemental Figure 6). The frequency distributions of patient characteristics according to HSCT administration and AMLlnc69 risk group are shown in supplemental Table 8.
Altogether, these results illustrated that AMLlnc69 use could improve traditional cytogenetic/molecular risk stratification, better defining low-risk and high-risk patients in terms of RFS.
AMLlnc69 is an independent prognostic factor
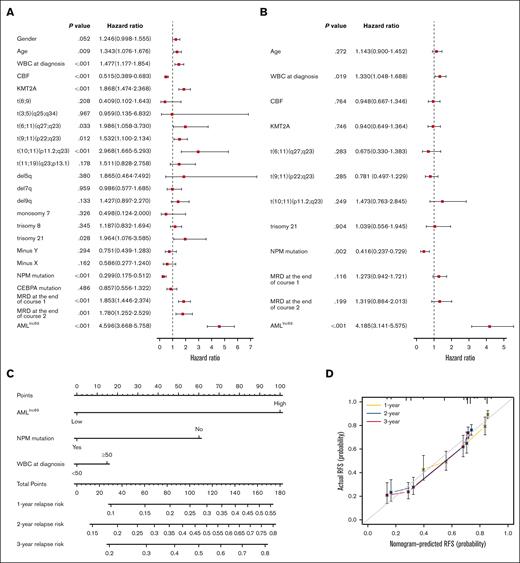
To evaluate whether the AMLlnc69 is an independent prognostic factor, we first performed Cox regression analysis and calculated c-indices for the discovery cohort. Due to the requirement for complete data, 704 patients were included in this analysis. Univariate Cox regression analysis indicated that age, white blood cell (WBC) count at the time of diagnosis, presence of core binding factor, KMT2A, t(6;11)(q27;q23), t(9;11)(p22;q23), t(10;11)(p11.2;q23), trisomy 21, nucleophosmin (NPM) mutation, MRD at the end of induction course 1, MRD at end of induction course 2, and the AMLlnc69 signature were prognostic factors (Figure 4A; supplemental Table 9). In this analysis, AMLlnc69 performed well in distinguishing low- and high-risk individuals in all subgroups (defined according to genetics and MRD; supplemental Figure 7), although some of these subgroups were small (n < 5), and these results should be interpreted with caution. Next, significant variables were examined further in a multivariate Cox analysis, which showed that the AMLlnc69 signature, NPM mutation, and WBC count at the time of diagnosis were independent prognostic factors (Figure 4B). C-indices indicated that AMLlnc69 had better RFS-predictive value than NPM mutation and WBC count at the time of diagnosis (supplemental Figure 8). Furthermore, AMLlnc69 was an independent prognostic factor in the validation cohort, as demonstrated by univariate (hazard ratio, 3.89; 95% confidence interval, 2.04-7.44; P < .001) and multivariate (hazard ratio, 3.56; 95% confidence interval, 1.85-6.84; P < .001) Cox regression analyses (supplemental Table 10).
Independent prognostic analysis of AMLlnc69. (A-B) Forest plots of univariate (A) and multivariate (B) independent Cox regression analyses of AMLlnc46 and other characters. (C) Nomogram model of the combined AMLlnc46, NPM mutation, and WBC at diagnosis for 1-, 2-, and 3-year relapse risk in patients with pedAML. (D) Calibration plot comparing nomogram predicted and actual RFS at 1, 2, and 3 years. CBF, core binding factor.
Independent prognostic analysis of AMLlnc69. (A-B) Forest plots of univariate (A) and multivariate (B) independent Cox regression analyses of AMLlnc46 and other characters. (C) Nomogram model of the combined AMLlnc46, NPM mutation, and WBC at diagnosis for 1-, 2-, and 3-year relapse risk in patients with pedAML. (D) Calibration plot comparing nomogram predicted and actual RFS at 1, 2, and 3 years. CBF, core binding factor.
Based on the multivariate Cox regression analysis results, a nomogram was established using the AMLlnc69, NPM mutation, and WBC count at the time of diagnosis (Figure 4C). Decision curve analysis indicated that the RFS-predictive effect of the 3 factors combined was superior to those of the individual factors (supplemental Figure 9A). AUCs for the prediction of 1-, 2-, and 3-year RFS were 0.76, 0.75, and 0.75, respectively, and the c-index was 0.80 (supplemental Figure 9B). Internal validation yielded a c-index of 0.71 and AUCs for the 1-, 2-, and 3-year RFS prediction of 0.76, 0.75, and 0.75, respectively. Calibration curves showed good consistency of these 3 nomogram predictions with actual observations (Figure 4D). Table 1 provides an overview of all combinations possible and 1-, 2, and 3-year cumulative pedAML relapse-risk prediction according to the nomogram. As an example, a patient with pedAML in the AMLlnc69 high-risk group (100 points) with no NPM mutation (60 points) and a WBC count <50 at the time of diagnosis (0 point) would have a 76.8% cumulative risk of relapse at 3 years.
Examples of 1-, 2-, and 3-year relapse-risk prediction for patients with pedAML using the nomogram prediction model
| AMLlnc69 . | NPM mutation . | WBC at diagnosis . | 1-y risk, % (95% CI) . | 2-y risk, % (95% CI) . | 3-y risk, % (95% CI) . |
|---|---|---|---|---|---|
| High | No | ≥50 | 56.8 (48.3-63.8) | 80.5 (72.9-86.0) | 83.6 (76.4-88.7) |
| High | Yes | ≥50 | 29.8 (14.2-42.6) | 49.9 (26.1-66.0) | 53.4 (28.5-69.7) |
| High | No | <50 | 49.2 (42.1-55.5) | 73.3 (66.1-79.0) | 76.8 (69.8-82.2) |
| High | Yes | <50 | 24.9 (11.7-36.2) | 42.8 (21.7-58.2) | 46.1 (23.8-61.9) |
| Low | No | ≥50 | 18.1 (13.8-22.3) | 32.3 (25.6-38.4) | 35.1 (28.0-41.4) |
| Low | Yes | ≥50 | 8.1 (3.7-12.4) | 15.2 (7.2-22.5) | 16.7 (7.9-24.6) |
| Low | No | <50 | 14.9 (11.9-17.9) | 27.1 (22.5-31.4) | 29.5 (24.7-34.0) |
| Low | Yes | <50 | 6.6 (3.0-10.0) | 12.5 (6.0-18.5) | 13.7 (6.6-20.3) |
| AMLlnc69 . | NPM mutation . | WBC at diagnosis . | 1-y risk, % (95% CI) . | 2-y risk, % (95% CI) . | 3-y risk, % (95% CI) . |
|---|---|---|---|---|---|
| High | No | ≥50 | 56.8 (48.3-63.8) | 80.5 (72.9-86.0) | 83.6 (76.4-88.7) |
| High | Yes | ≥50 | 29.8 (14.2-42.6) | 49.9 (26.1-66.0) | 53.4 (28.5-69.7) |
| High | No | <50 | 49.2 (42.1-55.5) | 73.3 (66.1-79.0) | 76.8 (69.8-82.2) |
| High | Yes | <50 | 24.9 (11.7-36.2) | 42.8 (21.7-58.2) | 46.1 (23.8-61.9) |
| Low | No | ≥50 | 18.1 (13.8-22.3) | 32.3 (25.6-38.4) | 35.1 (28.0-41.4) |
| Low | Yes | ≥50 | 8.1 (3.7-12.4) | 15.2 (7.2-22.5) | 16.7 (7.9-24.6) |
| Low | No | <50 | 14.9 (11.9-17.9) | 27.1 (22.5-31.4) | 29.5 (24.7-34.0) |
| Low | Yes | <50 | 6.6 (3.0-10.0) | 12.5 (6.0-18.5) | 13.7 (6.6-20.3) |
CI, confidence interval.
Comparison of characteristics and biological pathway enrichment
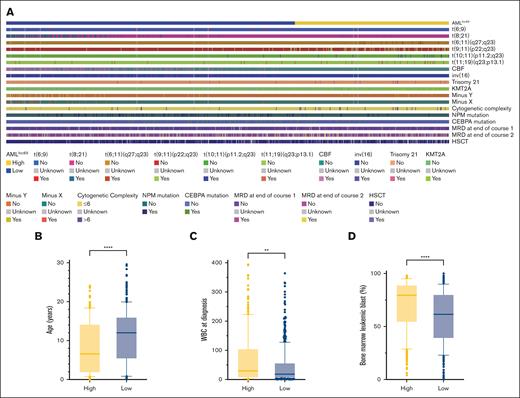
Differences in clinical characteristics between the low- and high-risk AMLlnc69 groups were examined (supplemental Table 11). Relative to the AMLlnc69 low-risk group, the AMLlnc69 high-risk group contained significantly more cases with t(6;11)(q27;q23), t(9;11)(p22;q23), t(10;11)(p11.2;q23), t(11;19)(q23;p13.1), trisomy 21, KMT2A, MRD at the end of courses 1 and 2, and HSCT, as well as greater cytogenetic complexity, a younger age at diagnosis, a higher WBC count at the time of diagnosis, and a higher percentage of leukemic bone marrow blasts (Figure 5).
Comparision of significant characters between AMLlnc69 low- and high-risk groups. (A) Heat map comparing the distribution of significant characters. (B-D) Comparison of age (B), WBC at diagnosis (C), and bone marrow leukemic blast (D).
Comparision of significant characters between AMLlnc69 low- and high-risk groups. (A) Heat map comparing the distribution of significant characters. (B-D) Comparison of age (B), WBC at diagnosis (C), and bone marrow leukemic blast (D).
To explore the biological pathways associated with the AMLlnc69 signature, we performed GSEA using the cancer gene ontology, Kyoto Encyclopedia of Genes and Genomes, and hallmark gene sets. The 5 most significant results are shown in supplemental Figure 10, and the clustered pathways are provided in supplemental Tables 12-14, encompassing all the clustered pathways. The GSEA analysis revealed the significant involvement of the Hedgehog and KRAS pathways and epithelial-mesenchymal transition (EMT) in AMLlnc69–based high risk.
Discussion
The survival of patients with pedAML has increased steadily in recent decades, largely due to the incorporation of risk stratification based on parameters such as cytogenetic and molecular abnormalities and MRD. Presently, pedAML relapse occurs in approximately one-third of patients, which is a major obstacle in treatment and adversely affects OS.9,10 AML relapse is attributable primarily to the poor responsiveness of therapy-resistant LSCs to common chemotherapeutic agents.34 Treatments for pedAML relapse include HSCT and reinduction regimens, but their inevitable generation of side effects remains a challenge.9
No robust prognostic model for the prediction of pedAML relapse is currently available. Several models based on EFS have been developed for pedAML risk stratification. However, due to the inclusion of relapse, induction failure, and death at first event, the LSC6, LSC17, and LSC47 signatures and lncScore predicted RFS in the discovery cohort inefficiently. Moreover, we argue that induction failure should be included in analyses as a binary variable, regardless of its temporal span. For these reasons, we excluded induction failure and death and focused solely on RFS.
In this study, we demonstrated that the AMLlnc69 prognostic signature strengthens risk prediction, even in multivariate analysis including established risk-stratifying factors. Among the lncRNAs constituting the signature, MIR100HG (ENSG00000255015),35 MIR17HG (ENSG00000215417),36 MALAT1 (ENSG00000251562),37-39 and ZFAS1 (ENSG00000177410)40-43 were previously shown to be associated with AML; the associations of the other 65 lncRNAs with AML are novel. In contrast to the use of median values as dichotomization standards in most studies, we used an outcome-oriented method to identify optimal cutoff values in this study, thereby maximizing the distinction between low- and high-risk groups in terms of RFS. The good RFS-predicting performance of AMLlnc69 was demonstrated through KM survival plots and ROC curves. Internal validation is crucial for the estimation of a model’s generalizability.44 Compared with other internal validation methods, bootstrap analysis not only enables the use of the entire sample for validation but also provides nearly unbiased estimates of model performance.45 The favorable results of bootstrap–based internal validation in this study provide strong evidence for the reliability of model construction. Because some high-risk patients underwent HSCT to improve RFS,46 we split patients in the discovery cohort into HSCT and no-HSCT groups. The model retained its predictive value in both scenarios. Moreover, external validation must be performed to determine a model’s reproducibility and generalizability to other samples.47 Thus, data from the AAML0531 study, distinct from the discovery cohort, were used for external validation in this study. Although this cohort was small and significantly more heterogeneous than the discovery cohort, the use of the AMLlnc69 to predict RFS in this cohort was successful. The AUCs obtained in this study were satisfactory, indicating the reproducibility of AMLlnc69 use in actual practice. The AMLlnc69 remained predictive independently of gemtuzumab ozogamicin administration in the AAML0531 cohort; no such analysis could be performed for sorafenib or bortezomib administration in the AAML1031 cohort due to the lack of information. With an ever-growing number of pedAML therapeutics available (including bcl-2 protein family and DNA methyltransferase inhibitors), the evaluation of the predictive value of AMLlnc69 in this evolving therapeutic landscape would be of interest.
Recently, Farrar et al14 constructed a lncRNA signature (lncScore) for pedAML with a completely different set of lncRNAs than that used in this study, which might be explained by some notable differences between the studies. First, lncScore was built based on EFS, whereas AMLlnc69 was developed using RFS, which thus limits the direct comparison of the performance of both signatures. Second, Farrar et al14 initially sought to identify lncRNAs that were differentially expressed in the bone marrow of patients with pedAML and that of healthy individuals to construct a model based on the upregulation of these lncRNAs in pedAML. However, the healthy individuals were actually postinduction patients, whose bone marrow may differ substantially from normal.48 Third, the selection of differentially expressed lncRNAs may have led to the overlooking of some lncRNAs that actually affect prognosis. Thus, instead of performing differential expression analysis, we directly used the entire lncRNAome for model construction. In addition, Farrar et al14 constructed a regression model based on EFS, whereas we focused on RFS, and they included samples with low-depth sequencing, whereas we excluded such samples due to the usually low expression of lncRNAs. Despite these differences in composition, construction criteria, and end points, the 2 signatures are associated with very similar pathways in GSEAs (data not shown).
Surprisingly, the standard-risk group (as defined in the TARGET cohort based on cytogenic and molecular characteristics) had worse RFS than the high-risk group. Aplenc et al26 reported that ∼78% of patients in the AAML1031 study were allocated to the low-risk group, with the remainder allocated to the high-risk group. However, a large number of these patients were reassigned to the standard-risk group in the TARGET database with the application of additional cytogenetic and molecular risk stratification criteria. In addition, AMLlnc69-Low_Risk group-High,”patients, the majority of whom underwent HSCT, had better RFS than AMLlnc69-Low_Risk group-Low/Standard” patients in this study. A portion of patients who were initially classified as “Risk group-Low/Standard,” missing HSCT, would have been reclassified as high risk, potentially explaining the superior RFS in AMLlnc69-Low_Risk group-High” patients relative to that of patients in the other 2 groups. Due to the retrospective nature of this study and the complexity of evolving risk stratification, complete clarification of these observed differences is difficult. Nevertheless, the results clearly suggest that all stratification schemes, including that used in the AAML1031 study and traditional cytogenetic and molecular risk stratification, have limitations, resulting in the underestimation or overestimation of the pedAML relapse risk. Importantly, the AMLlnc69 successfully stratified patients in the 3 TARGET risk groups (low, standard, and high) based on their RFS.
The AMLlnc69 was shown to be an independent predictor of RFS in the discovery and validation cohorts. With the continued undertaking of large-scale omics studies and identification of novel risk-stratifying parameters, such as upstream binding transcription factor-tandem duplications49 and GLIS2-fusions,50,51 the assessment of the predictive value of AMLlnc69 in prospective clinical studies52,53 documenting those molecular aberrations would be of great interest. Based on the results of the multivariate Cox regression analysis of discovery cohort data, we developed a nomogram for the intuitive prediction of the RFS of patients with pedAML that includes the AMLlnc69, NPM mutation, and WBC count at the time of diagnosis. Relative to the isolated application of each factor, the nomogram provides for more-refined estimation of 1-, 2-, and 3-year cumulative relapse risks. With further independent validation, this tool could offer valuable insights for prognostic assessment and therapeutic decision-making in the clinical context.
The GSEA performed in this study revealed that Hedgehog- and KRAS-associated pathways and EMT played important roles in the AMLlnc69-based estimation of high relapse risk. The Hedgehog signaling pathway plays a fundamental role in LSC quiescence and may be an effective target for the prevention of pedAML relapse.54,55 Similarly, KRAS contributes to the emergence of stemness traits,56 and KRAS mutations are frequent in patients with pedAML and associated with worse outcomes.57-59 EMT is a well-known cellular program that is crucial for the relapse and metastasis of various tumors, and it plays a key role in the progression and relapse of AML.60-62
In conclusion, we generated a comprehensive prognostic model including 69 lncRNAs for the prediction of the RFS of patients with pedAML. To our knowledge, this study was the first comprehensive evaluation of relationships between lncRNAs and RFS in this population. We provide evidence that our model could further refine current risk stratification. Its application requires the design of a microarray or targeted RNA-seq panel, which is currently the standard of practice in many clinical laboratories handling oncological diagnosis.63 Although AMLlnc69 is associated with several known predictive markers, it incorporates more information than that provided by these individual factors. Thus, AMLlnc69 use may avoid the need to perform numerous molecular and cellular assays, as is currently done for the full risk classification of patients with pedAML, and thereby be a great asset in resource-limited circumstances. Furthermore, samples from patients' bone marrow or peripheral blood, routinely collected during standard pedAML diagnosis, can be used. This lack of need for additional sample collection makes this method straightforward, cost effective, and easily implementable. Further validation of AMLlnc69 with large independent cohorts is needed to definitively confirm its clinical value.
Acknowledgments
This work was supported in part by vzw Kinderkankerfonds (T.L.) and Chinese Scholarship Council grant 202208340021 (Z.R.).
Authorship
Contribution: Z.R., J.V., and T.L. drafted the manuscript; Z.R., J.V., and T.L. designed the figures; and all authors critically revised the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tim Lammens, Department of Pediatric Hematology-Oncology and Stem Cell Transplantation, Ghent University Hospital, 3K12D C. Heymanslaan 10, 9000 Ghent, Belgium; email: tim.lammens@ugent.be.
References
Author notes
The results published here are, in whole or part, based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments initiative (phs000218). The data used for this analysis are available at the National Cancer Institute Genomic Data Commons (https://portal.gdc.cancer.gov).
The full-text version of this article contains a data supplement.